Liquid Biopsy, ctDNA Diagnosis through NGS
Abstract
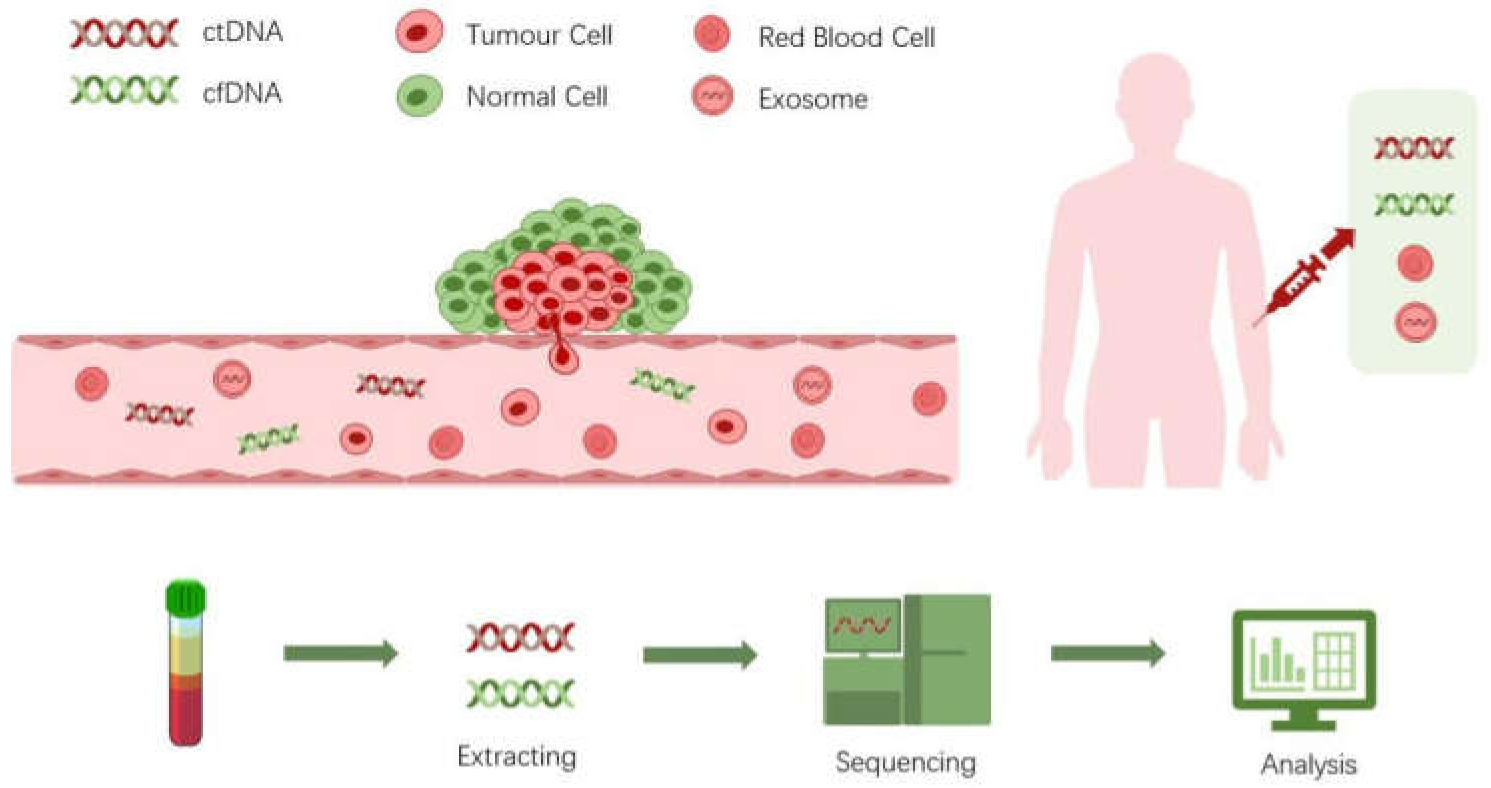
:1. Introduction
2. Biology of ctDNA
3. Next Generation Sequencing Technologies for ctDNA Detection
4. Strategies for True Mutation Detection Using NGS
5. Detection of ctDNA Mutations for Early Cancer Diagnosis
6. Detection of Therapeutically Targetable Mutations in ctDNA
7. Detection of ctDNA Mutations for Cancer Monitoring and Assessment of Minimal Residual Disease
8. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Junqueira-Neto, S.; Batista, I.A.; Costa, J.L.; Melo, S.A. Liquid Biopsy beyond Circulating Tumor Cells and Cell-Free DNA. Acta Cytol. 2019, 63, 479–488. [Google Scholar] [CrossRef] [PubMed]
- Poulet, G.; Massias, J.; Taly, V. Liquid Biopsy: General Concepts. Acta Cytol. 2019, 63, 449–455. [Google Scholar] [CrossRef]
- Dakubo, G.D. Cancer Biomarkers in Body Fluids; Springer International Publishing: Cham, Switzerland, 2017. [Google Scholar]
- Goodwin, S.; McPherson, J.D.; McCombie, W.R. Coming of age: Ten years of next-generation sequencing technologies. Nat. Rev. Genet. 2016, 17, 333–351. [Google Scholar] [CrossRef] [PubMed]
- Thierry, A.R.; El Messaoudi, S.; Gahan, P.B.; Anker, P.; Stroun, M. Origins, structures, and functions of circulating DNA in oncology. Cancer Metastasis Rev. 2016, 35, 347–376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mouliere, F.; Chandrananda, D.; Piskorz, A.M.; Moore, E.K.; Morris, J.; Ahlborn, L.B.; Mair, R.; Goranova, T.; Marass, F.; Heider, K.; et al. Enhanced detection of circulating tumor DNA by fragment size analysis. Sci. Transl. Med. 2018, 10, eaat4921. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fleischhacker, M.; Schmidt, B. Circulating nucleic acids (CNAs) and cancer—A survey. Biochim. Biophys. Acta BBA Rev. Cancer 2007, 1775, 181–232. [Google Scholar] [CrossRef] [PubMed]
- Aucamp, J.; Bronkhorst, A.J.; Badenhorst, C.P.S.; Pretorius, P.J. The diverse origins of circulating cell-free DNA in the human body: A critical re-evaluation of the literature. Biol. Rev. 2018, 93, 1649–1683. [Google Scholar] [CrossRef] [PubMed]
- Elazezy, M.; Joosse, S.A. Techniques of using circulating tumor DNA as a liquid biopsy component in cancer management. Comput. Struct. Biotechnol. J. 2018, 16, 370–378. [Google Scholar] [CrossRef]
- Cheng, F.; Su, L.; Qian, C. Circulating tumor DNA: A promising biomarker in the liquid biopsy of cancer. Oncotarget 2016, 7, 48832–48841. [Google Scholar] [CrossRef] [Green Version]
- Diehl, F.; Li, M.; Dressman, D.; He, Y.; Shen, D.; Szabo, S.; Diaz, L.A.; Goodman, S.N.; David, K.A.; Juhl, H.; et al. Detection and quantification of mutations in the plasma of patients with colorectal tumors. Proc. Natl. Acad. Sci. USA 2005, 102, 16368–16373. [Google Scholar] [CrossRef] [Green Version]
- Avanzini, S.; Kurtz, D.M.; Chabon, J.J.; Moding, E.J.; Hori, S.S.; Gambhir, S.S.; Alizadeh, A.A.; Diehn, M.; Reiter, J.G. A mathematical model of ctDNA shedding predicts tumor detection size. Sci. Adv. 2020, 6, eabc4308. [Google Scholar] [CrossRef]
- Stewart, C.M.; Kothari, P.D.; Mouliere, F.; Mair, R.; Somnay, S.; Benayed, R.; Zehir, A.; Weigelt, B.; Dawson, S.-J.; Arcila, M.E.; et al. The value of cell-free DNA for molecular pathology. J. Pathol. 2018, 244, 616–627. [Google Scholar] [CrossRef] [PubMed]
- Sueoka-Aragane, N.; Nakashima, C.; Yoshida, H.; Matsumoto, N.; Iwanaga, K.; Ebi, N.; Nishiyama, A.; Yatera, K.; Kuyama, S.; Fukuda, M.; et al. The role of comprehensive analysis with circulating tumor DNA in advanced non-small cell lung cancer patients considered for osimertinib treatment. Cancer Med. 2021, 10, 3873–3885. [Google Scholar] [CrossRef] [PubMed]
- Schwarzenbach, H.; Hoon, D.S.B.; Pantel, K. Cell-free nucleic acids as biomarkers in cancer patients. Nat. Rev. Cancer 2011, 11, 426–437. [Google Scholar] [CrossRef]
- Zaporozhchenko, I.A.; Ponomaryova, A.A.; Rykova, E.Y.; Laktionov, P.P. The potential of circulating cell-free RNA as a can-cer biomarker: Challenges and opportunities. Expert Rev. Mol. Diagn. 2018, 18, 133–145. [Google Scholar] [CrossRef]
- Keller, L.; Belloum, Y.; Wikman, H.; Pantel, K. Clinical relevance of blood-based ctDNA analysis: Mutation detection and beyond. Br. J. Cancer 2021, 124, 345–358. [Google Scholar] [CrossRef]
- Lu, L.; Bi, J.; Bao, L. Genetic profiling of cancer with circulating tumor DNA analysis. J. Genet. Genom. 2018, 45, 79–85. [Google Scholar] [CrossRef] [PubMed]
- Kastrisiou, M.; Zarkavelis, G.; Pentheroudakis, G.; Magklara, A. Clinical application of next-generation sequencing as a liq-uid biopsy technique in advanced colorectal cancer: A trick or a treat? Cancers 2019, 11, 1573. [Google Scholar] [CrossRef] [Green Version]
- Bai, Y.; Wang, Z.; Liu, Z.; Liang, G.; Gu, W.; Ge, Q. Technical progress in circulating tumor DNA analysis using next genera-tion sequencing. Mol. Cell Probes 2020, 49, 101480. [Google Scholar] [CrossRef] [PubMed]
- Levy, S.E.; Myers, R.M. Advancements in Next-Generation Sequencing. Annu. Rev. Genom. Hum. Genet. 2016, 17, 95–115. [Google Scholar] [CrossRef] [Green Version]
- Srivastava, V.K. Sequencing Technologies: Introduction and Applications. Int. J. Hum. Genet. 2019, 19. [Google Scholar] [CrossRef]
- Martignano, F.; Munagala, U.; Crucitta, S.; Mingrino, A.; Semeraro, R.; Del Re, M.; Petrini, I.; Magi, A.; Conticello, S.G. Na-nopore sequencing from liquid biopsy: Analysis of copy number variations from cell-free DNA of lung cancer patients. Mol. Cancer 2021, 20, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Liu, M.; Zhou, Y. Bioinformatics Analysis for Cell-Free Tumor DNA Sequencing Data. Methods Mol. Biol. 2018, 67–95. [Google Scholar] [CrossRef]
- Aparicio-Puerta, E.; Jáspez, D.; Lebrón, R.; Koppers-Lalic, D.; Marchal, J.A.; Hackenberg, M. liqDB: A small-RNAseq knowledge discovery database for liquid biopsy studies. Nucleic Acids Res. 2019, 47, D113–D120. [Google Scholar] [CrossRef] [Green Version]
- Zhao, L.; Wu, X.; Li, T.; Luo, J.; Dong, D. ctcRbase: The gene expression database of circulating tumor cells and microemboli. Database 2020, 2020. [Google Scholar] [CrossRef] [PubMed]
- Cheng, F.; Zhao, J.; Zhao, Z. Advances in computational approaches for prioritizing driver mutations and significantly mutated genes in cancer genomes. Brief. Bioinform. 2016, 17, 642–656. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bos, M.K.; Nasserinejad, K.; Jansen, M.P.H.M.; Angus, L.; Atmodimedjo, P.N.; de Jonge, E.; Dinjens, W.N.M.; van Schaik, R.H.N.; Del Re, M.; Dubbink, H.J.; et al. Comparison of variant allele frequency and number of mutant molecules as units of measurement for circulating tumor DNA. Mol. Oncol. 2021, 15, 57–66. [Google Scholar] [CrossRef]
- Forshew, T.; Murtaza, M.; Parkinson, C.; Gale, D.; Tsui, D.W.Y.; Kaper, F.; Dawson, S.-J.; Piskorz, A.M.; Jimenez-Linan, M.; Bentley, D.; et al. Noninvasive Identification and Monitoring of Cancer Mutations by Targeted Deep Sequencing of Plasma DNA. Sci. Transl. Med. 2012, 4, 136ra68. [Google Scholar] [CrossRef]
- Gale, D.; Lawson, A.R.J.; Howarth, K.; Madi, M.; Durham, B.; Smalley, S.; Calaway, J.; Blais, S.; Jones, G.; Clark, J.; et al. Development of a highly sensitive liquid biopsy platform to detect clinically-relevant cancer mutations at low allele fractions in cell-free DNA. PLoS ONE 2018, 13, e0194630. [Google Scholar] [CrossRef]
- Smith, T.; Heger, A.; Sudbery, I. UMI-tools: Modeling sequencing errors in Unique Molecular Identifiers to improve quantification accuracy. Genome Res. 2017, 27, 491–499. [Google Scholar] [CrossRef] [Green Version]
- Kinde, I.; Wu, J.; Papadopoulos, N.; Kinzler, K.W.; Vogelstein, B. Detection and quantification of rare mutations with massively parallel sequencing. Proc. Natl. Acad. Sci. USA 2011, 108, 9530–9535. [Google Scholar] [CrossRef] [Green Version]
- Newman, A.M.; Bratman, S.V.; To, J.; Wynne, J.F.; Eclov, N.C.W.; Modlin, L.A.; Liu, C.L.; Neal, J.W.; Wakelee, H.A.; Merritt, R.E.; et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat. Med. 2014, 20, 548–554. [Google Scholar] [CrossRef]
- Phallen, J.; Sausen, M.; Adleff, V.; Leal, A.; Hruban, C.; White, J.; Anagnostou, V.; Fiksel, J.; Cristiano, S.; Papp, E.; et al. Direct detection of early-stage cancers using circulating tumor DNA. Sci. Transl. Med. 2017, 9, eaan2415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takai, E.; Yachida, S. Circulating tumor DNA as a liquid biopsy target for detection of pancreatic cancer. World J. Gastroenterol. 2016, 22, 8480–8488. [Google Scholar] [CrossRef] [PubMed]
- Oshi, M.; Tokumaru, Y.; Patel, A.; Yan, L.; Matsuyama, R.; Endo, I.; Katz, M.H.G.; Takabe, K. A Novel Four-Gene Score to Predict Pathologically Complete (R0) Resection and Survival in Pancreatic Cancer. Cancers 2020, 12, 3635. [Google Scholar] [CrossRef]
- Zill, O.A.; Greene, C.; Sebisanovic, D.; Siew, L.M.; Leng, J.; Vu, M.; Hendifar, A.E.; Wang, Z.; Atreya, C.E.; Kelley, R.K.; et al. Cell-Free DNA Next-Generation Sequencing in Pancreatobiliary Carcinomas. Cancer Discov. 2015, 5, 1040–1048. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cohen, J.D.; Li, L.; Wang, Y.; Thoburn, C.; Afsari, B.; Danilova, L.; Douville, C.; Javed, A.A.; Wong, F.; Mattox, A.; et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science 2018, 359, 926–930. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Killock, D. Diagnosis: CancerSEEK and destroy-A blood test for early cancer detection. Nat. Rev. Clin. Oncol. 2018, 15, 133. [Google Scholar] [CrossRef]
- Bettegowda, C.; Sausen, M.; Leary, R.J.; Kinde, I.; Wang, Y.; Agrawal, N.; Bartlett, B.R.; Wang, H.; Luber, B.; Alani, R.M.; et al. Detection of Circulating Tumor DNA in Early- and Late-Stage Human Malignancies. Sci. Transl. Med. 2014, 6, 224ra24. [Google Scholar] [CrossRef] [Green Version]
- Cohen, J.D.; Javed, A.A.; Thoburn, C.; Wong, F.; Tie, J.; Gibbs, P.; Schmidt, C.M.; Yip-Schneider, M.T.; Allen, P.J.; Schattner, M.; et al. Combined circulating tumor DNA and protein biomarker-based liquid biopsy for the earlier detection of pancreatic cancers. Proc. Natl. Acad. Sci. USA 2017, 114, 10202–10207. [Google Scholar] [CrossRef] [Green Version]
- Fiala, C.; Diamandis, E.P. Utility of circulating tumor DNA in cancer diagnostics with emphasis on early detection. BMC Med. 2018, 16, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Liang, N.; Li, B.; Jia, Z.; Wang, C.; Wu, P.; Zheng, T.; Wang, Y.; Qiu, F.; Wu, Y.; Su, J.; et al. Ultrasensitive detection of circulating tumour DNA via deep methylation sequencing aided by machine learning. Nat. Biomed. Eng. 2021, 5, 586–599. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.; Diep, D.; Plongthongkum, N.; Fung, H.-L.; Zhang, K.Z.K.; Zhang, K. Identification of methylation haplotype blocks AIDS in deconvolution of heterogeneous tissue samples and tumor tissue-of-origin mapping from plasma DNA. Nat. Genet. 2017, 49, 635–642. [Google Scholar] [CrossRef]
- Moss, J.; Magenheim, J.; Neiman, D.; Zemmour, H.; Loyfer, N.; Korach, A.; Samet, Y.; Maoz, M.; Druid, H.; Arner, P.; et al. Comprehensive human cell-type methylation atlas reveals origins of circulating cell-free DNA in health and disease. Nat. Commun. 2018, 9, 5068. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.; Qiu, C.; Wang, B.; Bing, P.; Tian, G.; Zhang, X.; Ma, J.; He, B.; Yang, J. Evaluating DNA Methylation, Gene Expression, Somatic Mutation, and Their Combinations in Inferring Tumor Tissue-of-Origin. Front. Cell Dev. Biol. 2021, 9. [Google Scholar] [CrossRef]
- Lu, H.-Y.; Qin, J.; Han, N.; Lei, L.; Xie, F.; Li, C. EGFR, KRAS, BRAF, PTEN, and PIK3CA mutation in plasma of small cell lung cancer patients. Onco Targets Ther. 2018, 11, 2217–2226. [Google Scholar] [CrossRef] [Green Version]
- Jing, C.; Mao, X.; Wang, Z.; Sun, K.; Ma, R.; Wu, J.; Cao, H. Next-generation sequencing-based detection of EGFR, KRAS, BRAF, NRAS, PIK3CA, Her-2 and TP53 mutations in patients with non-small cell lung cancer. Mol. Med. Rep. 2018, 18, 2191–2197. [Google Scholar] [CrossRef]
- McCoach, C.E.; Blakely, C.M.; Banks, K.C.; Levy, B.; Chue, B.M.; Raymond, V.M.; Le, A.T.; Lee, C.E.; Diaz, J.; Waqar, S.N.; et al. Clinical Utility of Cell-Free DNA for the Detection of ALK Fusions and Genomic Mechanisms of ALK Inhibitor Resistance in Non–Small Cell Lung Cancer. Clin. Cancer Res. 2018, 24, 2758–2770. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hsiao, S.-Y.; He, H.-L.; Weng, T.-S.; Lin, C.-Y.; Chao, C.-M.; Huang, W.-T.; Tsao, C.-J. Colorectal Cancer with EML4-ALK Fusion Gene Response to Alectinib: A Case Report and Review of the Literature. Case Rep. Oncol. 2021, 14, 232–238. [Google Scholar] [CrossRef]
- Su, C.; Jiang, Y.; Jiang, W.; Wang, H.; Liu, S.; Shao, Y.; Zhao, W.; Ning, R.; Yu, Q. STRN-ALK Fusion in Lung Adenocarcinoma with Excellent Response Upon Alectinib Treatment: A Case Report and Literature Review. Onco Targets Ther. 2020, 13, 12515–12519. [Google Scholar] [CrossRef]
- Clifton, K.; Rich, T.A.; Parseghian, C.; Raymond, V.M.; Dasari, A.; Pereira, A.A.L.; Willis, J.; Loree, J.M.; Bauer, T.M.; Chae, Y.K.; et al. Identification of Actionable Fusions as an Anti-EGFR Resistance Mechanism Using a Circulating Tumor DNA Assay. JCO Precis. Oncol. 2019, 3, 1–15. [Google Scholar] [CrossRef]
- Ignatiadis, M.; Sledge, G.W.; Jeffrey, S.S. Liquid biopsy enters the clinic—implementation issues and future challenges. Nat. Rev. Clin. Oncol. 2021, 18, 297–312. [Google Scholar] [CrossRef]
- Chen, M.; Zhao, H. Next-generation sequencing in liquid biopsy: Cancer screening and early detection. Hum. Genom. 2019, 13, 34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alborelli, I.; Generali, D.; Jermann, P.; Cappelletti, M.R.; Ferrero, G.; Scaggiante, B.; Bortul, M.; Zanconati, F.; Nicolet, S.; Haegele, J.; et al. Cell-free DNA analysis in healthy individuals by next-generation sequencing: A proof of concept and technical validation study. Cell Death Dis. 2019, 10, 1–11. [Google Scholar] [CrossRef]
- Abbosh, C.; Birkbak, N.J.; Wilson, G.A.; Jamal-Hanjani, M.; Constantin, T.; Salari, R.; Le Quesne, J.; Moore, D.A.; Veeriah, S.; Rosenthal, R.; et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature 2017, 545, 446–451. [Google Scholar] [CrossRef] [PubMed]
- Tzanikou, E.; Markou, A.; Politaki, E.; Koutsopoulos, A.; Psyrri, A.; Mavroudis, D.; Georgoulias, V.; Lianidou, E. PIK3CAhotspot mutations in circulating tumor cells and paired circulating tumorDNAin breast cancer: A direct comparison study. Mol. Oncol. 2019, 13, 2515–2530. [Google Scholar] [CrossRef] [Green Version]
- Qin, Y.; Ekmekcioglu, S.; Forget, M.-A.; Szekvolgyi, L.; Hwu, P.; Grimm, E.A.; Jazaeri, A.A.; Roszik, J. Cervical Cancer Neoantigen Landscape and Immune Activity is Associated with Human Papillomavirus Master Regulators. Front. Immunol. 2017, 8, 689. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bao, C.; An, N.; Xie, H.; Xu, L.; Zhou, B.; Luo, J.; Huang, W.; Huang, J. Identifying Potential Neoantigens for Cervical Cancer Immunotherapy Using Comprehensive Genomic Variation Profiling of Cervical Intraepithelial Neoplasia and Cervical Cancer. Front. Oncol. 2021, 11. [Google Scholar] [CrossRef]
- Chen, C.; Liu, S.; Qu, R.; Li, B. Recurrent Neoantigens in Colorectal Cancer as Potential Immunotherapy Targets. BioMed Res. Int. 2020, 2020, 1–8. [Google Scholar] [CrossRef]
- Malekzadeh, P.; Pasetto, A.; Robbins, P.F.; Parkhurst, M.R.; Paria, B.C.; Jia, L.; Gartner, J.J.; Hill, V.; Yu, Z.; Restifo, N.P.; et al. Neoantigen screening identifies broad TP53 mutant immunogenicity in patients with epithelial cancers. J. Clin. Investig. 2019, 129, 1109–1114. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Wu, J.; Chen, S.; Zhou, Z. Shared neoantigens: Ideal targets for off-the-shelf cancer immunotherapy. Pharmacogenomics 2020, 21, 637–645. [Google Scholar] [CrossRef] [PubMed]
- Veatch, J.R.; Jesernig, B.L.; Kargl, J.; FitzGibbon, M.; Lee, S.M.; Baik, C.; Martins, R.; Houghton, A.M.G.; Riddell, S.R. Endogenous CD4+ T Cells Recognize Neoantigens in Lung Cancer Patients, Including Recurrent Oncogenic KRAS and ERBB2 (Her2) Driver Mutations. Cancer Immunol. Res. 2019, 7, 910–922. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Q.J.; Yu, Z.; Griffith, K.; Hanada, K.-I.; Restifo, N.P.; Yang, J.C. Identification of T-cell Receptors Targeting KRAS-Mutated Human Tumors. Cancer Immunol. Res. 2016, 4, 204–214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Skora, A.D.; Douglass, J.; Hwang, M.S.; Tam, A.J.; Blosser, R.L.; Gabelli, S.B.; Cao, J.; Diaz, L.A.; Papadopoulos, N.; Kinzler, K.W.; et al. Generation of MANAbodies specific to HLA-restricted epitopes encoded by somatically mutated genes. Proc. Natl. Acad. Sci. USA 2015, 112, 9967–9972. [Google Scholar] [CrossRef] [Green Version]
- Akazawa, Y.; Saito, Y.; Yoshikawa, T.; Saito, K.; Nosaka, K.; Shimomura, M.; Mizuno, S.; Nakamoto, Y.; Nakatsura, T. Efficacy of immunotherapy targeting the neoantigen derived from epidermal growth factor receptor T790M/C797S mutation in non–small cell lung cancer. Cancer Sci. 2020, 111, 2736–2746. [Google Scholar] [CrossRef]
- Dimou, A.; Grewe, P.; Sidney, J.; Sette, A.; Norman, P.J.; Doebele, R.C. HLA Class I Binding of Mutant EGFR Peptides in NSCLC Is Associated With Improved Survival. J. Thorac. Oncol. 2021, 16, 104–112. [Google Scholar] [CrossRef] [PubMed]
- Campbell, J.D.; Alexandrov, A.; Kim, J.; Wala, J.; Berger, A.H.; Pedamallu, C.S.; Shukla, S.A.; Guo, G.; Brooks, A.N.; Murray, B.A.; et al. Distinct patterns of somatic genome alterations in lung adenocarcinomas and squamous cell carcinomas. Nat. Genet. 2016, 48, 607–616. [Google Scholar] [CrossRef] [Green Version]
- Chang, G.A.; Tadepalli, J.S.; Shao, Y.; Zhang, Y.; Weiss, S.; Robinson, E.; Spittle, C.; Furtado, M.; Shelton, D.N.; Karlin-Neumann, G.; et al. Sensitivity of plasma BRAFmutantand NRASmutantcell-free DNA assays to detect metastatic melanoma in patients with low RECIST scores and non-RECIST disease progression. Mol. Oncol. 2016, 10, 157–165. [Google Scholar] [CrossRef] [Green Version]
- Prior, I.A.; Lewis, P.D.; Mattos, C. A Comprehensive Survey of Ras Mutations in Cancer. Cancer Res. 2012, 72, 2457–2467. [Google Scholar] [CrossRef] [Green Version]
- Ledet, E.M.; Lilly, M.B.; Sonpavde, G.; Lin, E.; Nussenzveig, R.H.; Barata, P.C.; Yandell, M.; Nagy, R.J.; Kiedrowski, L.; Agarwal, N.; et al. Comprehensive Analysis of AR Alterations in Circulating Tumor DNA from Patients with Advanced Prostate Cancer. Oncologist 2019, 25, 327–333. [Google Scholar] [CrossRef] [Green Version]
- Miller, M.S.; Douglass, J.; Hwang, M.S.; Skora, A.D.; Murphy, M.; Papadopoulos, N.; Kinzler, K.W.; Vogelstein, B.; Zhou, S.; Gabelli, S.B. An engineered antibody fragment targeting mutant β-catenin via major histocompatibility complex I neoantigen presentation. J. Biol. Chem. 2019, 294, 19322–19334. [Google Scholar] [CrossRef] [Green Version]
- Hattori, K.; Sakata-Yanagimoto, M.; Suehara, Y.; Yokoyama, Y.; Kato, T.; Kurita, N.; Nishikii, H.; Obara, N.; Takano, S.; Ishikawa, E.; et al. Clinical significance of disease-specificMYD88mutations in circulating DNA in primary central nervous system lymphoma. Cancer Sci. 2018, 109, 225–230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roschewski, M.; Dunleavy, K.; Pittaluga, S.; Moorhead, M.; Pepin, F.; Kong, K.; Shovlin, M.; Jaffe, E.S.; Staudt, L.M.; Lai, C.; et al. Circulating tumour DNA and CT monitoring in patients with untreated diffuse large B-cell lymphoma: A correlative biomarker study. Lancet Oncol. 2015, 16, 541–549. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Lu, M.; Qin, Y.; Gao, W.; Tao, L.; Su, W.; Zhong, J. Neoantigen: A New Breakthrough in Tumor Immunotherapy. Front. Immunol. 2021, 12. [Google Scholar] [CrossRef]
- Fritsch, E.F.; Burkhardt, U.E.; Hacohen, N.; Wu, C.J. Personal Neoantigen Cancer Vaccines: A Road Not Fully Paved. Cancer Immunol. Res. 2020, 8, 1465–1469. [Google Scholar] [CrossRef] [PubMed]
- Jia, Q.; Chiu, L.; Wu, S.; Bai, J.; Peng, L.; Zheng, L.; Zang, R.; Li, X.; Yuan, B.; Gao, Y.; et al. Tracking Neoantigens by Personalized Circulating Tumor DNA Sequencing during Checkpoint Blockade Immunotherapy in Non-Small Cell Lung Cancer. Adv. Sci. 2020, 7, 1903410. [Google Scholar] [CrossRef] [PubMed]
- Brannon, A.R.; Jayakumaran, G.; Diosdado, M.; Patel, J.; Razumova, A.; Hu, Y.; Meng, F.; Haque, M.; Sadowska, J.; Murphy, B.J.; et al. Enhanced specificity of clinical high-sensitivity tumor mutation profiling in cell-free DNA via paired normal sequencing using MSK-ACCESS. Nat. Commun. 2021, 12, 3770. [Google Scholar] [CrossRef]
- Fratte, C.D.; Guardascione, M.; De Mattia, E.; Borsatti, E.; Boschetto, R.; Farruggio, A.; Canzonieri, V.; Romanato, L.; Borsatti, R.; Gagno, S.; et al. Clonal Selection of a Novel Deleterious TP53 Somatic Mutation Discovered in ctDNA of a KIT/PDGFRA Wild-Type Gastrointestinal Stromal Tumor Resistant to Imatinib. Front. Pharmacol. 2020, 11, 36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patel, K.M.; Tsui, D.W.Y. The translational potential of circulating tumour DNA in oncology. Clin. Biochem. 2015, 48, 957–961. [Google Scholar] [CrossRef]
- Malone, E.R.; Oliva, M.; Sabatini, P.J.B.; Stockley, T.; Siu, L.L. Molecular profiling for precision cancer therapies. Genome Med. 2020, 12, 1–19. [Google Scholar] [CrossRef] [Green Version]
- Fujii, Y.; Ono, A.; Hayes, C.N.; Aikata, H.; Yamauchi, M.; Uchikawa, S.; Kodama, K.; Teraoka, Y.; Fujino, H.; Nakahara, T.; et al. Identification and monitoring of mutations in circulating cell-free tumor DNA in hepatocellular carcinoma treated with lenvatinib. J. Exp. Clin. Cancer Res. 2021, 40, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Raja, R.; Kuziora, M.; Brohawn, P.Z.; Higgs, B.W.; Gupta, A.; Dennis, P.A.; Ranade, K. Early reduction in ctDNA predicts sur-vival in patients with lung and bladder cancer treated with durvalumab. Clin. Cancer Res. 2018, 24, 6212–6222. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Magbanua, M.J.M.; Li, W.; Wolf, D.M.; Yau, C.; Hirst, G.L.; Swigart, L.B.; Newitt, D.C.; Gibbs, J.; Delson, A.L.; Kalashnikova, E.; et al. Circulating tumor DNA and magnetic resonance imaging to predict neoadjuvant chemotherapy response and recurrence risk. NPJ Breast Cancer 2021, 7, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Xu, Y.; Wang, C.; Gong, Y.; Zhang, Y.; Yao, R.; Li, P.; Zhu, X.; Bai, J.; Guan, Y.; et al. Serial circulating tumor DNA identification associated with the efficacy and prognosis of neoadjuvant chemotherapy in breast cancer. Breast Cancer Res. Treat. 2021, 1–13. [Google Scholar] [CrossRef]
- Madsen, A.T.; Winther-Larsen, A.; McCulloch, T.; Meldgaard, P.; Sorensen, B.S. Genomic Profiling of Circulating Tumor DNA Predicts Outcome and Demonstrates Tumor Evolution in ALK-Positive Non-Small Cell Lung Cancer Patients. Cancers 2020, 12, 947. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Huang, J.; Wang, T.; Zhou, J.; Zheng, J.; Feng, J.; Zhuang, W.; Chen, J.; Zhao, J.; Zhong, W.; et al. Decoding the Evolutionary Response to Ensartinib in Patients With ALK-Positive NSCLC by Dynamic Circulating Tumor DNA Sequencing. J. Thorac. Oncol. 2021, 16, 827–839. [Google Scholar] [CrossRef]
- Annala, M.; Taavitsainen, S.; Khalaf, D.J.; Vandekerkhove, G.; Beja, K.; Sipola, J.; Warner, E.W.; Herberts, C.; Wong, A.; Fu, S.; et al. Evolution of Castration-Resistant Prostate Cancer in CtDNA during Sequential Androgen Receptor Pathway Inhibition. Clin. Cancer Res. 2021. [Google Scholar] [CrossRef]
- Blatter, S.; Rottenberg, S. Minimal residual disease in cancer therapy—Small things make all the difference. Drug Resist. Updat. 2015, 21–22, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Tachtsidis, A.; McInnes, L.M.; Jacobsen, N.; Thompson, E.W.; Saunders, C.M. Minimal residual disease in breast cancer: An overview of circulating and disseminated tumour cells. Clin. Exp. Metastasis 2016, 33, 521–550. [Google Scholar] [CrossRef] [Green Version]
- Chin, R.-I.; Chen, K.; Usmani, A.; Chua, C.; Harris, P.K.; Binkley, M.S.; Azad, T.D.; Dudley, J.C.; Chaudhuri, A.A. Detection of Solid Tumor Molecular Residual Disease (MRD) Using Circulating Tumor DNA (ctDNA). Mol. Diagn. Ther. 2019, 23, 311–331. [Google Scholar] [CrossRef]
- Abbosh, C.; Birkbak, N.J.; Swanton, C. Early stage NSCLC—challenges to implementing ctDNA-based screening and MRD detection. Nat. Rev. Clin. Oncol. 2018, 15, 577–586. [Google Scholar] [CrossRef] [PubMed]
| Cancer | Gene | Mutation | Clinical Implication | Clinical Trials or Related Studies |
|---|---|---|---|---|
| Breast cancer | ESR1 | D538G/Y537S/Y537C/Y538G | Biomarker of tumor evolution and prognostic impact | [56] |
| PIK3CA | E545K/E542K/H1047R | Biomarker of tumor evolution and prognostic impact | [57] | |
| Cervical cancer | ERBB2/HER2 | S310F/V104M | Biomarker of tumor evolution and prognostic impact | [58] |
| PIK3CA | R88Q | Biomarker of tumor evolution and prognostic impact | [59] | |
| Colorectal cancer | KRAS | G12C/G12V/G12D/G12A/G13D/G12R | Biomarker of tumor evolution and prognostic impact | NCT04117087 |
| PIK3CA | E545K | Biomarker of tumor evolution and prognostic impact | [60] | |
| Epithelial cancers | TP53 | R175H/R249S/R273C/R273H | Potential targets for immunotherapy | [61] |
| Gliomas | IDH1 | R132H | Biomarker of tumor evolution and prognostic impact | NCT 02454634 |
| Kidney renal papillary cell carcinoma | MET | M1250T | Potential target for immunotherapy | [62] |
| Lung cancer | ERBB2 | Internal tandem duplication | Augment CD4 + neoantigen-specific T cells | [63] |
| KRAS | G12D | T cells target mutated antigen | [64] | |
| KRAS | G12V | Potential target for immunotherapy | [63,65] | |
| Non-small cell lung cancer | CTNNB1 | S37F/S45F/S45P/T41A | Biomarker of tumor evolution | NCT 03953235 |
| ERBB2/HER2 | Y772_A775dup | Biomarker of tumor evolution and prognostic impact | NCT 03953235 | |
| EGFR | T790M/C797S | Resistance of EGFR-TKIs treatment | [66] | |
| KRAS | G12C | Biomarker of tumor evolution | NCT 04685135 | |
| Lung adenocarcinomas | EGFR | L858R/E746_A750del | Prognostic impact | [67] |
| EGFR | L858R | Potential target for immunotherapy | [65] | |
| Lung Squamous cell carcinomas | EGFR | G719A | Assessment of neoantigen load and recurrence | [68] |
| PIK3CA | E542K | Assessment of neoantigen load and recurrence | [68] | |
| TP53 | V157F/G154V/R175G/P278A | Assessment of neoantigen load and recurrence | [68] | |
| Melanoma | BRAF | V600E/V600K | Immunotherapy treatment monitoring | NCT 04364230 |
| NRAS | Q61K/Q61R/Q61L/Q61H | Early biomarker of tumor evolution | [69] | |
| Ovarian cancer | TP53 | R175H/C176S/R196P/R273H | Biomarker of tumor evolution and prognostic impact | [29] |
| Pancreatic cancer | KRAS | G12V | Biomarker of tumor evolution and prognostic impact | NCT 04146298 |
| KRAS | G12D | Biomarker of tumor evolution and Potential drug targets | NCT 03608631 | |
| KRAS | G12R | Biomarker of tumor evolution and prognostic impact | [70] | |
| Prostate cancer | AR | L720H/H874Y/T877A/T878A | Resistance of CYP17 inhibitor treatment | [71] |
| Pan-cancer | CTNNB1 | S45F | Potential target for immunotherapy | [72] |
| Lymphoma | MYD88 | L265P | Biomarker of tumor evolution and prognostic impact | [73] |
| VDJ | rearrangement | Biomarker of tumor evolution and prognostic impact | [74] | |
| Other solid tumor | BRAF | G466V | Biomarker of tumor evolution | NCT 03953235 |
| KRAS | G12C/G12D/G12V/G13D/Q61H/Q61K/Q61L/Q61R | Biomarker of tumor evolution | NCT 03953235 | |
| TP53 | K132E/K132N/R213L/R249M/S127Y | Biomarker of tumor evolution | NCT 03953235 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, C.; Liu, X.; Zheng, B.; Ke, R.; Tzeng, C.-M. Liquid Biopsy, ctDNA Diagnosis through NGS. Life 2021, 11, 890. https://doi.org/10.3390/life11090890
Lin C, Liu X, Zheng B, Ke R, Tzeng C-M. Liquid Biopsy, ctDNA Diagnosis through NGS. Life. 2021; 11(9):890. https://doi.org/10.3390/life11090890
Chicago/Turabian StyleLin, Chen, Xuzhu Liu, Bingyi Zheng, Rongqin Ke, and Chi-Meng Tzeng. 2021. "Liquid Biopsy, ctDNA Diagnosis through NGS" Life 11, no. 9: 890. https://doi.org/10.3390/life11090890
APA StyleLin, C., Liu, X., Zheng, B., Ke, R., & Tzeng, C.-M. (2021). Liquid Biopsy, ctDNA Diagnosis through NGS. Life, 11(9), 890. https://doi.org/10.3390/life11090890






