Identifying MicroRNA Markers That Predict COVID-19 Severity Using Machine Learning Methods
Abstract
:1. Introduction
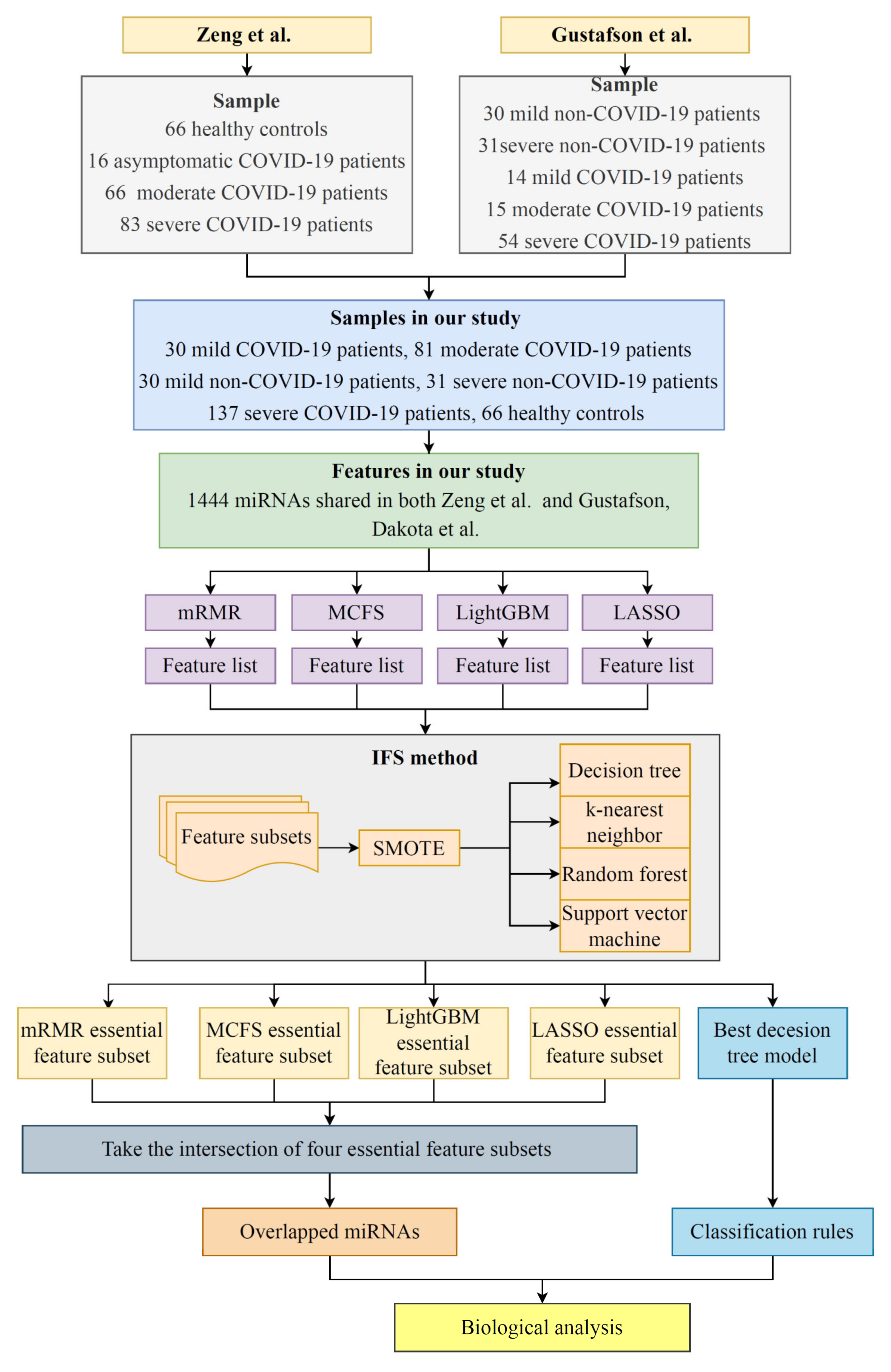
2. Materials and Methods
2.1. Data and Preprocessing
2.2. Feature Ranking Algorithms
2.2.1. LASSO
2.2.2. LightGBM
2.2.3. MCFS
2.2.4. mRMR
2.3. Incremental Feature Selection
- 1.
- Each feature matrix was constructed using the top features from the four feature ranking algorithms, where is the total number of features;
- 2.
- The 10-fold cross-validation was performed on each feature matrix to evaluate the performance of the classification model.
- 3.
- The most effective classification model and its feature subset were selected for each of the four feature rankings.
2.4. Synthetic Minority Oversampling Technique
2.5. Classification Algorithm
2.6. Performance Evaluation
3. Results
3.1. Results of the Feature Ranking Algorithms
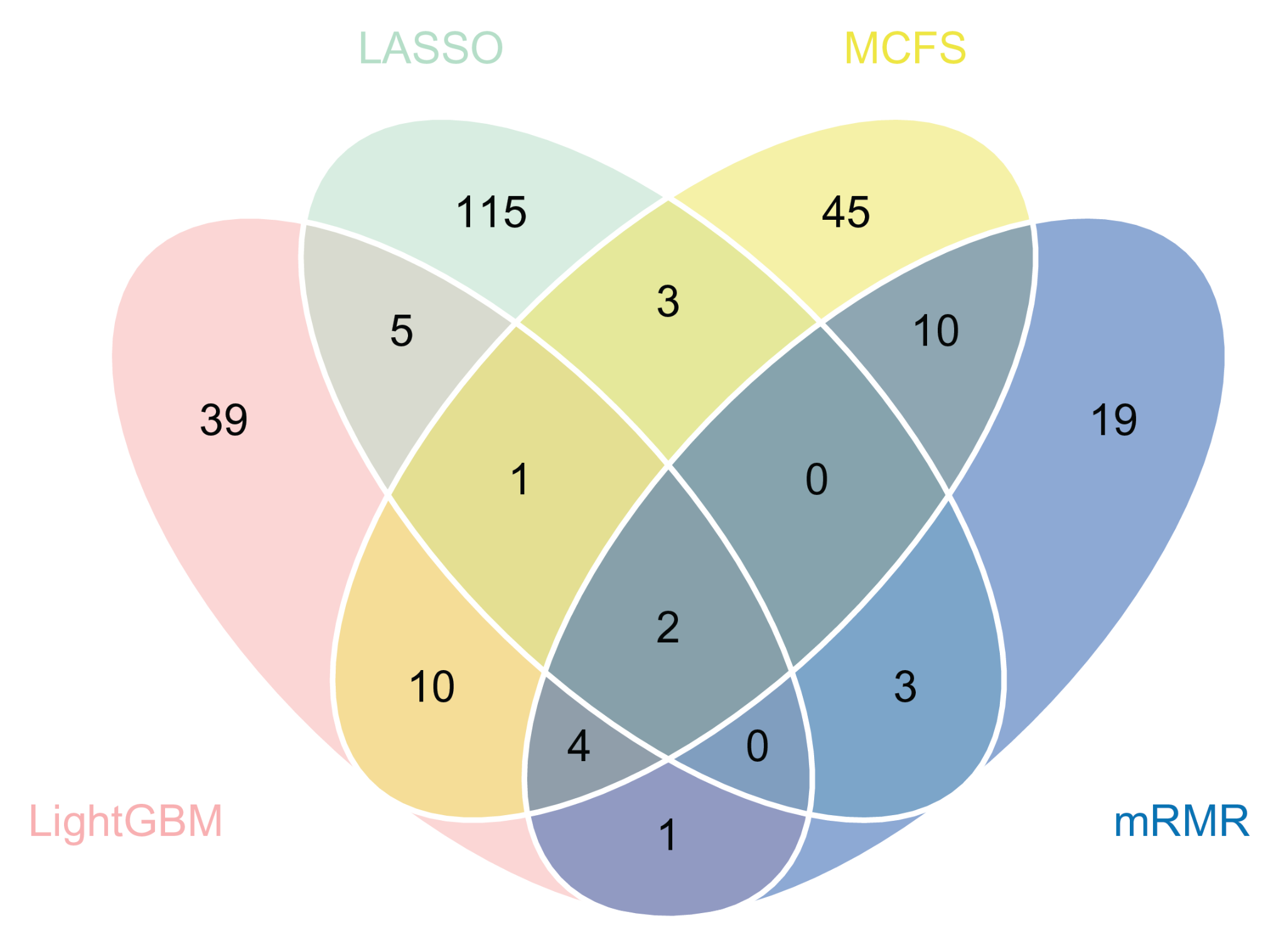
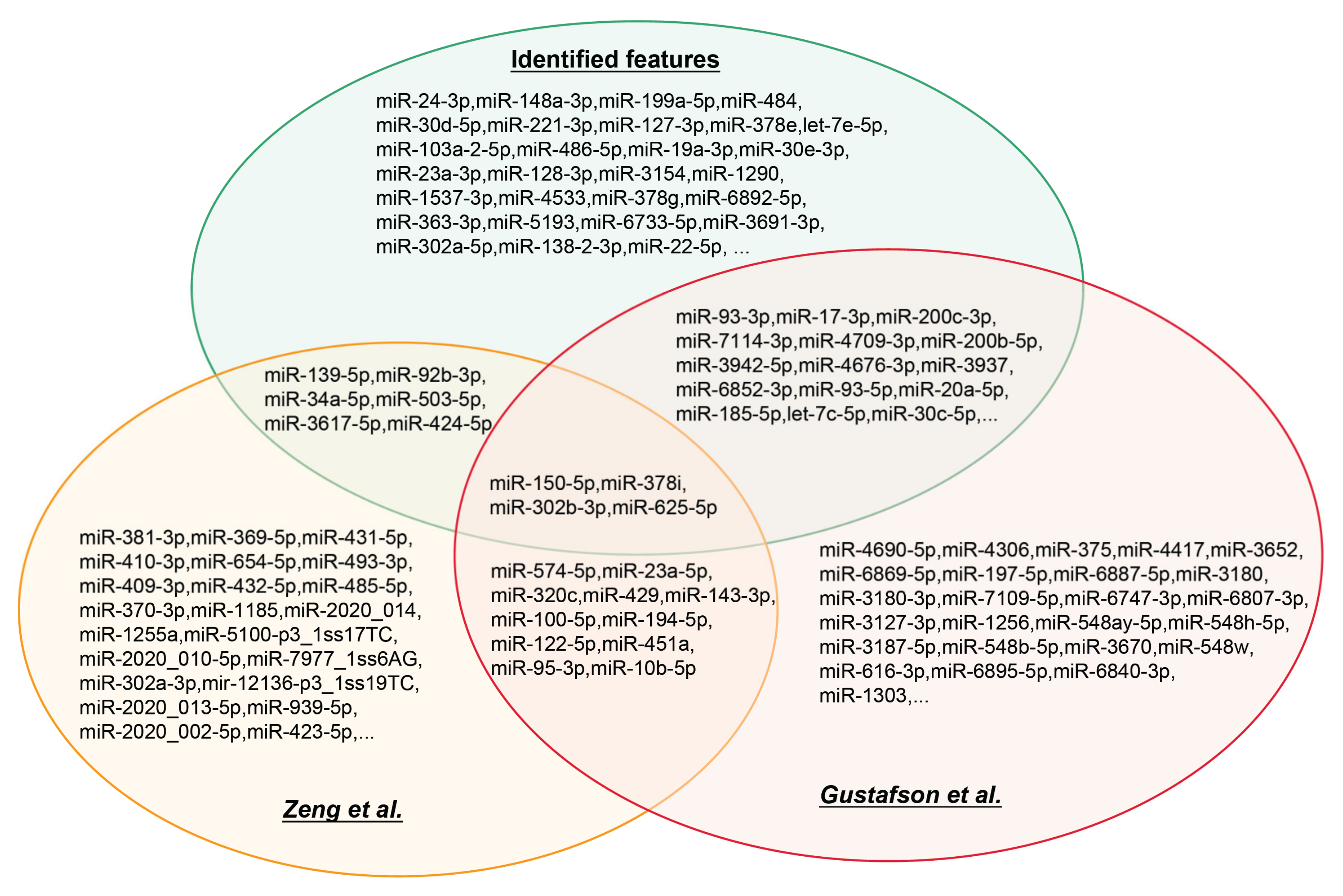
3.2. IFS Results and Feature Intersections
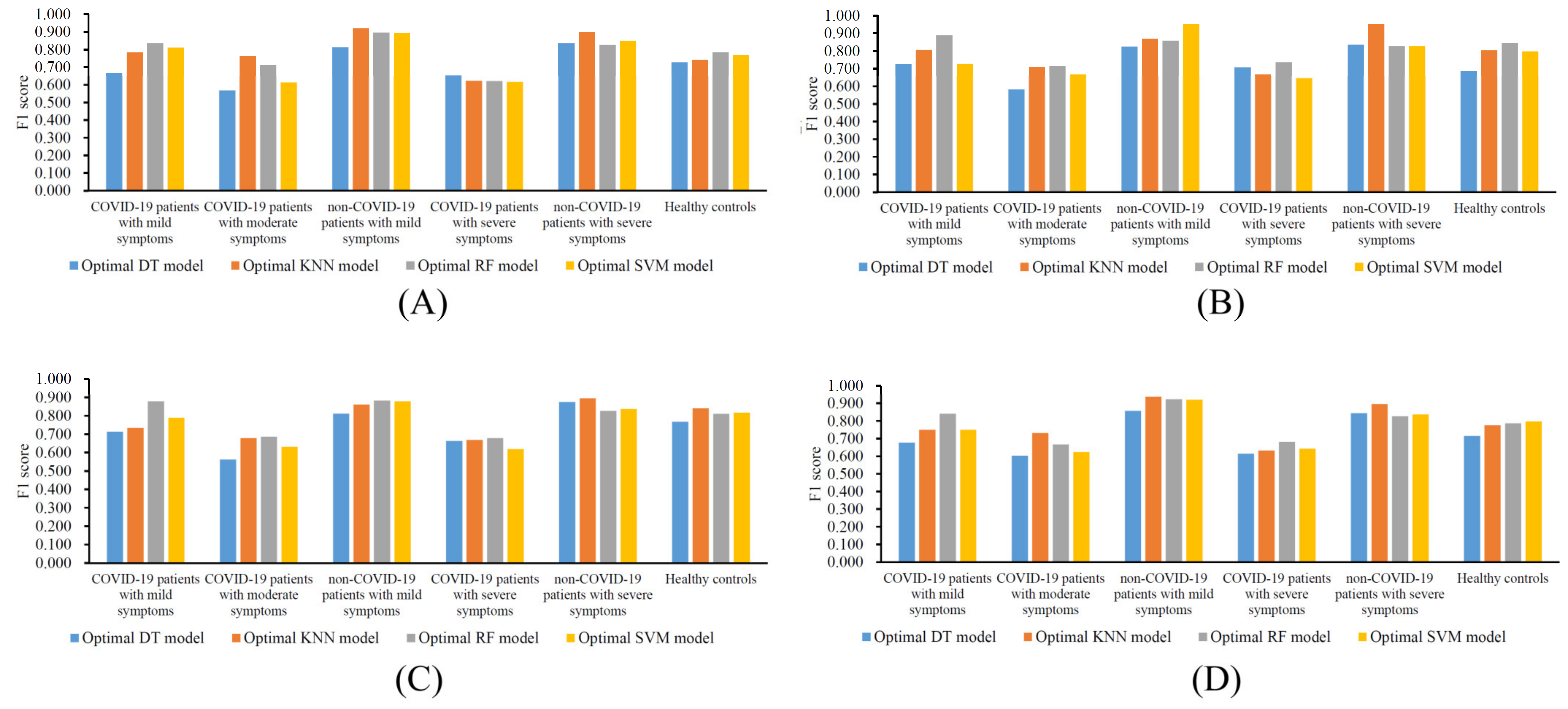
3.3. Classification Rules
4. Discussion
4.1. Analysis of the Key Biomarkers
4.2. Analysis of the Classification Rules
4.3. The Advantages and Limitations of the Proposed Method
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- WHO Coronavirus (COVID-19) Dashboard with Vaccination Data. Available online: https://covid19.who.int/ (accessed on 23 June 2022).
- Wu, C.; Chen, X.; Cai, Y.; Xia, J.; Zhou, X.; Xu, S.; Huang, H.; Zhang, L.; Zhou, X.; Du, C.; et al. Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan, China. JAMA Intern. Med. 2020, 180, 934–943. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brodin, P. Why is COVID-19 so mild in children? Acta Paediatr. 2020, 109, 1082–1083. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Han, T.; Chen, J.; Hou, C.; Hua, L.; He, S.; Guo, Y.; Zhang, S.; Wang, Y.; Yuan, J.; et al. Clinical and autoimmune characteristics of severe and critical cases of COVID-19. Clin. Transl. Sci. 2020, 13, 1077–1086. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frater, J.L.; Zini, G.; d’Onofrio, G.; Rogers, H.J. Covid-19 and the clinical hematology laboratory. Int. J. Lab. Hematol. 2020, 42, 11–18. [Google Scholar] [CrossRef] [Green Version]
- Zhao, W.; Zha, X.; Wang, N.; Li, D.; Li, A.; Yu, S. Clinical characteristics and durations of hospitalized patients with COVID-19 in beijing: A retrospective cohort study. Cardiovasc. Innov. Appl. 2021, 6, 33–44. [Google Scholar] [CrossRef]
- Perez, L. Acute phase protein response to viral infection and vaccination. Arch. Biochem. Biophys. 2019, 671, 196–202. [Google Scholar] [CrossRef]
- Chen, X.; Zhao, B.; Qu, Y.; Chen, Y.; Xiong, J.; Feng, Y.; Men, D.; Huang, Q.; Liu, Y.; Yang, B.; et al. Detectable serum severe acute respiratory syndrome coronavirus 2 viral load (rnaemia) is closely correlated with drastically elevated interleukin 6 level in critically ill patients with coronavirus disease 2019. Clin. Infect. Dis 2020, 71, 1937–1942. [Google Scholar] [CrossRef]
- Yang, Y.; Shen, C.; Li, J.; Yuan, J.; Wei, J.; Huang, F.; Wang, F.; Li, G.; Li, Y.; Xing, L.; et al. Plasma ip-10 and mcp-3 levels are highly associated with disease severity and predict the progression of COVID-19. J. Allergy Clin. Immunol 2020, 146, 119–127.e114. [Google Scholar] [CrossRef]
- Henry, B.M.; de Oliveira, M.H.S.; Benoit, S.; Plebani, M.; Lippi, G. Hematologic, biochemical and immune biomarker abnormalities associated with severe illness and mortality in coronavirus disease 2019 (COVID-19): A meta-analysis. Clin. Chem. Lab. Med. 2020, 58, 1021–1028. [Google Scholar] [CrossRef] [Green Version]
- Borges do Nascimento, I.J.; Cacic, N.; Abdulazeem, H.M.; von Groote, T.C.; Jayarajah, U.; Weerasekara, I.; Esfahani, M.A.; Civile, V.T.; Marusic, A.; Jeroncic, A.; et al. Novel coronavirus infection (COVID-19) in humans: A scoping review and meta-analysis. J. Clin. Med. 2020, 9, 941. [Google Scholar] [CrossRef]
- Rupaimoole, R.; Slack, F.J. Microrna therapeutics: Towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 2017, 16, 203–222. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.S.; Jin, J.P.; Wang, J.Q.; Zhang, Z.G.; Freedman, J.H.; Zheng, Y.; Cai, L. Mirnas in cardiovascular diseases: Potential biomarkers, therapeutic targets and challenges. Acta Pharm. Sin. 2018, 39, 1073–1084. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lodge, R.; Gilmore, J.C.; Ferreira Barbosa, J.A.; Lombard-Vadnais, F.; Cohen, É.A. Regulation of cd4 receptor and hiv-1 entry by micrornas-221 and -222 during differentiation of thp-1 cells. Viruses 2017, 10, 13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lodge, R.; Bellini, N.; Laporte, M.; Salahuddin, S.; Routy, J.P.; Ancuta, P.; Costiniuk, C.T.; Jenabian, M.A.; Cohen, É.A. Interleukin-1β triggers p53-mediated downmodulation of ccr5 and hiv-1 entry in macrophages through micrornas 103 and 107. mBio 2020, 11, e02314–e02320. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.; Ouyang, Y.; Mo, J.; Li, R.; Fu, L.; Peng, S. Upregulation of microrna-328-3p by hepatitis b virus contributes to thle-2 cell injury by downregulating foxo4. J. Transl. Med. 2020, 18, 143. [Google Scholar] [CrossRef]
- Fu, Y.; Xu, W.; Chen, D.; Feng, C.; Zhang, L.; Wang, X.; Lv, X.; Zheng, N.; Jin, Y.; Wu, Z. Enterovirus 71 induces autophagy by regulating has-mir-30a expression to promote viral replication. Antivir. Res. 2015, 124, 43–53. [Google Scholar] [CrossRef] [Green Version]
- Li, C.; Hu, X.; Li, L.; Li, J.H. Differential microrna expression in the peripheral blood from human patients with COVID-19. J. Clin. Lab. Anal. 2020, 34, e23590. [Google Scholar] [CrossRef]
- Farr, R.J.; Rootes, C.L.; Rowntree, L.C.; Nguyen, T.H.O.; Hensen, L.; Kedzierski, L.; Cheng, A.C.; Kedzierska, K.; Au, G.G.; Marsh, G.A.; et al. Altered microrna expression in COVID-19 patients enables identification of SARS-CoV-2 infection. PLoS Pathog. 2021, 17, e1009759. [Google Scholar] [CrossRef]
- Khan, M.A.; Sany, M.R.U.; Islam, M.S.; Islam, A. Epigenetic regulator mirna pattern differences among SARS-CoV, SARS-CoV-2, and SARS-CoV-2 world-wide isolates delineated the mystery behind the epic pathogenicity and distinct clinical characteristics of pandemic COVID-19. Front. Genet. 2020, 11, 765. [Google Scholar] [CrossRef]
- Saçar Demirci, M.D.; Adan, A. Computational analysis of microrna-mediated interactions in SARS-CoV-2 infection. PeerJ 2020, 8, e9369. [Google Scholar] [CrossRef]
- Hosseini Rad Sm, A.; McLellan, A.D. Implications of SARS-CoV-2 mutations for genomic rna structure and host microrna targeting. Int. J. Mol. Sci. 2020, 21, 4807. [Google Scholar] [CrossRef] [PubMed]
- Girardi, E.; López, P.; Pfeffer, S. On the importance of host micrornas during viral infection. Front. Genet. 2018, 9, 439. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bartel, D.P. Micrornas: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilson, L.; McKinlay, C.; Gage, P.; Ewart, G. Sars coronavirus e protein forms cation-selective ion channels. Virology 2004, 330, 322–331. [Google Scholar] [CrossRef] [Green Version]
- Masters, P.S. The molecular biology of coronaviruses. Adv. Virus Res. 2006, 66, 193–292. [Google Scholar]
- Tang, H.; Gao, Y.; Li, Z.; Miao, Y.; Huang, Z.; Liu, X.; Xie, L.; Li, H.; Wen, W.; Zheng, Y.; et al. The noncoding and coding transcriptional landscape of the peripheral immune response in patients with COVID-19. Clin. Transl. Med. 2020, 10, e200. [Google Scholar] [CrossRef]
- Gustafson, D.; Ngai, M.; Wu, R.; Hou, H.; Schoffel, A.C.; Erice, C.; Mandla, S.; Billia, F.; Wilson, M.D.; Radisic, M.; et al. Cardiovascular signatures of COVID-19 predict mortality and identify barrier stabilizing therapies. EBioMedicine 2022, 78, 103982. [Google Scholar] [CrossRef]
- Zeng, Q.; Qi, X.; Ma, J.; Hu, F.; Wang, X.; Qin, H.; Li, M.; Huang, S.; Yang, Y.; Li, Y.; et al. Distinct mirnas associated with various clinical presentations of SARS-CoV-2 infection. Iscience 2022, 25, 104309. [Google Scholar] [CrossRef]
- Breiman, L. Better subset regression using the nonnegative garrote. Technometrics 1995, 37, 373–384. [Google Scholar] [CrossRef]
- Tibshirani, R.J. Regression shrinkage and selection via the lasso. J. R. Stat. Soc. Ser. B Methodol. 1996, 73, 273–282. [Google Scholar] [CrossRef]
- Ke, G.; Meng, Q.; Finely, T.; Wang, T.; Chen, W.; Ma, W.; Ye, Q.; Liu, T.-Y. Lightgbm: A highly Efficient Gradient Boosting Decision Tree. In Advances in Neural Information Processing Systems 30 (NIP 2017); Curran Associates Inc.: Red Hook, NY, USA, 2017. [Google Scholar]
- Micha, D.; Rada-Iglesias, A.; Enroth, S.; Wadelius, C.; Koronacki, J.; Komorowski, J. Monte carlo feature selection for supervised classification. Bioinformatics 2008, 24, 110–117. [Google Scholar]
- Peng, H.; Fulmi, L.; Ding, C. Feature selection based on mutual information criteria of max-dependency, max-relevance, and min-redundancy. Pattern Anal. Mach. Intell. IEEE Trans. 2005, 27, 1226–1238. [Google Scholar] [CrossRef] [PubMed]
- Safavian, S.R.; Landgrebe, D. A survey of decision tree classifier methodology. IEEE Trans. Syst. Man Cybern. 1991, 21, 660–674. [Google Scholar] [CrossRef] [Green Version]
- Cover, T.; Hart, P. Nearest neighbor pattern classification. IEEE Trans. Inf. Theory 1967, 13, 21–27. [Google Scholar] [CrossRef] [Green Version]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef] [Green Version]
- Cortes, C.; Vapnik, V. Support-vector networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Liu, H.; Setiono, R. Incremental feature selection. Appl. Intell. 1998, 9, 217–230. [Google Scholar] [CrossRef]
- Chen, L.; Li, J.; Zhang, Y.H.; Feng, K.; Wang, S.; Zhang, Y.; Huang, T.; Kong, X.; Cai, Y.D. Identification of gene expression signatures across different types of neural stem cells with the monte-carlo feature selection method. J. Cell. Biochem. 2018, 119, 3394–3403. [Google Scholar] [CrossRef]
- Chen, X.; Jin, Y.; Feng, Y. Evaluation of plasma extracellular vesicle microrna signatures for lung adenocarcinoma and granuloma with monte-carlo feature selection method. Front. Genet. 2019, 10, 367. [Google Scholar] [CrossRef]
- Zhang, Y.H.; Guo, W.; Zeng, T.; Zhang, S.; Chen, L.; Gamarra, M.; Mansour, R.F.; Escorcia-Gutierrez, J.; Huang, T.; Cai, Y.D. Identification of microbiota biomarkers with orthologous gene annotation for type 2 diabetes. Front. Microbiol. 2021, 12, 711244. [Google Scholar] [CrossRef]
- Zhang, Y.H.; Li, Z.; Zeng, T.; Pan, X.; Chen, L.; Liu, D.; Li, H.; Huang, T.; Cai, Y.D. Distinguishing glioblastoma subtypes by methylation signatures. Front. Genet. 2020, 11, 604336. [Google Scholar] [CrossRef] [PubMed]
- Chawla, N.V.; Bowyer, K.W.; Hall, L.O.; Kegelmeyer, W.P. Smote: Synthetic minority over-sampling technique. J. Artif. Intell. Res. 2002, 16, 321–357. [Google Scholar] [CrossRef]
- Pan, X.; Chen, L.; Liu, I.; Niu, Z.; Huang, T.; Cai, Y.D. Identifying protein subcellular locations with embeddings-based node2loc. IEEE/ACM Trans. Comput. Biol. Bioinform. 2022, 19, 666–675. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Li, Z.; Zhang, S.; Zhang, Y.-H.; Huang, T.; Cai, Y.-D. Predicting rna 5-methylcytosine sites by using essential sequence features and distributions. BioMed. Res. Int. 2022, 2022, 4035462. [Google Scholar] [CrossRef] [PubMed]
- Ding, S.; Wang, D.; Zhou, X.; Chen, L.; Feng, K.; Xu, X.; Huang, T.; Li, Z.; Cai, Y. Predicting heart cell types by using transcriptome profiles and a machine learning method. Life 2022, 12, 228. [Google Scholar] [CrossRef]
- Zhou, X.; Ding, S.; Wang, D.; Chen, L.; Feng, K.; Huang, T.; Li, Z.; Cai, Y.-D. Identification of cell markers and their expression patterns in skin based on single-cell rna-sequencing profiles. Life 2022, 12, 550. [Google Scholar] [CrossRef]
- Tang, S.; Chen, L. Iatc-nfmlp: Identifying classes of anatomical therapeutic chemicals based on drug networks, fingerprints and multilayer perceptron. Curr. Bioinform. 2022, 17, 814–824. [Google Scholar]
- Wu, C.; Chen, L. A model with deep analysis on a large drug network for drug classification. Math. Biosci. Eng. 2022, 20, 383–401. [Google Scholar] [CrossRef]
- Yang, Y.; Chen, L. Identification of drug–disease associations by using multiple drug and disease networks. Curr. Bioinform. 2022, 17, 48–59. [Google Scholar] [CrossRef]
- Ran, B.; Chen, L.; Li, M.; Han, Y.; Dai, Q. Drug-drug interactions prediction using fingerprint only. Comput. Math. Methods Med. 2022, 2022, 7818480. [Google Scholar] [CrossRef]
- Wang, Y.; Zhu, X.; Jiang, X.M.; Guo, J.; Fu, Z.; Zhou, Z.; Yang, P.; Guo, H.; Guo, X.; Liang, G.; et al. Decreased inhibition of exosomal mirnas on SARS-CoV-2 replication underlies poor outcomes in elderly people and diabetic patients. Signal. Transduct. Target. 2021, 6, 300. [Google Scholar] [CrossRef] [PubMed]
- García-Hidalgo, M.C.; González, J.; Benítez, I.D.; Carmona, P.; Santisteve, S.; Pérez-Pons, M.; Moncusí-Moix, A.; Gort-Paniello, C.; Rodríguez-Jara, F.; Molinero, M.; et al. Identification of circulating microrna profiles associated with pulmonary function and radiologic features in survivors of SARS-CoV-2-induced ards. Emerg. Microbes. Infect. 2022, 11, 1537–1549. [Google Scholar] [CrossRef] [PubMed]
- Guzik, T.J.; Mohiddin, S.A.; Dimarco, A.; Patel, V.; Savvatis, K.; Marelli-Berg, F.M.; Madhur, M.S.; Tomaszewski, M.; Maffia, P.; D’Acquisto, F.; et al. Covid-19 and the cardiovascular system: Implications for risk assessment, diagnosis, and treatment options. Cardiovasc. Res. 2020, 116, 1666–1687. [Google Scholar] [CrossRef] [PubMed]
- Souchelnytskyi, S.; Nera, A.; Souchelnytskyi, N. Covid-19 engages clinical markers for the management of cancer and cancer-relevant regulators of cell proliferation, death, migration, and immune response. Sci. Rep. 2021, 11, 5228. [Google Scholar] [CrossRef] [PubMed]
- de Gonzalo-Calvo, D.; Benítez, I.D.; Pinilla, L.; Carratalá, A.; Moncusí-Moix, A.; Gort-Paniello, C.; Molinero, M.; González, J.; Torres, G.; Bernal, M.; et al. Circulating microrna profiles predict the severity of COVID-19 in hospitalized patients. Transl. Res. 2021, 236, 147–159. [Google Scholar] [CrossRef]
- Mallick, B.; Ghosh, Z.; Chakrabarti, J. Micrornome analysis unravels the molecular basis of sars infection in bronchoalveolar stem cells. PLoS ONE 2009, 4, e7837. [Google Scholar] [CrossRef]
- Chow, J.T.; Salmena, L. Prediction and analysis of SARS-CoV-2-targeting microrna in human lung epithelium. Genes 2020, 11, 1002. [Google Scholar] [CrossRef]
- Li, C.; Wu, A.; Song, K.; Gao, J.; Huang, E.; Bai, Y.; Liu, X. Identifying putative causal links between micrornas and severe COVID-19 using mendelian randomization. Cells 2021, 10, 3504. [Google Scholar] [CrossRef]
- Molinero, M.; Benítez, I.D.; González, J.; Gort-Paniello, C.; Moncusí-Moix, A.; Rodríguez-Jara, F.; García-Hidalgo, M.C.; Torres, G.; Vengoechea, J.J.; Gómez, S.; et al. Bronchial aspirate-based profiling identifies microrna signatures associated with COVID-19 and fatal disease in critically ill patients. Front. Med. 2021, 8, 756517. [Google Scholar] [CrossRef]
- Kobayashi, K.; Suemasa, F.; Sagara, H.; Nakamura, S.; Ino, Y.; Kobayashi, K.; Hiramatsu, H.; Haraguchi, T.; Kurokawa, K.; Todo, T.; et al. Mir-199a inhibits secondary envelopment of herpes simplex virus-1 through the downregulation of cdc42-specific gtpase activating protein localized in golgi apparatus. Sci. Rep. 2017, 7, 6650. [Google Scholar] [CrossRef] [Green Version]
- Keikha, R.; Hashemi-Shahri, S.M.; Jebali, A. The relative expression of mir-31, mir-29, mir-126, and mir-17 and their mrna targets in the serum of COVID-19 patients with different grades during hospitalization. Eur. J. Med. Res. 2021, 26, 75. [Google Scholar] [CrossRef] [PubMed]
- Sardar, R.; Satish, D.; Gupta, D. Identification of novel SARS-CoV-2 drug targets by host micrornas and transcription factors co-regulatory interaction network analysis. Front. Genet. 2020, 11, 571274. [Google Scholar] [CrossRef] [PubMed]
- Banaganapalli, B.; Al-Rayes, N.; Awan, Z.A.; Alsulaimany, F.A.; Alamri, A.S.; Elango, R.; Malik, M.Z.; Shaik, N.A. Multilevel systems biology analysis of lung transcriptomics data identifies key mirnas and potential mirna target genes for SARS-CoV-2 infection. Comput. Biol. Med. 2021, 135, 104570. [Google Scholar] [CrossRef]
- Fayyad-Kazan, M.; Makki, R.; Skafi, N.; El Homsi, M.; Hamade, A.; El Majzoub, R.; Hamade, E.; Fayyad-Kazan, H.; Badran, B. Circulating mirnas: Potential diagnostic role for coronavirus disease 2019 (COVID-19). Infect. Genet. Evol. 2021, 94, 105020. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Li, D.; Fiselier, A.; Kovalchuk, I.; Kovalchuk, O. New akt-dependent mechanisms of anti-COVID-19 action of high-cbd cannabis sativa extracts. Cell Death Discov. 2022, 8, 110. [Google Scholar] [CrossRef] [PubMed]
- Bozgeyik, I. Therapeutic potential of mirnas targeting SARS-CoV-2 host cell receptor ace2. Meta Gene 2021, 27, 100831. [Google Scholar] [CrossRef]
- Pimenta, R.; Viana, N.I.; Dos Santos, G.A.; Candido, P.; Guimarães, V.R.; Romão, P.; Silva, I.A.; de Camargo, J.A.; Hatanaka, D.M.; Queiroz, P.G.S.; et al. Mir-200c-3p expression may be associated with worsening of the clinical course of patients with COVID-19. Mol. Biol. Res. Commun. 2021, 10, 141–147. [Google Scholar]
- Abdolahi, S.; Hosseini, M.; Rezaei, R.; Mohebbi, S.R.; Rostami-Nejad, M.; Mojarad, E.N.; Mirjalali, H.; Yadegar, A.; Asadzadeh Aghdaei, H.; Zali, M.R.; et al. Evaluation of mir-200c-3p and mir-421-5p levels during immune responses in the admitted and recovered COVID-19 subjects. Infect. Genet. Evol. 2022, 98, 105207. [Google Scholar] [CrossRef]
- Soltani, S.; Zandi, M. Mir-200c-3p upregulation and ace2 downregulation via bacterial lps and lta as interesting aspects for COVID-19 treatment and immunity. Mol. Biol. Rep. 2021, 48, 5809–5810. [Google Scholar] [CrossRef]
- Wang, N.; Bu, R.; Duan, Z.; Zhang, X.; Chen, P.; Li, Z.; Wu, J.; Cai, G.; Chen, X. Profiling and initial validation of urinary micrornas as biomarkers in iga nephropathy. PeerJ 2015, 3, e990. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Jiang, M.; Deng, S.; Lu, J.; Huang, H.; Zhang, Y.; Gong, P.; Shen, X.; Ruan, H.; Jin, M.; et al. Mir-93-5p-containing exosomes treatment attenuates acute myocardial infarction-induced myocardial damage. Mol. Ther. Nucleic Acids 2018, 11, 103–115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Centa, A.; Fonseca, A.S.; Ferreira, S.; Azevedo, M.L.V.; Vaz de Paula, C.B.; Nagashima, S.; Machado-Souza, C.; Miggiolaro, A.; Baena, C.P.; de Noronha, L.; et al. Deregulated mirna expression is associated with endothelial dysfunction in post-mortem lung biopsies of COVID-19 patients. Am. J. Physiol. Lung Cell Mol. Physiol. 2020, 320, L405–L412. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Shuang, O.; Li, J.; Cai, Z.; Wu, C.; Wang, W. Mir-34a alleviates spinal cord injury via tlr4 signaling by inhibiting hmgb-1. Exp. Ther. Med. 2019, 17, 1912–1918. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, H.J.; Joe, Y.; Yu, J.K.; Chen, Y.; Jeong, S.O.; Mani, N.; Cho, G.J.; Pae, H.O.; Ryter, S.W.; Chung, H.T. Carbon monoxide protects against hepatic ischemia/reperfusion injury by modulating the mir-34a/sirt1 pathway. Biochim. Biophys. Acta 2015, 1852, 1550–1559. [Google Scholar] [CrossRef]
- Yuan, J.; Zhang, Y. Sevoflurane reduces inflammatory factor expression, increases viability and inhibits apoptosis of lung cells in acute lung injury by microrna-34a-3p upregulation and stat1 downregulation. Chem. Biol. Interact. 2020, 322, 109027. [Google Scholar] [CrossRef]
- Li, C.X.; Chen, J.; Lv, S.K.; Li, J.H.; Li, L.L.; Hu, X. Whole-transcriptome rna sequencing reveals significant differentially expressed mrnas, mirnas, and lncrnas and related regulating biological pathways in the peripheral blood of COVID-19 patients. Mediat. Inflamm. 2021, 2021, 6635925. [Google Scholar] [CrossRef]
- Xiao, J.; Tang, J.; Chen, Q.; Tang, D.; Liu, M.; Luo, M.; Wang, Y.; Wang, J.; Zhao, Z.; Tang, C.; et al. Mir-429 regulates alveolar macrophage inflammatory cytokine production and is involved in lps-induced acute lung injury. Biochem. J. 2015, 471, 281–291. [Google Scholar] [CrossRef]
- Lv, H.; Zhang, S.; Hao, X. Swainsonine protects h9c2 cells against lipopolysaccharide-induced apoptosis and inflammatory injury via down-regulating mir-429. Cell Cycle 2020, 19, 207–217. [Google Scholar] [CrossRef]
- Yu, X.; Chen, X.; Sun, T. Microrna-205-5p targets hmgb1 to suppress inflammatory responses during lung injury after hip fracture. Biomed. Res. Int. 2019, 2019, 7304895. [Google Scholar] [CrossRef] [Green Version]
- Peng, F.; He, J.; Loo, J.F.; Yao, J.; Shi, L.; Liu, C.; Zhao, C.; Xie, W.; Shao, Y.; Kong, S.K.; et al. Identification of micrornas in throat swab as the biomarkers for diagnosis of influenza. Int. J. Med. Sci. 2016, 13, 77–84. [Google Scholar] [CrossRef] [Green Version]
- Martínez-González, E.; Brochado-Kith, Ó.; Gómez-Sanz, A.; Martín-Carbonero, L.; Jimenez-Sousa, M.; Martínez-Román, P.; Resino, S.; Briz, V.; Fernández-Rodríguez, A. Comparison of methods and characterization of small rnas from plasma extracellular vesicles of hiv/hcv coinfected patients. Sci. Rep. 2020, 10, 11140. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Tussy, P.; Fernández-Ramos, D.; Lopitz-Otsoa, F.; Simón, J.; Barbier-Torres, L.; Gomez-Santos, B.; Nuñez-Garcia, M.; Azkargorta, M.; Gutiérrez-de Juan, V.; Serrano-Macia, M.; et al. Mir-873-5p targets mitochondrial gnmt-complex ii interface contributing to non-alcoholic fatty liver disease. Mol. Metab. 2019, 29, 40–54. [Google Scholar] [CrossRef] [PubMed]
- Mone, P.; Gambardella, J.; Wang, X.; Jankauskas, S.S.; Matarese, A.; Santulli, G. Mir-24 targets the transmembrane glycoprotein neuropilin-1 in human brain microvascular endothelial cells. Non-Coding RNA 2021, 7, 9. [Google Scholar] [CrossRef] [PubMed]
- Daly, J.L.; Simonetti, B.; Klein, K.; Chen, K.E.; Williamson, M.K.; Antón-Plágaro, C.; Shoemark, D.K.; Simón-Gracia, L.; Bauer, M.; Hollandi, R.; et al. Neuropilin-1 is a host factor for SARS-CoV-2 infection. Science 2020, 370, 861–865. [Google Scholar] [CrossRef] [PubMed]
- Cantuti-Castelvetri, L.; Ojha, R.; Pedro, L.D.; Djannatian, M.; Franz, J.; Kuivanen, S.; van der Meer, F.; Kallio, K.; Kaya, T.; Anastasina, M.; et al. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science 2020, 370, 856–860. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Mansouri, M.; Rizk, A.; Berger, P. Regulation of vegfr2 trafficking and signaling by rab gtpase-activating proteins. Sci. Rep. 2019, 9, 13342. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, B.; Xuan, L.; Tang, M.; Wang, H.; Zhou, J.; Liu, J.; Wu, S.; Li, M.; Wang, X.; Zhang, H. Mir-93-3p alleviates lipopolysaccharide-induced inflammation and apoptosis in h9c2 cardiomyocytes by inhibiting toll-like receptor 4. Pathol. Res. Pract. 2018, 214, 1686–1693. [Google Scholar] [CrossRef]
- Płóciennikowska, A.; Hromada-Judycka, A.; Borzęcka, K.; Kwiatkowska, K. Co-operation of tlr4 and raft proteins in lps-induced pro-inflammatory signaling. Cell Mol. Life Sci. 2015, 72, 557–581. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, Y.; Jin, H.; Yang, X.; Wang, L.; Su, L.; Liu, K.; Gu, Q.; Xu, X. Microrna-93 inhibits inflammatory cytokine production in lps-stimulated murine macrophages by targeting irak4. FEBS Lett. 2014, 588, 1692–1698. [Google Scholar] [CrossRef] [Green Version]
- Yan, X.T.; Ji, L.J.; Wang, Z.; Wu, X.; Wang, Q.; Sun, S.; Lu, J.M.; Zhang, Y. Microrna-93 alleviates neuropathic pain through targeting signal transducer and activator of transcription 3. Int. Immunopharmacol. 2017, 46, 156–162. [Google Scholar] [CrossRef]
- Xu, Z.K.; Asahchop, E.L.; Branton, W.G.; Gelman, B.B.; Power, C.; Hobman, T.C. Micrornas upregulated during hiv infection target peroxisome biogenesis factors: Implications for virus biology, disease mechanisms and neuropathology. PLoS Pathog. 2017, 13, e1006360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, Q.; Yu, Z.; Lu, Y.; Fan, J.; Ni, Y.; Ma, L. Microrna-148a-3p inhibited the proliferation and epithelial-mesenchymal transition progression of non-small-cell lung cancer via modulating ras/mapk/erk signaling. J. Cell Physiol. 2019, 234, 12786–12799. [Google Scholar] [CrossRef] [PubMed]
- Porstner, M.; Winkelmann, R.; Daum, P.; Schmid, J.; Pracht, K.; Côrte-Real, J.; Schreiber, S.; Haftmann, C.; Brandl, A.; Mashreghi, M.F.; et al. Mir-148a promotes plasma cell differentiation and targets the germinal center transcription factors mitf and bach2. Eur. J. Immunol. 2015, 45, 1206–1215. [Google Scholar] [CrossRef] [PubMed]
- Miyashita, Y.; Yoshida, T.; Takagi, Y.; Tsukamoto, H.; Takashima, K.; Kouwaki, T.; Makino, K.; Fukushima, S.; Nakamura, K.; Oshiumi, H. Circulating extracellular vesicle micrornas associated with adverse reactions, proinflammatory cytokine, and antibody production after COVID-19 vaccination. NPJ Vaccines 2022, 7, 16. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Wang, M.; Sun, Z.; Zhuang, S.; Zhang, W.; Yang, Z.; Han, X.; Nie, S. Microrna-139-5p improves sepsis-induced lung injury by targeting rho-kinase1. Exp. Ther. Med. 2021, 22, 1059. [Google Scholar] [CrossRef]
- Zhang, X.; Liu, X.; Chang, R.; Li, Y. Mir-139-5p protects septic mice with acute lung injury by inhibiting toll-like receptor 4/myeloid differentiation factor 88/nuclear factor-&mac_kgr;b signaling pathway. Clinics 2021, 76, e2484. [Google Scholar]
- Katsumi, T.; Ninomiya, M.; Nishina, T.; Mizuno, K.; Tomita, K.; Haga, H.; Okumoto, K.; Saito, T.; Shimosegawa, T.; Ueno, Y. Mir-139-5p is associated with inflammatory regulation through c-fos suppression, and contributes to the progression of primary biliary cholangitis. Lab. Investig. A J. Technol. Methods Pathol. 2016, 96, 1165–1177. [Google Scholar] [CrossRef] [Green Version]
- Zou, F.; Mao, R.; Yang, L.; Lin, S.; Lei, K.; Zheng, Y.; Ding, Y.; Zhang, P.; Cai, G.; Liang, X.; et al. Targeted deletion of mir-139-5p activates mapk, nf-κb and stat3 signaling and promotes intestinal inflammation and colorectal cancer. Febs. J. 2016, 283, 1438–1452. [Google Scholar] [CrossRef]
- Song, R.; Liu, Q.; Liu, T.; Li, J. Connecting rules from paired mirna and mrna expression data sets of hcv patients to detect both inverse and positive regulatory relationships. BMC Genom. 2015, 16, S11. [Google Scholar] [CrossRef] [Green Version]
- Du, W.W.; Li, X.; Li, T.; Li, H.; Khorshidi, A.; Liu, F.; Yang, B.B. The microrna mir-17-3p inhibits mouse cardiac fibroblast senescence by targeting par4. J. Cell Sci. 2015, 128, 293–304. [Google Scholar] [CrossRef]
- Yang, X.; Du, W.W.; Li, H.; Liu, F.; Khorshidi, A.; Rutnam, Z.J.; Yang, B.B. Both mature mir-17-5p and passenger strand mir-17-3p target timp3 and induce prostate tumor growth and invasion. Nucleic. Acids Res. 2013, 41, 9688–9704. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cai, Y.; Zhang, Y.; Chen, H.; Sun, X.H.; Zhang, P.; Zhang, L.; Liao, M.Y.; Zhang, F.; Xia, Z.Y.; Man, R.Y.; et al. Microrna-17-3p suppresses nf-κb-mediated endothelial inflammation by targeting nik and ikkβ binding protein. Acta Pharm. Sin. 2021, 42, 2046–2057. [Google Scholar] [CrossRef] [PubMed]
- Van der Goten, J.; Vanhove, W.; Lemaire, K.; Van Lommel, L.; Machiels, K.; Wollants, W.J.; De Preter, V.; De Hertogh, G.; Ferrante, M.; Van Assche, G.; et al. Integrated mirna and mrna expression profiling in inflamed colon of patients with ulcerative colitis. PLoS ONE 2014, 9, e116117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bao, Z.; Wang, L.J.; He, K.; Lin, X.; Yu, T.; Li, J.; Gong, J.; Xiang, G. High expression of ace2 in the human lung leads to the release of il6 by suppressing cellular immunity: Il6 plays a key role in COVID-19. Eur. Rev. Med. Pharmacol. Sci. 2021, 25, 527–540. [Google Scholar]
- Farr, R.J.; Rootes, C.L.; Stenos, J.; Foo, C.H.; Cowled, C.; Stewart, C.R. Detection of SARS-CoV-2 infection by microrna profiling of the upper respiratory tract. PLoS ONE 2022, 17, e0265670. [Google Scholar] [CrossRef]
- Fabbri, E.; Borgatti, M.; Montagner, G.; Bianchi, N.; Finotti, A.; Lampronti, I.; Bezzerri, V.; Dechecchi, M.C.; Cabrini, G.; Gambari, R. Expression of microrna-93 and interleukin-8 during pseudomonas aeruginosa-mediated induction of proinflammatory responses. Am. J. Respir Cell Mol. Biol. 2014, 50, 1144–1155. [Google Scholar] [CrossRef]
- Gasparello, J.; d’Aversa, E.; Breveglieri, G.; Borgatti, M.; Finotti, A.; Gambari, R. In vitro induction of interleukin-8 by SARS-CoV-2 spike protein is inhibited in bronchial epithelial ib3-1 cells by a mir-93-5p agomir. Int. Immunopharmacol. 2021, 101, 108201. [Google Scholar] [CrossRef]
- Ma, Q.; Pan, W.; Li, R.; Liu, B.; Li, C.; Xie, Y.; Wang, Z.; Zhao, J.; Jiang, H.; Huang, J.; et al. Liu shen capsule shows antiviral and anti-inflammatory abilities against novel coronavirus SARS-CoV-2 via suppression of nf-κb signaling pathway. Pharm. Res. 2020, 158, 104850. [Google Scholar] [CrossRef]
- Hirano, T.; Murakami, M. Covid-19: A new virus, but a familiar receptor and cytokine release syndrome. Immunity 2020, 52, 731–733. [Google Scholar] [CrossRef]
- Chi, Y.; Ge, Y.; Wu, B.; Zhang, W.; Wu, T.; Wen, T.; Liu, J.; Guo, X.; Huang, C.; Jiao, Y.; et al. Serum cytokine and chemokine profile in relation to the severity of coronavirus disease 2019 in china. J. Infect. Dis. 2020, 222, 746–754. [Google Scholar] [CrossRef]

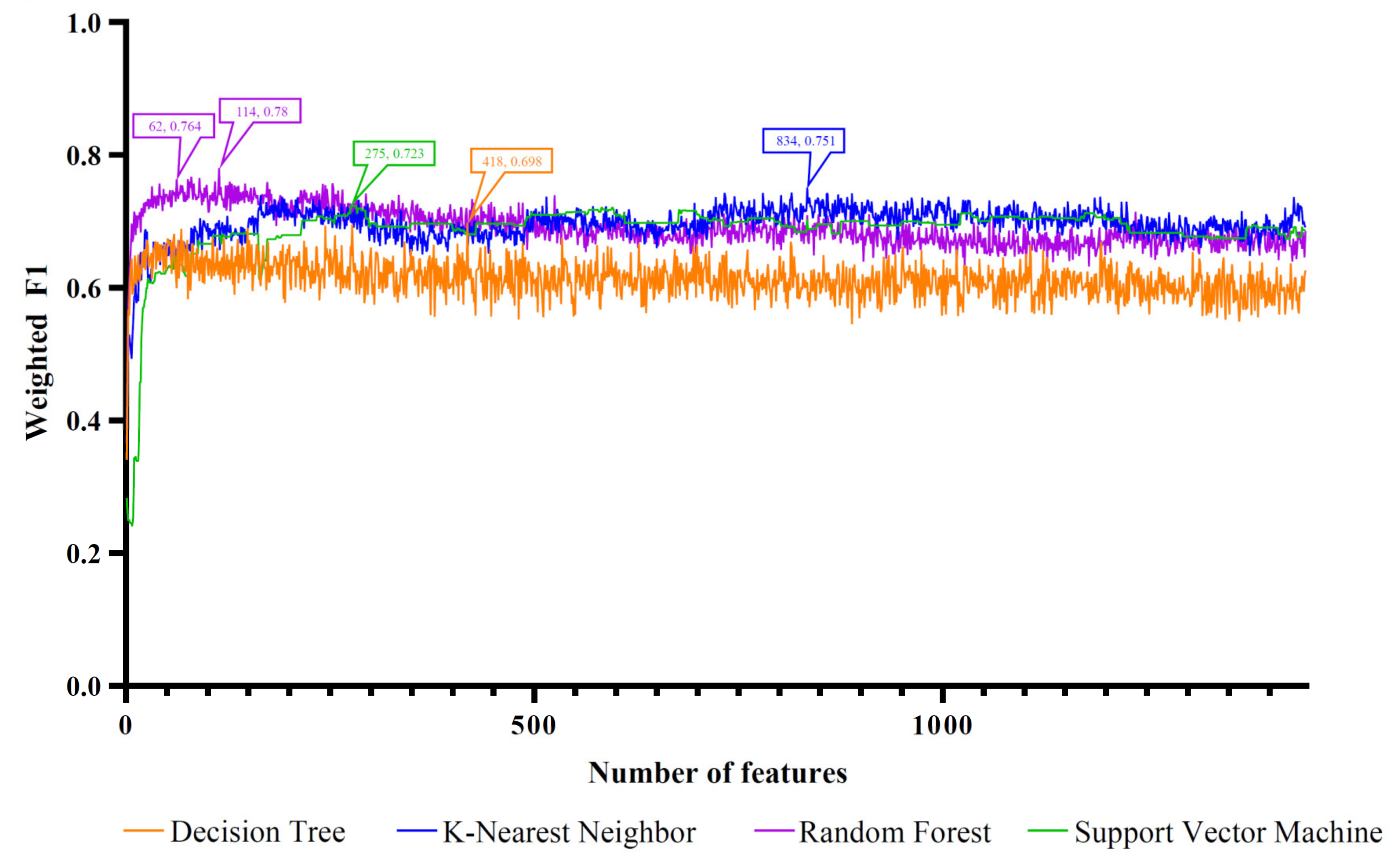


| Feature Ranking Algorithm | Classification Algorithm | Number of Features | ACC | MCC | Macro F1 | Weighed F1 |
|---|---|---|---|---|---|---|
| LASSO | DT | 1335 | 0.680 | 0.595 | 0.710 | 0.677 |
| KNN | 734 | 0.747 | 0.700 | 0.788 | 0.733 | |
| RF | 300 | 0.739 | 0.686 | 0.779 | 0.725 | |
| SVM | 406 | 0.709 | 0.644 | 0.759 | 0.700 | |
| LightGBM | DT | 418 | 0.699 | 0.618 | 0.727 | 0.698 |
| KNN | 834 | 0.760 | 0.711 | 0.801 | 0.751 | |
| RF | 114 | 0.787 | 0.741 | 0.812 | 0.780 | |
| SVM | 275 | 0.731 | 0.673 | 0.769 | 0.723 | |
| MCFS | DT | 639 | 0.696 | 0.616 | 0.733 | 0.694 |
| KNN | 55 | 0.747 | 0.689 | 0.780 | 0.741 | |
| RF | 239 | 0.757 | 0.707 | 0.794 | 0.748 | |
| SVM | 381 | 0.720 | 0.659 | 0.762 | 0.709 | |
| mRMR | DT | 249 | 0.677 | 0.594 | 0.719 | 0.673 |
| KNN | 823 | 0.747 | 0.699 | 0.787 | 0.735 | |
| RF | 583 | 0.749 | 0.699 | 0.788 | 0.741 | |
| SVM | 141 | 0.720 | 0.659 | 0.762 | 0.713 |
| Feature Ranking Algorithm | Classification Algorithm | Number of Features | ACC | MCC | Macro F1 | Weighed F1 |
|---|---|---|---|---|---|---|
| LASSO | KNN | 129 | 0.707 | 0.645 | 0.740 | 0.696 |
| LightGBM | RF | 62 | 0.771 | 0.721 | 0.803 | 0.764 |
| MCFS | RF | 75 | 0.731 | 0.679 | 0.774 | 0.714 |
| mRMR | RF | 39 | 0.725 | 0.669 | 0.768 | 0.712 |
| miRNA | Target Gene | Expression Level | Predicted Class | Ref. |
|---|---|---|---|---|
| miR-24-3p | NRP-1 | Upregulated | Healthy | [53,54] |
| miR-93-3p | TLR4 | Upregulated | Severe COVID-19 | [55,56] |
| miR-148a-3p | SOS2, BACH2, MITF | Upregulated | Severe COVID-19 | [57,58] |
| miR-139-5p | MYD88, c-FOS, RAP1B | Downregulated | Non-COVID-19-mild | [59,60] |
| miR-199a-5p | ARHGAP21 | Upregulated | Healthy | [61,62] |
| miR-17-3p | NIBP | Upregulated | Severe COVID-19 | [54,63,64,65,66] |
| miR-200c-3p | ACE2, IL8 | Downregulated | Non-COVID-19-severe | [67,68,69,70,71] |
| miR-6750-5p | POU2F2 | Downregulated | Severe COVID-19 | [72] |
| miR-93-5p | PDCD1LG2 | Downregulated | Severe COVID-19 | [73] |
| miR-34a-5p | SDK2 | Upregulated | Severe COVID-19 | [61,74,75,76,77] |
| miR-29b-2-5p | POU2F2 | Upregulated | Severe COVID-19 | [78] |
| miR-429 | NR5A2 | Downregulated | Severe COVID-19 | [79,80] |
| miR-205-5p | MOSMO | Downregulated | Severe COVID-19 | [81,82,83] |
| miR-873-5p | PHF6 | Downregulated | Severe COVID-19 | [84] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ren, J.; Guo, W.; Feng, K.; Huang, T.; Cai, Y. Identifying MicroRNA Markers That Predict COVID-19 Severity Using Machine Learning Methods. Life 2022, 12, 1964. https://doi.org/10.3390/life12121964
Ren J, Guo W, Feng K, Huang T, Cai Y. Identifying MicroRNA Markers That Predict COVID-19 Severity Using Machine Learning Methods. Life. 2022; 12(12):1964. https://doi.org/10.3390/life12121964
Chicago/Turabian StyleRen, Jingxin, Wei Guo, Kaiyan Feng, Tao Huang, and Yudong Cai. 2022. "Identifying MicroRNA Markers That Predict COVID-19 Severity Using Machine Learning Methods" Life 12, no. 12: 1964. https://doi.org/10.3390/life12121964
APA StyleRen, J., Guo, W., Feng, K., Huang, T., & Cai, Y. (2022). Identifying MicroRNA Markers That Predict COVID-19 Severity Using Machine Learning Methods. Life, 12(12), 1964. https://doi.org/10.3390/life12121964








