Shewanella oneidensis arcA Mutation Impairs Aerobic Growth Mainly by Compromising Translation
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Plasmids, and Culture Conditions
2.2. Gene Knock In and Expression
2.3. Droplet Assays
2.4. Microscopic Analysis
2.5. Analysis of Gene Expression
2.6. Purification of Ribosomes
2.7. Electrophoretic Motility Shift Assay (EMSA)
2.8. Other Analyses
3. Results
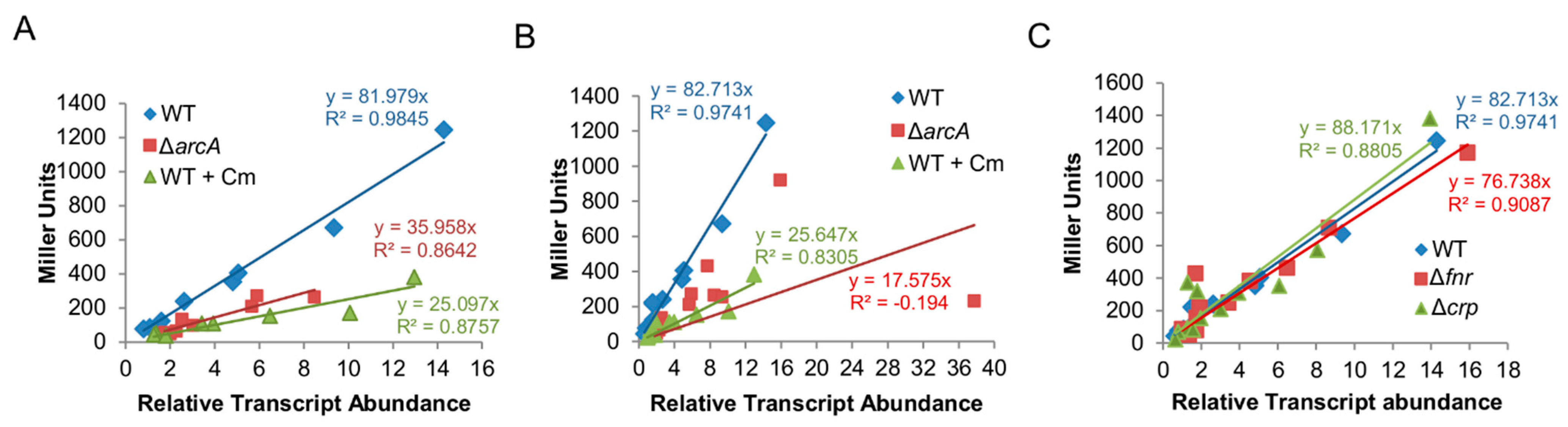
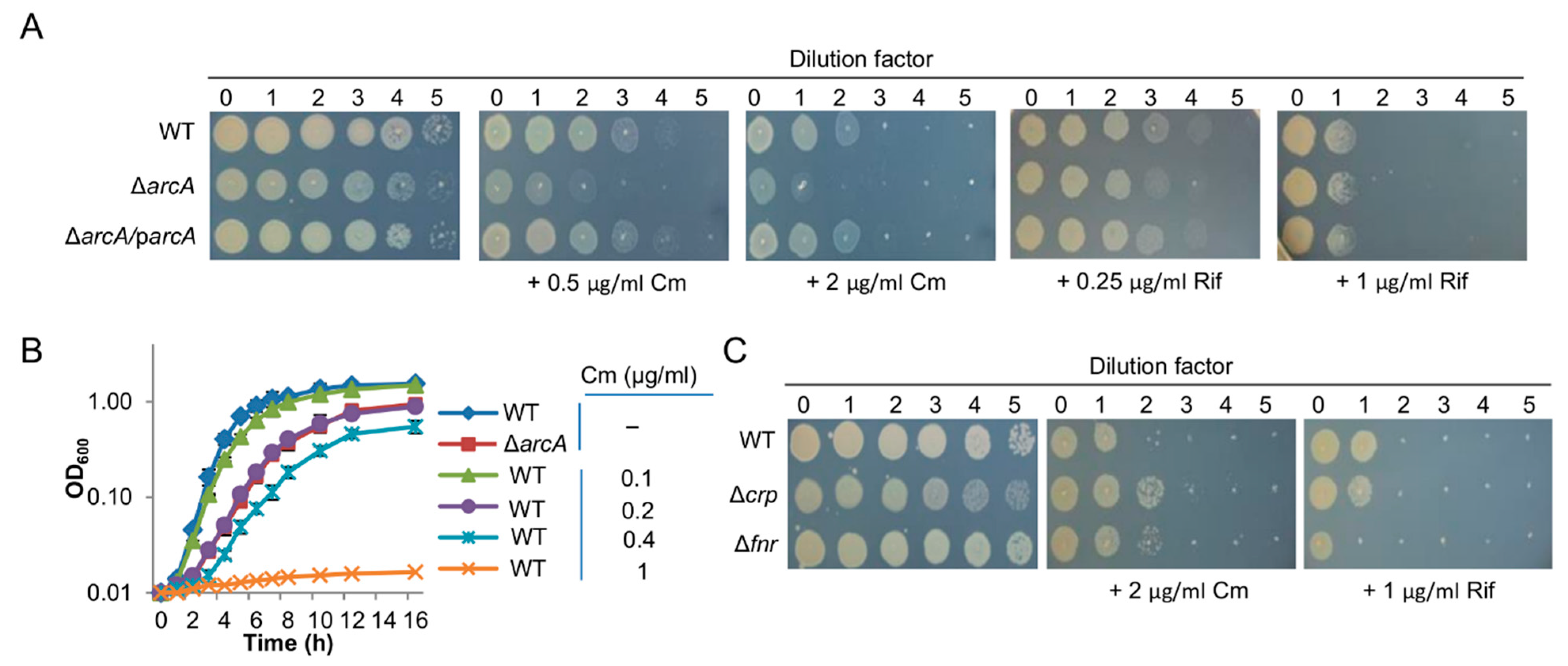
3.1. The arcA Mutation Impairs Translation Efficacy
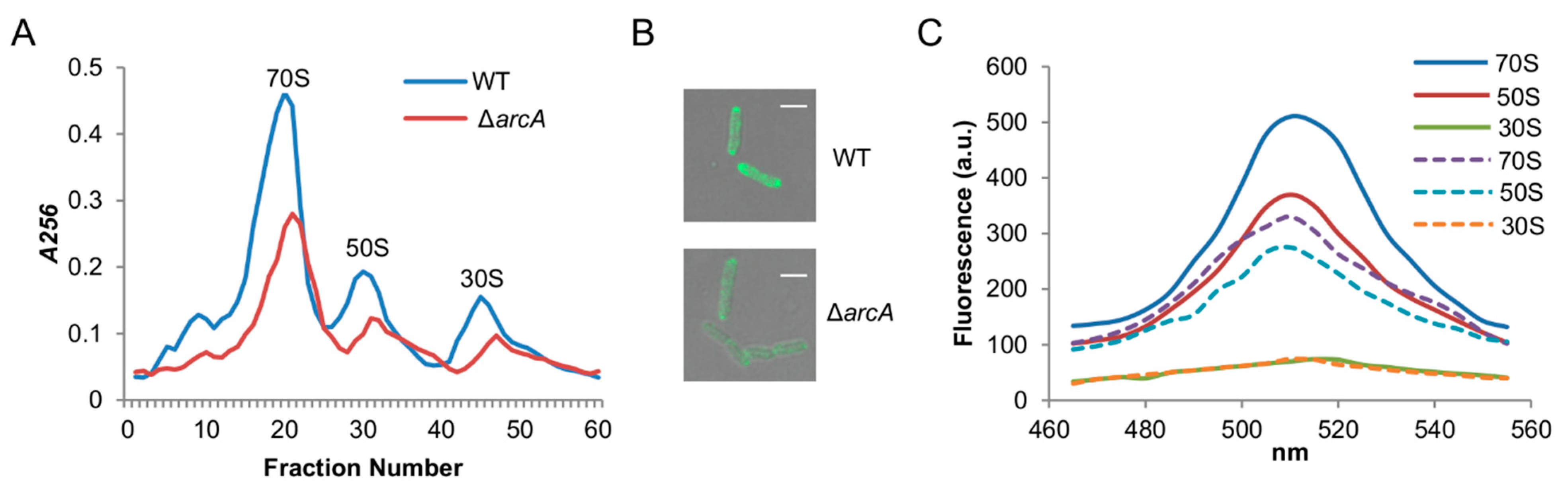
3.2. Reduced Ribosome Biosynthesis May Underlie Translation and Growth Defects of the S. oneidensis arcA Mutant
3.3. ArcA (but Not Fnr or Crp) Impacts Translation Efficiency
3.4. The ArcA Loss Reduces Ribosome Biosynthesis but Does Not Affect Ribosome Assembly
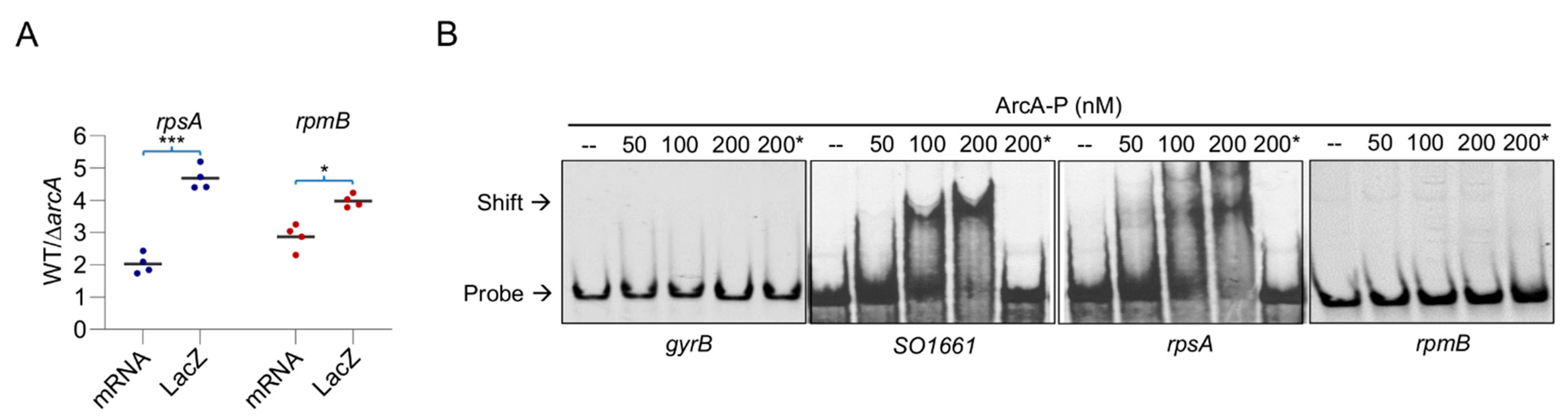
3.5. rpsA Is under the Direct Control of ArcA
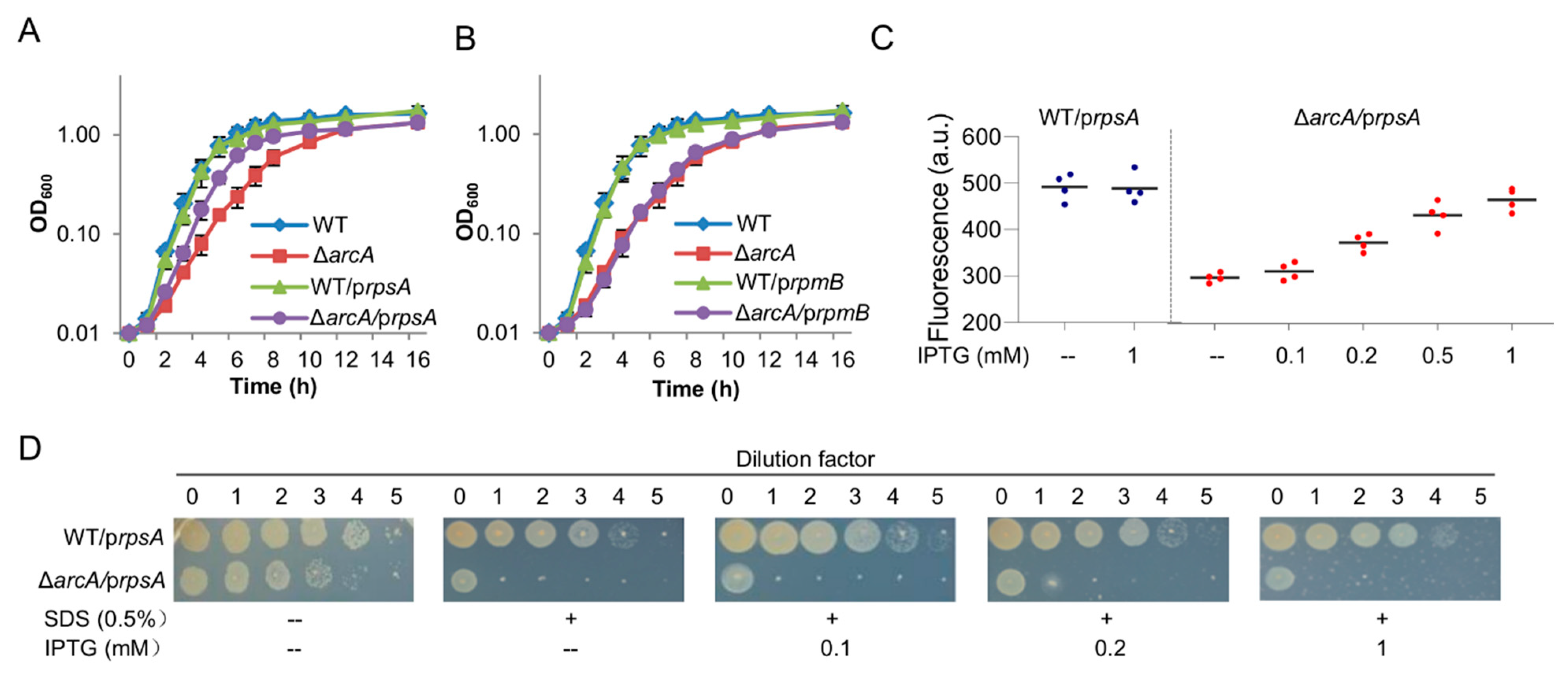
3.6. Reduced Expression of rpsA Is, at Least in Part, Accountable for Defects in Aerobic Growth but Not in the Cell Envelope Integrity Caused by the ArcA Loss
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Groisman, E.A. Feedback Control of Two-Component Regulatory Systems. Annu. Rev. Microbiol. 2016, 70, 103–124. [Google Scholar] [CrossRef]
- Lynch, A.S.; Lin, E.C. Transcriptional control mediated by the ArcA two-component response regulator protein of Escherichia coli: Characterization of DNA binding at target promoters. J. Bacteriol. 1996, 178, 6238–6249. [Google Scholar] [CrossRef]
- Park, D.M.; Akhtar, M.S.; Ansari, A.Z.; Landick, R.; Kiley, P.J. The bacterial response regulator ArcA uses a diverse binding site architecture to regulate carbon oxidation globally. PLoS Genet. 2013, 9, e1003839. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Wan, F.; Yin, J.; Gao, H. Ecological roles of Arc signal transduction system revealed by evolutionary genetics analysis. J. Bacteriol. Mycol. 2014, 1, 8. [Google Scholar]
- Basan, M.; Hui, S.; Williamson, J.R. ArcA overexpression induces fermentation and results in enhanced growth rates of E. coli. Sci. Rep. 2017, 7, 11866. [Google Scholar] [CrossRef]
- Georgellis, D.; Kwon, O.; Lin, E.C. Quinones as the redox signal for the arc two-component system of bacteria. Science 2001, 292, 2314–2316. [Google Scholar] [CrossRef]
- Salmon, K.A.; Hung, S.P.; Steffen, N.R.; Krupp, R.; Baldi, P.; Hatfield, G.W.; Gunsalus, R.P. Global gene expression profiling in Escherichia coli K12: Effects of oxygen availability and ArcA. J. Biol. Chem. 2005, 280, 15084–15096. [Google Scholar] [CrossRef]
- Jiang, F.; An, C.; Bao, Y.; Zhao, X.; Jernigan, R.L.; Lithio, A.; Nettleton, D.; Li, L.; Wurtele, E.S.; Nolan, L.K.; et al. ArcA Controls Metabolism, Chemotaxis, and Motility Contributing to the Pathogenicity of Avian Pathogenic Escherichia coli. Infect. Immun. 2015, 83, 3545–3554. [Google Scholar] [CrossRef]
- Wang, Z.; Sun, J.; Xia, T.; Liu, Y.; Fu, J.; Lo, Y.K.; Chang, C.; Yan, A.; Liu, X. Proteomic Delineation of the ArcA Regulon in Salmonella Typhimurium during Anaerobiosis. Mol. Cell Proteom. 2018, 17, 1937–1947. [Google Scholar] [CrossRef]
- Green, J.; Paget, M.S. Bacterial redox sensors. Nat. Rev. Microbiol. 2004, 2, 954–966. [Google Scholar] [CrossRef]
- Jiang, F.; Huang, X.; Barbieri, N.L.; Logue, C.M.; Nolan, L.K.; Li, G. Citrate utilization under anaerobic environment in Escherichia coli is under direct control of Fnr and indirect control of ArcA and Fnr via CitA-CitB system. Environ. Microbiol. 2021, 23, 1496–1509. [Google Scholar] [CrossRef]
- Klumpp, S.; Hwa, T. Bacterial growth: Global effects on gene expression, growth feedback and proteome partition. Curr. Opin. Biotechnol. 2014, 28, 96–102. [Google Scholar] [CrossRef]
- You, C.; Okano, H.; Hui, S.; Zhang, Z.; Kim, M.; Gunderson, C.W.; Wang, Y.P.; Lenz, P.; Yan, D.; Hwa, T. Coordination of bacterial proteome with metabolism by cyclic AMP signalling. Nature 2013, 500, 301–306. [Google Scholar] [CrossRef]
- Lemaire, O.N.; Méjean, V.; Iobbi-Nivol, C. The Shewanella genus: Ubiquitous organisms sustaining and preserving aquatic ecosystems. FEMS Microbiol. Rev. 2020, 44, 155–170. [Google Scholar] [CrossRef]
- Gao, H.; Wang, X.; Yang, Z.K.; Chen, J.; Liang, Y.; Chen, H.; Palzkill, T.; Zhou, J. Physiological roles of ArcA, Crp, and EtrA and their interactive control on aerobic and anaerobic respiration in Shewanella oneidensis. PLoS ONE 2010, 5, e15295. [Google Scholar] [CrossRef]
- Cruz-García, C.; Murray, A.E.; Rodrigues, J.L.; Gralnick, J.A.; McCue, L.A.; Romine, M.F.; Löffler, F.E.; Tiedje, J.M. Fnr (EtrA) acts as a fine-tuning regulator of anaerobic metabolism in Shewanella oneidensis MR-1. BMC Microbiol. 2011, 11, 64. [Google Scholar] [CrossRef] [PubMed]
- Charania, M.A.; Brockman, K.L.; Zhang, Y.; Banerjee, A.; Pinchuk, G.E.; Fredrickson, J.K.; Beliaev, A.S.; Saffarini, D.A. Involvement of a membrane-bound class III adenylate cyclase in regulation of anaerobic respiration in Shewanella oneidensis MR-1. J. Bacteriol. 2009, 191, 4298–4306. [Google Scholar] [CrossRef]
- Fu, H.; Chen, H.; Wang, J.; Zhou, G.; Zhang, H.; Zhang, L.; Gao, H. Crp-dependent cytochrome bd oxidase confers nitrite resistance to Shewanella oneidensis. Environ. Microbiol. 2013, 15, 2198–2212. [Google Scholar] [CrossRef] [PubMed]
- Zhou, G.; Yin, J.; Chen, H.; Hua, Y.; Sun, L.; Gao, H. Combined effect of loss of the caa3 oxidase and Crp regulation drives Shewanella to thrive in redox-stratified environments. ISME J. 2013, 7, 1752–1763. [Google Scholar] [CrossRef]
- Wu, G.; Li, N.; Mao, Y.; Zhou, G.; Gao, H. Endogenous generation of hydrogen sulfide and its regulation in Shewanella oneidensis. Front. Microbiol. 2015, 6, 374. [Google Scholar] [CrossRef]
- Yin, J.; Meng, Q.; Fu, H.; Gao, H. Reduced expression of cytochrome oxidases largely explains cAMP inhibition of aerobic growth in Shewanella oneidensis. Sci. Rep. 2016, 6, 24449. [Google Scholar] [CrossRef]
- Jin, M.; Fu, H.; Yin, J.; Yuan, J.; Gao, H. Molecular Underpinnings of Nitrite Effect on CymA-Dependent Respiration in Shewanella oneidensis. Front. Microbiol. 2016, 7, 1154. [Google Scholar] [CrossRef]
- Gralnick, J.A.; Brown, C.T.; Newman, D.K. Anaerobic regulation by an atypical Arc system in Shewanella oneidensis. Mol. Microbiol. 2005, 56, 1347–1357. [Google Scholar] [CrossRef]
- Gao, H.; Wang, X.; Yang, Z.K.; Palzkill, T.; Zhou, J. Probing regulon of ArcA in Shewanella oneidensis MR-1 by integrated genomic analyses. BMC Genom. 2008, 9, 42. [Google Scholar] [CrossRef]
- Lassak, J.; Henche, A.L.; Binnenkade, L.; Thormann, K.M. ArcS, the cognate sensor kinase in an atypical Arc system of Shewanella oneidensis MR-1. Appl. Environ. Microbiol. 2010, 76, 3263–3274. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Gao, H.; Shen, Y.; Weinstock, G.M.; Zhou, J.; Palzkill, T. A high-throughput percentage-of-binding strategy to measure binding energies in DNA-protein interactions: Application to genome-scale site discovery. Nucleic Acids Res. 2008, 36, 4863–4871. [Google Scholar] [CrossRef]
- Wan, F.; Mao, Y.; Dong, Y.; Ju, L.; Wu, G.; Gao, H. Impaired cell envelope resulting from arcA mutation largely accounts for enhanced sensitivity to hydrogen peroxide in Shewanella oneidensis. Sci. Rep. 2015, 5, 10228. [Google Scholar] [CrossRef]
- Liang, H.; Zhang, Y.; Wang, S.; Gao, H. Mutual interplay between ArcA and σ(E) orchestrates envelope stress response in Shewanella oneidensis. Environ. Microbiol. 2021, 23, 652–668. [Google Scholar] [CrossRef]
- Xie, P.; Liang, H.; Wang, J.; Huang, Y.; Gao, H. Lipopolysaccharide Transport System Links Physiological Roles of σ(E) and ArcA in the Cell Envelope Biogenesis in Shewanella oneidensis. Microbiol. Spectr. 2021, 9, e00690-21. [Google Scholar] [CrossRef]
- Guo, M.S.; Gross, C.A. Stress-induced remodeling of the bacterial proteome. Curr. Biol. 2014, 24, R424–R434. [Google Scholar] [CrossRef]
- Meng, Q.; Liang, H.; Gao, H. Roles of multiple KASIII homologues of Shewanella oneidensis in initiation of fatty acid synthesis and in cerulenin resistance. Biochim. Biophys. Acta. Mol. Cell Biol. Lipids 2018, 1863, 1153–1163. [Google Scholar] [CrossRef] [PubMed]
- Fu, H.; Liu, L.; Dong, Z.; Guo, S.; Gao, H. Dissociation between Iron and Heme Biosyntheses Is Largely Accountable for Respiration Defects of Shewanella oneidensis fur Mutants. Appl. Environ. Microbiol. 2018, 84, e00039-18. [Google Scholar] [CrossRef] [PubMed]
- Luo, Q.; Dong, Y.; Chen, H.; Gao, H. Mislocalization of Rieske protein PetA predominantly accounts for the aerobic growth defect of Tat mutants in Shewanella oneidensis. PLoS ONE 2013, 8, e62064. [Google Scholar] [CrossRef]
- Jin, M.; Jiang, Y.; Sun, L.; Yin, J.; Fu, H.; Wu, G.; Gao, H. Unique organizational and functional features of the cytochrome c maturation system in Shewanella oneidensis. PLoS ONE 2013, 8, e75610. [Google Scholar] [CrossRef]
- Shi, M.; Wan, F.; Mao, Y.; Gao, H. Unraveling the Mechanism for the Viability Deficiency of Shewanella oneidensis oxyR Null Mutant. J. Bacteriol. 2015, 197, 2179–2189. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Dong, Y.; Luo, Q.; Li, N.; Wu, G.; Gao, H. Protection from oxidative stress relies mainly on derepression of OxyR-dependent KatB and Dps in Shewanella oneidensis. J. Bacteriol. 2014, 196, 445–458. [Google Scholar] [CrossRef]
- Yuan, J.; Wei, B.; Shi, M.; Gao, H. Functional assessment of EnvZ/OmpR two-component system in Shewanella Oneidensis. PLoS ONE 2011, 6, e23701. [Google Scholar] [CrossRef] [PubMed]
- Fu, H.; Jin, M.; Ju, L.; Mao, Y.; Gao, H. Evidence for function overlapping of CymA and the cytochrome bc1 complex in the Shewanella oneidensis nitrate and nitrite respiration. Environ. Microbiol. 2014, 16, 3181–3195. [Google Scholar] [CrossRef]
- Charollais, J.; Pflieger, D.; Vinh, J.; Dreyfus, M.; Iost, I. The DEAD-box RNA helicase SrmB is involved in the assembly of 50S ribosomal subunits in Escherichia coli. Mol. Microbiol. 2003, 48, 1253–1265. [Google Scholar] [CrossRef]
- Liang, H.; Mao, Y.; Sun, Y.; Gao, H. Transcriptional regulator ArcA mediates expression of oligopeptide transport systems both directly and indirectly in Shewanella oneidensis. Sci. Rep. 2019, 9, 13839. [Google Scholar] [CrossRef] [PubMed]
- Scott, M.; Gunderson, C.W.; Mateescu, E.M.; Zhang, Z.; Hwa, T. Interdependence of cell growth and gene expression: Origins and consequences. Science 2010, 330, 1099–1102. [Google Scholar] [CrossRef]
- Yuan, J.; Wei, B.; Lipton, M.S.; Gao, H. Impact of ArcA loss in Shewanella oneidensis revealed by comparative proteomics under aerobic and anaerobic conditions. Proteomics 2012, 12, 1957–1969. [Google Scholar] [CrossRef]
- Yin, J.; Sun, L.; Dong, Y.; Chi, X.; Zhu, W.; Qi, S.H.; Gao, H. Expression of blaA underlies unexpected ampicillin-induced cell lysis of Shewanella oneidensis. PLoS ONE 2013, 8, e60460. [Google Scholar] [CrossRef] [PubMed]
- Chai, Q.; Singh, B.; Peisker, K.; Metzendorf, N.; Ge, X.; Dasgupta, S.; Sanyal, S. Organization of ribosomes and nucleoids in Escherichia coli cells during growth and in quiescence. J. Biol. Chem. 2014, 289, 11342–11352. [Google Scholar] [CrossRef] [PubMed]
- Bakshi, S.; Siryaporn, A.; Goulian, M.; Weisshaar, J.C. Superresolution imaging of ribosomes and RNA polymerase in live Escherichia coli cells. Mol. Microbiol. 2012, 85, 21–38. [Google Scholar] [CrossRef]
- Salah, P.; Bisaglia, M.; Aliprandi, P.; Uzan, M.; Sizun, C.; Bontems, F. Probing the relationship between Gram-negative and Gram-positive S1 proteins by sequence analysis. Nucleic Acids Res. 2009, 37, 5578–5588. [Google Scholar] [CrossRef]
- Sengupta, J.; Agrawal, R.K.; Frank, J. Visualization of protein S1 within the 30S ribosomal subunit and its interaction with messenger RNA. Proc. Natl. Acad. Sci. USA 2001, 98, 11991–11996. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Zhu, M. Coupling of Ribosome Synthesis and Translational Capacity with Cell Growth. Trends Biochem. Sci. 2020, 45, 681–692. [Google Scholar] [CrossRef]
- Dai, X.; Zhu, M.; Warren, M.; Balakrishnan, R.; Patsalo, V.; Okano, H.; Williamson, J.R.; Fredrick, K.; Wang, Y.P.; Hwa, T. Reduction of translating ribosomes enables Escherichia coli to maintain elongation rates during slow growth. Nat. Microbiol. 2016, 2, 16231. [Google Scholar] [CrossRef]
- Serres, M.H.; Riley, M. Genomic analysis of carbon source metabolism of Shewanella oneidensis MR-1: Predictions versus experiments. J. Bacteriol. 2006, 188, 4601–4609. [Google Scholar] [CrossRef]
- Sørensen, M.A.; Fricke, J.; Pedersen, S. Ribosomal protein S1 is required for translation of most, if not all, natural mRNAs in Escherichia coli in vivo. J. Mol. Biol. 1998, 280, 561–569. [Google Scholar] [CrossRef]
- Wimberly, B.T.; Brodersen, D.E.; Clemons, W.M., Jr.; Morgan-Warren, R.J.; Carter, A.P.; Vonrhein, C.; Hartsch, T.; Ramakrishnan, V. Structure of the 30S ribosomal subunit. Nature 2000, 407, 327–339. [Google Scholar] [CrossRef]
- Schuwirth, B.S.; Borovinskaya, M.A.; Hau, C.W.; Zhang, W.; Vila-Sanjurjo, A.; Holton, J.M.; Cate, J.H. Structures of the bacterial ribosome at 3.5 A resolution. Science 2005, 310, 827–834. [Google Scholar] [CrossRef] [PubMed]
- Gualerzi, C.O.; Pon, C.L. Initiation of mRNA translation in bacteria: Structural and dynamic aspects. Cell Mol. Life Sci. 2015, 72, 4341–4367. [Google Scholar] [CrossRef]
- Rodnina, M.V.; Fischer, N.; Maracci, C.; Stark, H. Ribosome dynamics during decoding. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2017, 372, 20160182. [Google Scholar] [CrossRef] [PubMed]
- Lassak, J.; Bubendorfer, S.; Thormann, K.M. Domain analysis of ArcS, the hybrid sensor kinase of the Shewanella oneidensis MR-1 Arc two-component system, reveals functional differentiation of its two receiver domains. J. Bacteriol. 2013, 195, 482–492. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Alvarez, A.F.; Malpica, R.; Contreras, M.; Escamilla, E.; Georgellis, D. Cytochrome d but not cytochrome o rescues the toluidine blue growth sensitivity of arc mutants of Escherichia Coli. J. Bacteriol. 2010, 192, 391–399. [Google Scholar] [CrossRef] [PubMed]

| Strain or Plasmid | Description | Reference or Source |
|---|---|---|
| E. coli strains | ||
| DH5α | Host for cloning | Lab stock |
| WM3064 | ΔdapA, donor strain for conjugation | W. Metcalf, UIUC |
| S. oneidensis strains | ||
| MR-1 | Wild type | ATCC 700550 |
| HG0624 | ∆crp derived from MR-1 | [15] |
| HG2356 | ∆fnr derived from MR-1 | [15] |
| HG3988 | ∆arcA derived from MR-1 | [24] |
| HG3927W | eGFP knock-in at rplI derived from MR-1 | This study |
| HG3927A | eGFP knock-in at rplI derived from ∆arcA | This study |
| Plasmid | ||
| pHGM01 | Apr Gmr Cmr suicide vector | [31] |
| pHGEI01 | Kmr, integrative lacZ reporter vector | [32] |
| pHGEN–Ptac | Kmr, IPTG-inducible expression vector | [33] |
| pBBR–Cre | Spr, helper plasmid for antibiotic cassette removal | [18] |
| pHG101–ArcA | Complementation vector carrying arcA | [29] |
| pHGEI01–PaprE | PaprE–lacZ fusion within pHGEI01 | [29] |
| pHGEI01–Psap | Psap–lacZ fusion within pHGEI01 | This study |
| pHGEI01–PdtpA | PdtpA–lacZ fusion within pHGEI01 | This study |
| pHGEI01–PdtpB | PdtpB–lacZ fusion within pHGEI01 | This study |
| pHGEI01–PSO1505 | PSO1505–lacZ fusion within pHGEI01 | This study |
| pHGEI01–PSO3195 | PSO3195–lacZ fusion within pHGEI01 | This study |
| pHGEI01–PrplK | PrplK–lacZ fusion within pHGEI01 | This study |
| pHGEI01–PsspA | PsspA–lacZ fusion within pHGEI01 | This study |
| pHGEI01–PSO0783 | PSO0783–lacZ fusion within pHGEI01 | This study |
| pHGEI01–PbtuD | PbtuD–lacZ fusion within pHGEI01 | This study |
| pHGEI01–PnuoA | PnuoA–lacZ fusion within pHGEI01 | This study |
| pHGEI01–PSO2061 | PSO2016–lacZ fusion within pHGEI01 | This study |
| pHGEI01–PSO3282 | PSO3282–lacZ fusion within pHGEI01 | This study |
| pHGEI01–PSO4542 | PSO4542–lacZ fusion within pHGEI01 | This study |
| pHGEI01–PpspF | PpspF–lacZ fusion within pHGEI01 | This study |
| pHGEI01–PhoxK | PhoxK–lacZ fusion within pHGEI01 | This study |
| pHGEI01–PcsgB | PcsgB–lacZ fusion within pHGEI01 | This study |
| pHGEI01–PrpsA | PrpsA–lacZ fusion within pHGEI01 | This study |
| pHGEI01–PrpmB | PrpmB–lacZ fusion within pHGEI01 | This study |
| pHGEN-Ptac-rpsA | Ptac-rpsA within pHGE-Ptac | This study |
| pHGEN-Ptac-rpmB | Ptac-rpmB within pHGE-Ptac | This study |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xie, P.; Wang, J.; Liang, H.; Gao, H. Shewanella oneidensis arcA Mutation Impairs Aerobic Growth Mainly by Compromising Translation. Life 2021, 11, 926. https://doi.org/10.3390/life11090926
Xie P, Wang J, Liang H, Gao H. Shewanella oneidensis arcA Mutation Impairs Aerobic Growth Mainly by Compromising Translation. Life. 2021; 11(9):926. https://doi.org/10.3390/life11090926
Chicago/Turabian StyleXie, Peilu, Jiahao Wang, Huihui Liang, and Haichun Gao. 2021. "Shewanella oneidensis arcA Mutation Impairs Aerobic Growth Mainly by Compromising Translation" Life 11, no. 9: 926. https://doi.org/10.3390/life11090926
APA StyleXie, P., Wang, J., Liang, H., & Gao, H. (2021). Shewanella oneidensis arcA Mutation Impairs Aerobic Growth Mainly by Compromising Translation. Life, 11(9), 926. https://doi.org/10.3390/life11090926







