Dynamin Inhibitors Prevent the Establishment of the Cytomegalovirus Assembly Compartment in the Early Phase of Infection
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Lines, Viruses, and Infection Conditions
2.2. Antibodies and Reagents
2.3. Immunofluorescence, Confocal Microscopy, and Image Analysis
2.4. Flow Cytometry and Quantification of Total M55/gB Protein
2.5. Western Blot
2.6. Quantification of Virus Growth by the Plaque Assay
2.7. Cell Viability
2.8. Data Presentation and Statistical Analysis
3. Results
3.1. The Time Course of the preAC Establishment
3.2. Dynasore Inhibits Establishment of the preAC
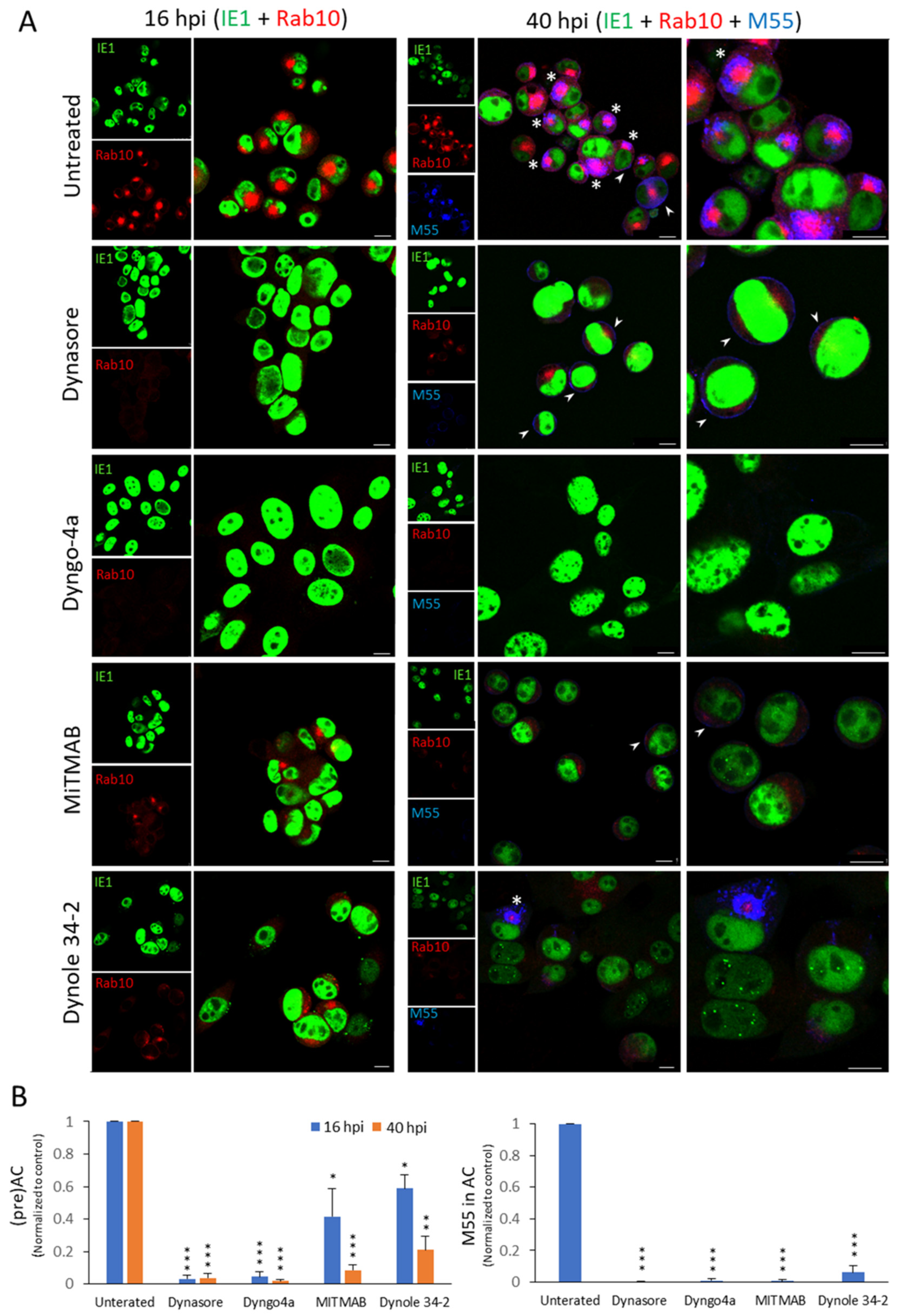
3.3. Other Dynamin Inhibitors, but Not Clathrin Inhibitor and Cholesterol Depletion, Prevent the Establishment of the preAC
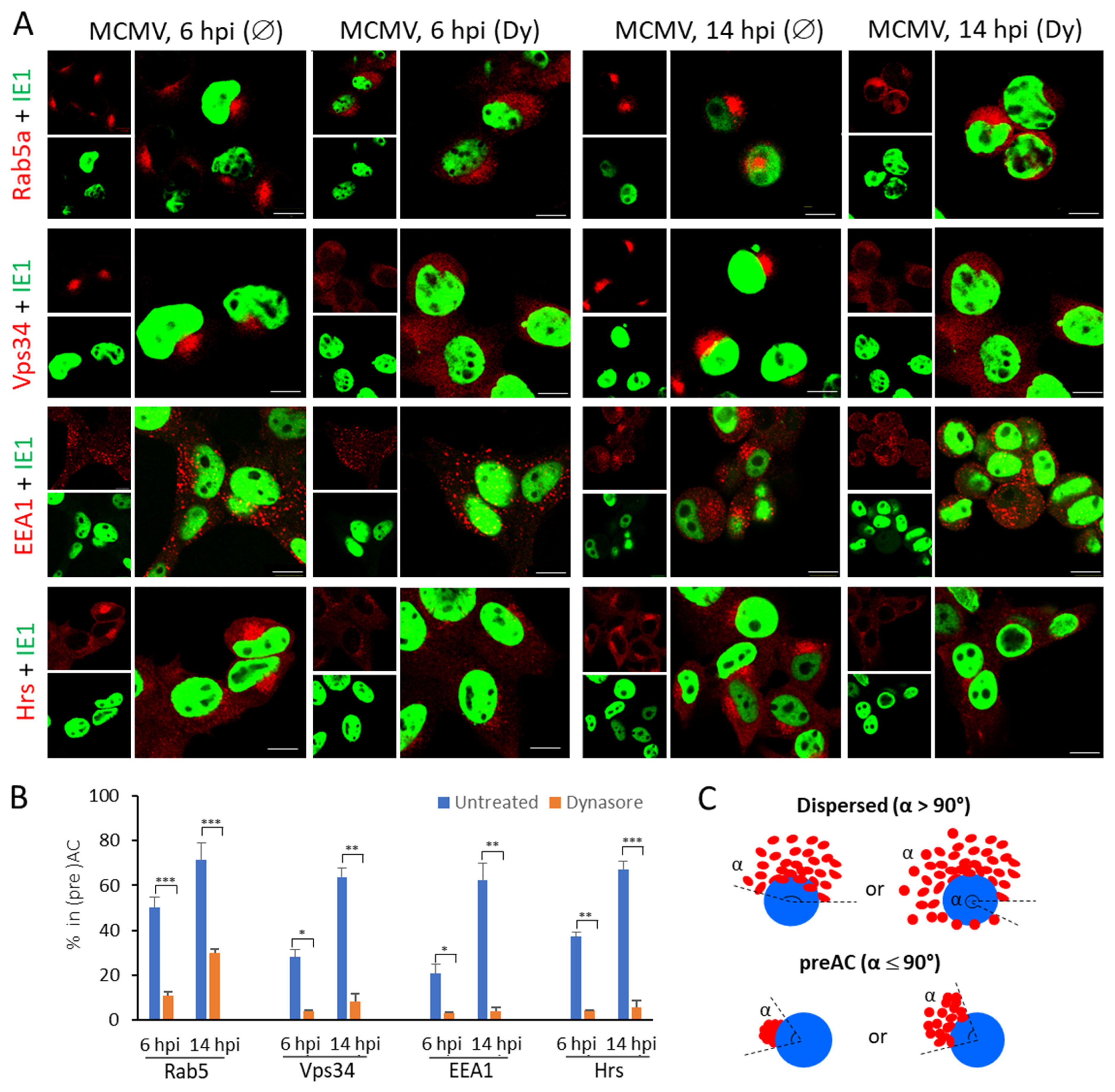
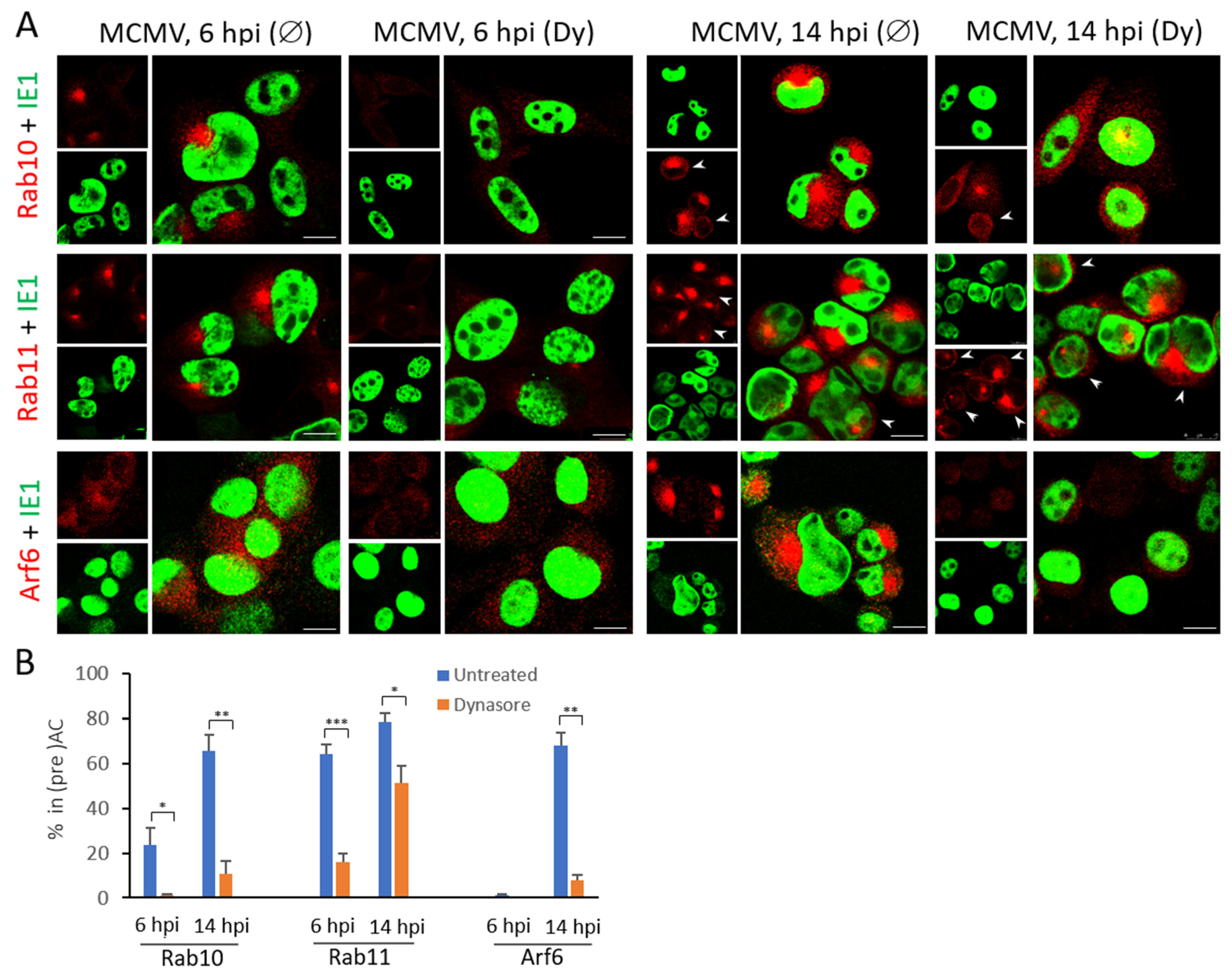
3.4. The Dynasore Treatment Affects Pericentriolar Recruitment of EE-ERC-Derived Organelles and Reorganization of the Entire Golgi Stack in the Early Phase of MCMV Infection
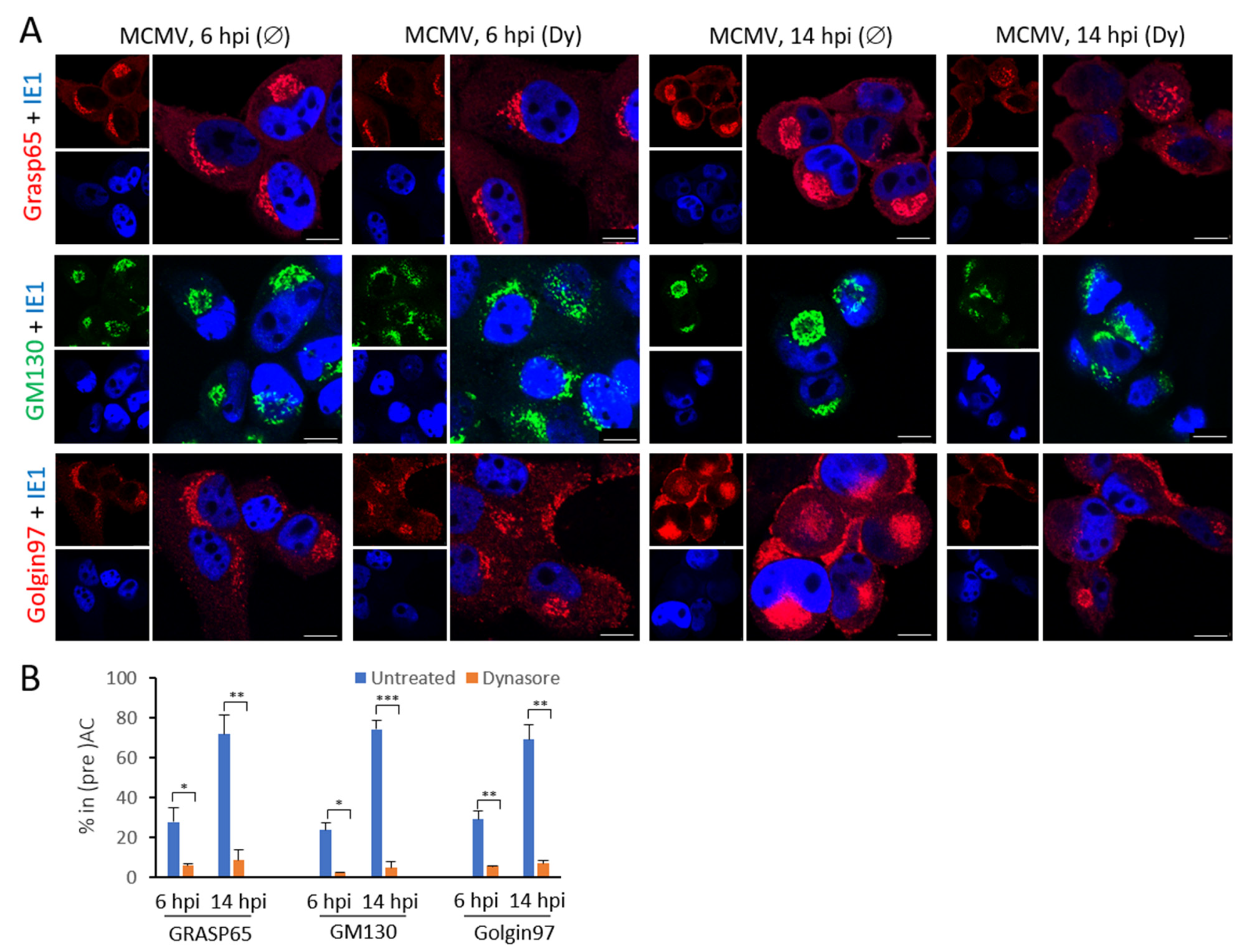
3.5. Dynasore Affects Localization of Entire Golgi
3.6. Dynasore Does Not Affect Late Endosomes
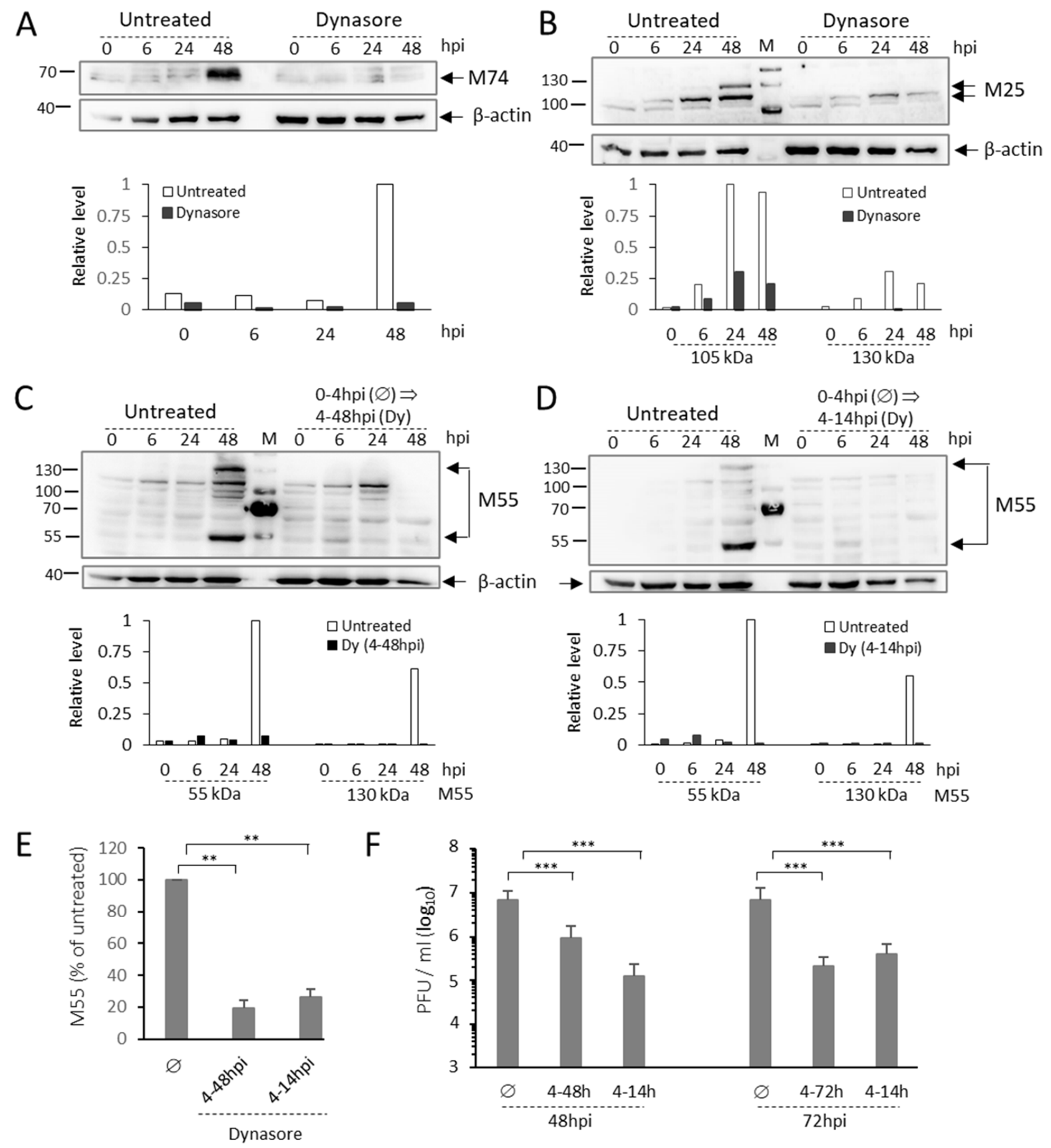
3.7. Dynasore Prevents AC Formation When Present Only in the Early Phase of MCMV Infection
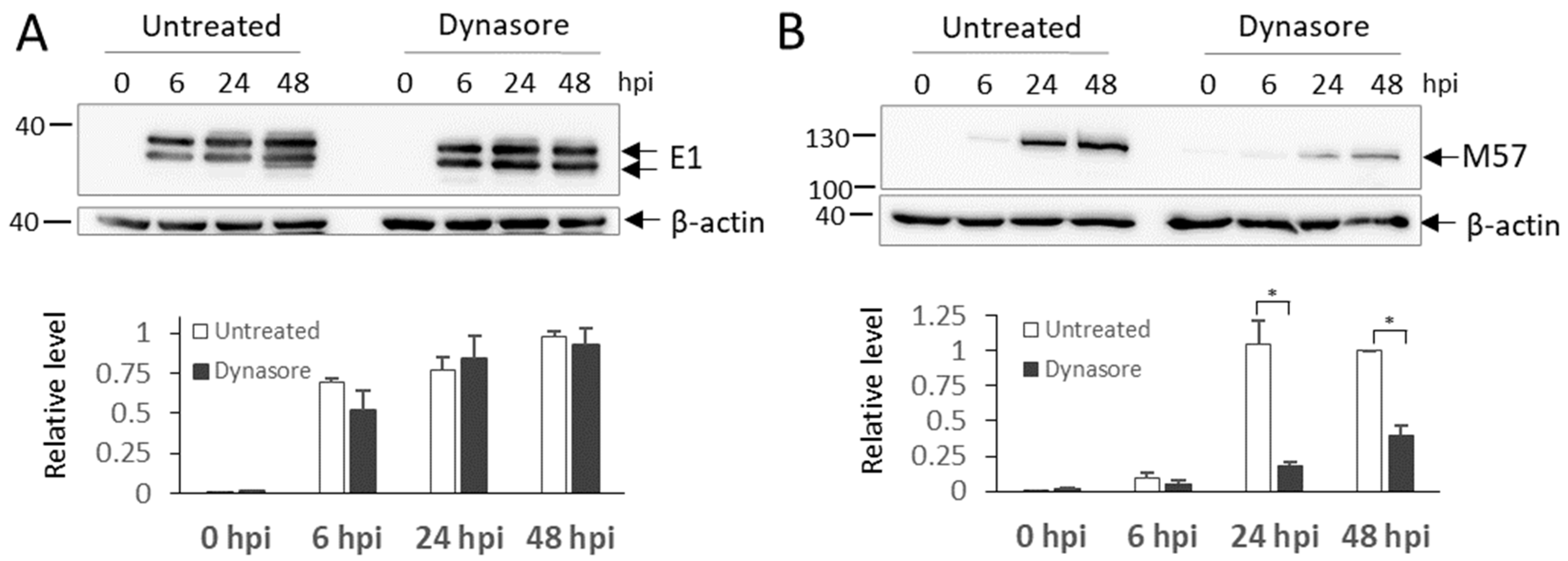
3.8. Dynasore Treatment in the Early Phase of Infection Blocks the Synthesis of Late MCMV Proteins and Inhibits the Release of Infective Virions
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Griffiths, P.; Baraniak, I.; Reeves, M. The Pathogenesis of Human Cytomegalovirus. J. Pathol. 2015, 235, 288–297. [Google Scholar] [CrossRef]
- Chen, S.J.; Wang, S.C.; Chen, Y.C. Antiviral Agents as Therapeutic Strategies against Cytomegalovirus Infections. Viruses 2019, 12, 21. [Google Scholar] [CrossRef]
- Tandon, R.; Mocarski, E.S. Viral and Host Control of Cytomegalovirus Maturation. Trends Microbiol. 2012, 20, 392–401. [Google Scholar] [CrossRef]
- Close, W.L.; Anderson, A.N.; Pellett, P.E. Betaherpesvirus virion assembly and egress. In Advances in Experimental Medicine and Biology; Springer: New York, NY, USA, 2018; Volume 1045, pp. 167–207. [Google Scholar]
- Alwine, J.C. The Human Cytomegalovirus Assembly Compartment: A Masterpiece of Viral Manipulation of Cellular Processes That Facilitates Assembly and Egress. PLoS Pathog. 2012, 8, e1002878. [Google Scholar] [CrossRef]
- Das, S.; Pellett, P.E. Spatial Relationships between Markers for Secretory and Endosomal Machinery in Human Cytomegalovirus-Infected Cells versus Those in Uninfected Cells. J. Virol. 2011, 85, 5864–5879. [Google Scholar] [CrossRef]
- Das, S.; Vasanji, A.; Pellett, P.E. Three-Dimensional Structure of the Human Cytomegalovirus Cytoplasmic Virion Assembly Complex Includes a Reoriented Secretory Apparatus. J. Virol. 2007, 81, 11861–11869. [Google Scholar] [CrossRef]
- Cepeda, V.; Esteban, M.; Fraile-Ramos, A. Human Cytomegalovirus Final Envelopment on Membranes Containing Both Trans-Golgi Network and Endosomal Markers. Cell. Microbiol. 2010, 12, 386–404. [Google Scholar] [CrossRef]
- Karleuša, L.; Mahmutefendić, H.; Tomaš, M.I.; Zagorac, G.B.; Lučin, P. Landmarks of Endosomal Remodeling in the Early Phase of Cytomegalovirus Infection. Virology 2018, 515, 108–122. [Google Scholar] [CrossRef] [PubMed]
- Lučin, P.; Jug Vučko, N.; Karleuša, L.; Mahmutefendić Lučin, H.; Blagojević Zagorac, G.; Lisnić, B.; Pavišić, V.; Marcelić, M.; Grabušić, K.; Brizić, I.; et al. Cytomegalovirus Generates Assembly Compartment in the Early Phase of Infection by Perturbation of Host-Cell Factors Recruitment at the Early Endosome/Endosomal Recycling Compartment/Trans-Golgi Interface. Front. Cell Dev. Biol. 2020, 8, 914. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Ortiz, D.A.; Gurczynski, S.J.; Khan, F.; Pellett, P.E. Identification of Human Cytomegalovirus Genes Important for Biogenesis of the Cytoplasmic Virion Assembly Complex. J. Virol. 2014, 88, 9086–9099. [Google Scholar] [CrossRef] [PubMed]
- Wandinger-Ness, A.; Zerial, M. Rab Proteins and the Compartmentalization of the Endosomal System. Cold Spring Harb. Perspect. Biol. 2014, 6, a022616. [Google Scholar] [CrossRef]
- Taisne, C.; Lussignol, M.; Hernandez, E.; Moris, A.; Mouna, L.; Esclatine, A. Human Cytomegalovirus Hijacks the Autophagic Machinery and LC3 Homologs in Order to Optimize Cytoplasmic Envelopment of Mature Infectious Particles. Sci. Rep. 2019, 9, 4560. [Google Scholar] [CrossRef]
- Lučin, P.; Kareluša, L.; Zagorac, G.B.; Lučin, H.M.; Pavišić, V.; Vučko, N.J.; Jurić, S.L.; Marcelić, M.; Lisnić, B.; Jonjić, S. Cytomegaloviruses Exploit Recycling Rab Proteins in the Sequential Establishment of the Assembly Compartment. Front. Cell Dev. Biol. 2018, 6, 165. [Google Scholar] [CrossRef]
- Kučić, N.; Ilić Tomaš, M.; Mahmutefendić, H.; Blagojević, G.; Lučin, P. Early Endosomal Retention of Murine Cytomegalovirus M06 Protein. Croat. Chem. Acta 2012, 85, 213–221. [Google Scholar] [CrossRef]
- Naslavsky, N.; Caplan, S. The Enigmatic Endosome—Sorting the Ins and Outs of Endocytic Trafficking. J. Cell Sci. 2018, 131, jcs216499. [Google Scholar] [CrossRef]
- Cullen, P.J.; Steinberg, F. To Degrade or Not to Degrade: Mechanisms and Significance of Endocytic Recycling. Nat. Rev. Mol. Cell Biol. 2018, 19, 679–696. [Google Scholar] [CrossRef] [PubMed]
- Jimah, J.R.; Hinshaw, J.E. Structural Insights into the Mechanism of Dynamin Superfamily Proteins. Trends Cell Biol. 2019, 29, 257–273. [Google Scholar] [CrossRef]
- Urrutia, R.; Henley, J.R.; Cook, T.; McNiven, M.A. The Dynamins: Redundant or Distinct Functions for an Expanding Family of Related GTPases? Proc. Natl. Acad. Sci. USA 1997, 94, 377–384. [Google Scholar] [CrossRef] [PubMed]
- Cao, H.; Garcia, F.; McNiven, M.A. Differential Distribution of Dynamin Isoforms in Mammalian Cells. Mol. Biol. Cell 1998, 9, 2595–2609. [Google Scholar] [CrossRef] [PubMed]
- Damke, H.; Baba, T.; Warnock, D.E.; Schmid, S.L. Induction of Mutant Dynamin Specifically Blocks Endocytic Coated Vesicle Formation. J. Cell Biol. 1994, 127, 915–934. [Google Scholar] [CrossRef] [PubMed]
- Takei, K.; McPherson, P.S.; Schmid, S.L.; de Camilli, P. Tubular Membrane Invaginations Coated by Dynamin Rings Are Induced by GTP-ΓS in Nerve Terminals. Nature 1995, 374, 186–190. [Google Scholar] [CrossRef]
- Sever, S.; Damke, H.; Schmid, S.L. Dynamin: GTP Controls the Formation of Constricted Coated Pits, the Rate Limiting Step in Clathrin-Mediated Endocytosis. J. Cell Biol. 2000, 150, 1137–1147. [Google Scholar] [CrossRef] [PubMed]
- Jones, S.M.; Howell, K.E.; Henley, J.R.; Cao, H.; McNiven, M.A. Role of Dynamin in the Formation of Transport Vesicles from the Trans-Golgi Network. Science 1998, 279, 573–577. [Google Scholar] [CrossRef] [PubMed]
- Kreitzer, G.; Marmorstein, A.; Okamoto, P.; Vallee, R.; Rodriguez-Boulan, E. Kinesin and Dynamin Are Required for Post-Golgi Transport of a Plasma-Membrane Protein. Nat. Cell Biol. 2000, 2, 125–127. [Google Scholar] [CrossRef]
- McNiven, M.A.; Thompson, H.M. Vesicle Formation at the Plasma Membrane and Trans-Golgi Network: The Same but Different. Science 2006, 313, 1591–1594. [Google Scholar] [CrossRef]
- Anantharam, A.; Bittner, M.A.; Aikman, R.L.; Stuenkel, E.L.; Schmid, S.L.; Axelrod, D.; Holz, R.W. A New Role for the Dynamin GTPase in the Regulation of Fusion Pore Expansion. Mol. Biol. Cell 2011, 22, 1907. [Google Scholar] [CrossRef] [PubMed]
- Maeda, K.; Nakata, T.; Noda, Y.; Sato-Yoshitake, R.; Hirokawa, N. Interaction of Dynamin with Microtubules: Its Structure and GTPase Activity Investigated by Using Highly Purified Dynamin. Mol. Biol. Cell 1992, 3, 1181–1194. [Google Scholar] [CrossRef]
- Ochoa, G.-C.; Slepnev, V.I.; Neff, L.; Ringstad, N.; Takei, K.; Daniell, L.; Kim, W.; Cao, H.; McNiven, M.; Baron, R.; et al. A Functional Link between Dynamin and the Actin Cytoskeleton at Podosomes. J. Cell Biol. 2000, 150, 377–390. [Google Scholar] [CrossRef]
- Gonzalez-Jamett, A.; Baez-Matus, X.; Olivares, M.; Hinostroza, F.; Guerra-Fernández, M.; Vasquez-Navarrete, J.; Bui, M.; Guicheney, P.; Romero, N.B.; Bevilacqua, J.; et al. Dynamin-2 Mutations Linked to Centronuclear Myopathy Impair Actin-Dependent Trafficking in Muscle Cells. Sci. Rep. 2017, 7, 4580. [Google Scholar] [CrossRef] [PubMed]
- Tanabe, K.; Takei, K. Dynamic Instability of Microtubules Requires Dynamin 2 and Is Impaired in a Charcot-Marie-Tooth Mutant. J. Cell Biol. 2009, 185, 939–948. [Google Scholar] [CrossRef]
- Ishida, N.; Nakamura, Y.; Tanabe, K.; Li, S.-A.; Takei, K. Dynamin 2 Associates with Microtubules at Mitosis and Regulates Cell Cycle Progression. Cell Struct. Funct. 2011, 36, 145–154. [Google Scholar] [CrossRef]
- Mesaki, K.; Tanabe, K.; Obayashi, M.; Oe, N.; Takei, K. Fission of Tubular Endosomes Triggers Endosomal Acidification and Movement. PLoS ONE 2011, 6, e19764. [Google Scholar] [CrossRef]
- Damke, H.; Binns, D.D.; Ueda, H.; Schmid, S.L.; Baba, T. Dynamin GTPase Domain Mutants Block Endocytic Vesicle Formation at Morphologically Distinct Stages. Mol. Biol. Cell 2017, 12, 2578–2589. [Google Scholar] [CrossRef]
- Loerke, D.; Mettlen, M.; Yarar, D.; Jaqaman, K.; Jaqaman, H.; Danuser, G.; Schmid, S.L. Cargo and Dynamin Regulate Clathrin-Coated Pit Maturation. PLoS Biol. 2009, 7, e1000057. [Google Scholar] [CrossRef]
- Weller, S.G.; Capitani, M.; Cao, H.; Micaroni, M.; Luini, A.; Sallese, M.; McNiven, M.A. Src Kinase Regulates the Integrity and Function of the Golgi Apparatus via Activation of Dynamin 2. Proc. Natl. Acad. Sci. USA 2010, 107, 5863–5868. [Google Scholar] [CrossRef] [PubMed]
- Park, R.J.; Shen, H.; Liu, L.; Liu, X.; Ferguson, S.M.; de Camilli, P. Dynamin Triple Knockout Cells Reveal off Target Effects of Commonly Used Dynamin Inhibitors. J. Cell Sci. 2013, 126, 5305–5312. [Google Scholar] [CrossRef] [PubMed]
- Hasan, M.H.; Davis, L.E.; Bollavarapu, R.K.; Mitra, D.; Parmar, R.; Tandon, R. Dynamin Is Required for Efficient Cytomegalovirus Maturation and Envelopment. J. Virol. 2018, 92, e01418-18. [Google Scholar] [CrossRef] [PubMed]
- Harper, C.B.; Popoff, M.R.; McCluskey, A.; Robinson, P.J.; Meunier, F.A. Targeting Membrane Trafficking in Infection Prophylaxis: Dynamin Inhibitors. Trends Cell Biol. 2013, 23, 90–101. [Google Scholar] [CrossRef]
- Eschenburg, S.; Reubold, T.F. Modulation of Dynamin Function by Small Molecules. Biol. Chem. 2018, 399, 1421–1432. [Google Scholar] [CrossRef] [PubMed]
- Macia, E.; Ehrlich, M.; Massol, R.; Boucrot, E.; Brunner, C.; Kirchhausen, T. Dynasore, a Cell-Permeable Inhibitor of Dynamin. Dev. Cell 2006, 10, 839–850. [Google Scholar] [CrossRef]
- McCluskey, A.; Daniel, J.A.; Hadzic, G.; Chau, N.; Clayton, E.L.; Mariana, A.; Whiting, A.; Gorgani, N.N.; Lloyd, J.; Quan, A.; et al. Building a Better Dynasore: The Dyngo Compounds Potently Inhibit Dynamin and Endocytosis. Traffic 2013, 14, 1272–1289. [Google Scholar] [CrossRef] [PubMed]
- Daniel, J.A.; Malladi, C.S.; Kettle, E.; McCluskey, A.; Robinson, P.J. Analysis of Synaptic Vesicle Endocytosis in Synaptosomes by High-Content Screening. Nat. Protoc. 2012, 7, 1439–1455. [Google Scholar] [CrossRef]
- Hill, T.A.; Gordon, C.P.; McGeachie, A.B.; Venn-Brown, B.; Odell, L.R.; Chau, N.; Quan, A.; Mariana, A.; Sakoff, J.A.; Fabbro, M.C.; et al. Inhibition of Dynamin Mediated Endocytosis by the Dynoles—Synthesis and Functional Activity of a Family of Indoles. J. Med. Chem. 2009, 52, 3762–3773. [Google Scholar] [CrossRef] [PubMed]
- Hill, T.A.; Odell, L.R.; Quan, A.; Abagyan, R.; Ferguson, G.; Robinson, P.J.; McCluskey, A. Long Chain Amines and Long Chain Ammonium Salts as Novel Inhibitors of Dynamin GTPase Activity. Bioorg. Med. Chem. Lett. 2004, 14, 3275–3278. [Google Scholar] [CrossRef]
- Abdulkarim, A.S.; Cao, H.; Huang, B.; McNiven, M.A. The Large GTPase Dynamin Is Required for Hepatitis B Virus Protein Secretion from Hepatocytes. J. Hepatol. 2003, 38, 76–83. [Google Scholar] [CrossRef]
- Albecka, A.; Laine, R.F.; Janssen, A.F.J.; Kaminski, C.F.; Crump, C.M. HSV-1 Glycoproteins Are Delivered to Virus Assembly Sites through Dynamin-Dependent Endocytosis. Traffic 2016, 17, 21–39. [Google Scholar] [CrossRef] [PubMed]
- Archer, M.A.; Brechtel, T.M.; Davis, L.E.; Parmar, R.C.; Hasan, M.H.; Tandon, R. Inhibition of Endocytic Pathways Impacts Cytomegalovirus Maturation. Sci. Rep. 2017, 7, 46069. [Google Scholar] [CrossRef] [PubMed]
- Joshi, S.; Perera, S.; Gilbert, J.; Smith, C.M.; Mariana, A.; Gordon, C.P.; Sakoff, J.A.; McCluskey, A.; Robinson, P.J.; Braithwaite, A.W.; et al. The Dynamin Inhibitors MiTMAB and OcTMAB Induce Cytokinesis Failure and Inhibit Cell Proliferation in Human Cancer Cells. Mol. Cancer Ther. 2010, 9, 1995–2006. [Google Scholar] [CrossRef]
- Tremblay, C.S.; Chiu, S.K.; Saw, J.; McCalmont, H.; Litalien, V.; Boyle, J.; Sonderegger, S.E.; Chau, N.; Evans, K.; Cerruti, L.; et al. Small Molecule Inhibition of Dynamin-Dependent Endocytosis Targets Multiple Niche Signals and Impairs Leukemia Stem Cells. Nat. Commun. 2020, 11, 6211. [Google Scholar] [CrossRef] [PubMed]
- Brizić, I.; Lisnić, B.; Brune, W.; Hengel, H.; Jonjić, S. Cytomegalovirus Infection: Mouse Model. Curr. Protoc. Immunol. 2018, 122, e51. [Google Scholar] [CrossRef]
- Crnković-Mertens, I.; Messerle, M.; Milotić, I.; Szepan, U.; Kučić, N.; Krmpotić, A.; Jonjić, S.; Koszinowski, U.H. Virus Attenuation after Deletion of the Cytomegalovirus Fc Receptor Gene Is Not Due to Antibody Control. J. Virol. 1998, 72, 1377–1382. [Google Scholar] [CrossRef]
- Strober, W. Trypan Blue Exclusion Test of Cell Viability. Curr. Protoc. Immunol. 1997, 21, A.3B.1–A.3B.2. [Google Scholar] [CrossRef]
- Babbey, C.M.; Ahktar, N.; Wang, E.; Chen, C.C.-H.; Grant, B.D.; Dunn, K.W. Rab10 Regulates Membrane Transport through Early Endosomes of Polarized Madin-Darby Canine Kidney Cells. Mol. Biol. Cell 2006, 17, 3156–3175. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Liu, H.; Wang, Y.; Liu, O.; Zhang, J.; Gleason, A.; Yang, Z.; Wang, H.; Shi, A.; Grant, B.D. RAB-10 Promotes EHBP-1 Bridging of Filamentous Actin and Tubular Recycling Endosomes. PLoS Genet. 2016, 12, e1006093. [Google Scholar] [CrossRef]
- Rebmann, G.M.; Grabski, R.; Sanchez, V.; Britt, W.J. Phosphorylation of Golgi Peripheral Membrane Protein Grasp65 Is an Integral Step in the Formation of the Human Cytomegalovirus Cytoplasmic Assembly Compartment. mBio 2016, 7, e01554-16. [Google Scholar] [CrossRef]
- Wollrab, V.; Caballero, D.; Thiagarajan, R.; Riveline, D. Ordering Single Cells and Single Embryos in 3D Confinement: A New Device for High Content Screening. JoVE J. Vis. Exp. 2016, 2016, e51880. [Google Scholar] [CrossRef]
- Saraste, J.; Prydz, K. A New Look at the Functional Organization of the Golgi Ribbon. Front. Cell Dev. Biol. 2019, 7, 171. [Google Scholar] [CrossRef]
- Carcedo, C.H.; Bonazzi, M.; Spanò, S.; Turacchio, G.; Colanzi, A.; Luini, A.; Corda, D. Mitotic Golgi Partitioning Is Driven by the Membrane-Fissioning Protein CtBP3/BARS. Science 2004, 305, 93–96. [Google Scholar] [CrossRef]
- Colanzi, A.; Carcedo, C.H.; Persico, A.; Cericola, C.; Turacchio, G.; Bonazzi, M.; Luini, A.; Corda, D. The Golgi Mitotic Checkpoint Is Controlled by BARS-Dependent Fission of the Golgi Ribbon into Separate Stacks in G2. EMBO J. 2007, 26, 2465–2476. [Google Scholar] [CrossRef]
- Rapp, M.; Messerle, M.; Bühler, B.; Tannheimer, M.; Keil, G.M.; Koszinowski, U.H. Identification of the Murine Cytomegalovirus Glycoprotein B Gene and Its Expression by Recombinant Vaccinia Virus. J. Virol. 1992, 66, 4399–4406. [Google Scholar] [CrossRef] [PubMed]
- Traub, L.M. Common Principles in Clathrin-Mediated Sorting at the Golgi and the Plasma Membrane. Biochim. Biophys. Acta Mol. Cell Res. 2005, 1744, 415–437. [Google Scholar] [CrossRef]
- Von Kleist, L.; Stahlschmidt, W.; Bulut, H.; Gromova, K.; Puchkov, D.; Robertson, M.J.; MacGregor, K.A.; Tomilin, N.; Pechstein, A.; Chau, N.; et al. Role of the Clathrin Terminal Domain in Regulating Coated Pit Dynamics Revealed by Small Molecule Inhibition. Cell 2011, 146, 841. [Google Scholar] [CrossRef]
- Wu, X.; Zhao, X.; Puertollano, R.; Bonifacino, J.S.; Eisenberg, E.; Greene, L.E. Adaptor and Clathrin Exchange at the Plasma Membrane and Trans-Golgi Network. Mol. Biol. Cell 2003, 14, 516–528. [Google Scholar] [CrossRef] [PubMed]
- Girard, E.; Paul, J.L.; Fournier, N.; Beaune, P.; Johannes, L.; Lamaze, C.; Védie, B. The Dynamin Chemical Inhibitor Dynasore Impairs Cholesterol Trafficking and Sterol-Sensitive Genes Transcription in Human HeLa Cells and Macrophages. PLoS ONE 2011, 6, e29042. [Google Scholar] [CrossRef] [PubMed]
- Preta, G.; Cronin, J.G.; Sheldon, I.M. Dynasore—Not Just a Dynamin Inhibitor. Cell Commun. Signal. 2015, 13, 24. [Google Scholar] [CrossRef] [PubMed]
- Neufeld, E.B.; Cooney, A.M.; Pitha, J.; Dawidowicz, E.A.; Dwyer, N.K.; Pentchev, P.G.; Joan Blanchette-Mackie, E. Intracellular Trafficking of Cholesterol Monitored with a Cyclodextrin. J. Biol. Chem. 1996, 271, 21604–21613. [Google Scholar] [CrossRef] [PubMed]
- Rink, J.; Ghigo, E.; Kalaidzidis, Y.; Zerial, M. Rab Conversion as a Mechanism of Progression from Early to Late Endosomes. Cell 2005, 122, 735–749. [Google Scholar] [CrossRef]
- Murray, J.T.; Panaretou, C.; Stenmark, H.; Miaczynska, M.; Backer, J.M. Role of Rab5 in the Recruitment of HVps34/P150 to the Early Endosome. Traffic 2002, 3, 416–427. [Google Scholar] [CrossRef]
- Simonsen, A.; Lippe, R.; Christoforidis, S.; Gaullier, J.M.; Brech, A.; Callaghan, J.; Toh, B.-H.; Murphy, C.; Zerial, M.; Stenmark, H. EEA1 Links PI(3)K Function to Rab5 Regulation of Endosome Fusion. Nature 1998, 394, 494–498. [Google Scholar] [CrossRef]
- Raiborg, C.; Wesche, J.; Malerød, L.; Stenmark, H. Flat Clathrin Coats on Endosomes Mediate Degradative Protein Sorting by Scaffolding Hrs in Dynamic Microdomains. J. Cell Sci. 2006, 119, 2414–2424. [Google Scholar] [CrossRef]
- Ullrich, O.; Reinsch, S.; Urbé, S.; Zerial, M.; Parton, R.G. Rab11 Regulates Recycling through the Pericentriolar Recycling Endosome. J. Cell Biol. 1996, 135, 913–924. [Google Scholar] [CrossRef] [PubMed]
- Naslavsky, N.; Weigert, R.; Donaldson, J.G. Convergence of Non-Clathrin- and Clathrin-Derived Endosomes Involves Arf6 Inactivation and Changes in Phosphoinositides. Mol. Biol. Cell 2002, 14, 417–431. [Google Scholar] [CrossRef]
- Grant, B.D.; Donaldson, J.G. Pathways and Mechanisms of Endocytic Recycling. Nat. Rev. Mol. Cell Biol. 2009, 10, 597–608. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, S.; Kubo, K.; Waguri, S.; Yabashi, A.; Shin, H.-W.; Katoh, Y.; Nakayama, K. Rab11 Regulates Exocytosis of Recycling Vesicles at the Plasma Membrane. J. Cell Sci. 2012, 125, 4049–4057. [Google Scholar] [CrossRef] [PubMed]
- Homma, Y.; Fukuda, M. Rabin8 Regulates Neurite Outgrowth in Both GEF Activity–Dependent and –Independent Manners. Mol. Biol. Cell 2016, 27, 2107–2118. [Google Scholar] [CrossRef] [PubMed]
- De Matteis, M.A.; Mironov, A.A.; Beznoussenko, G.V. The Golgi Ribbon and the Function of the Golgins; Springer: Vienna, Austria, 2008; pp. 223–246. [Google Scholar] [CrossRef]
- Barr, F.A.; Nakamura, N.; Warren, G. Mapping the Interaction between GRASP65 and GM130, Components of a Protein Complex Involved in the Stacking of Golgi Cisternae. EMBO J. 1998, 17, 3258–3268. [Google Scholar] [CrossRef]
- Marra, P.; Maffucci, T.; Daniele, T.; di Tullio, G.; Ikehara, Y.; Chan, E.K.L.; Luini, A.; Beznoussenko, G.; Mironov, A.; de Matteis, M.A. The GM130 and GRASP65 Golgi Proteins Cycle through and Define a Subdomain of the Intermediate Compartment. Nat. Cell Biol. 2001, 3, 1101–1113. [Google Scholar] [CrossRef]
- Cepeda, V.; Fraile-Ramos, A. A Role for the SNARE Protein Syntaxin 3 in Human Cytomegalovirus Morphogenesis. Cell. Microbiol. 2011, 13, 846–858. [Google Scholar] [CrossRef]
- Rohrer, J.; Schweizer, A.; Russell, D.; Kornfeld, S. The Targeting of Lamp1 to Lysosomes Is Dependent on the Spacing of Its Cytoplasmic Tail Tyrosine Sorting Motif Relative to the Membrane. J. Cell Biol. 1996, 132, 565–576. [Google Scholar] [CrossRef]
- Mahmutefendić, H.; Zagorac, G.B.; Grabušić, K.; Karleuša, L.; Maćešić, S.; Momburg, F.; Lučin, P. Late Endosomal Recycling of Open MHC-I Conformers. J. Cell. Physiol. 2017, 232, 872–887. [Google Scholar] [CrossRef] [PubMed]
- Marcinowski, L.; Lidschreiber, M.; Windhager, L.; Rieder, M.; Bosse, J.B.; Rädle, B.; Bonfert, T.; Györy, I.; Graaf, M.d.; Costa, O.P.; et al. Real-Time Transcriptional Profiling of Cellular and Viral Gene Expression during Lytic Cytomegalovirus Infection. PLoS Pathog. 2012, 8, e1002908. [Google Scholar] [CrossRef] [PubMed]
- Scrivano, L.; Esterlechner, J.; Mühlbach, H.; Ettischer, N.; Hagen, C.; Grünewald, K.; Mohr, C.A.; Ruzsics, Z.; Koszinowski, U.; Adler, B. The M74 Gene Product of Murine Cytomegalovirus (MCMV) Is a Functional Homolog of Human CMV GO and Determines the Entry Pathway of MCMV. J. Virol. 2010, 84, 4469. [Google Scholar] [CrossRef]
- Hanson, L.K.; Slater, J.S.; Cavanaugh, V.J.; Newcomb, W.W.; Bolin, L.L.; Nelson, C.N.; Fetters, L.D.; Tang, Q.; Brown, J.C.; Maul, G.G.; et al. Murine Cytomegalovirus Capsid Assembly Is Dependent on US22 Family Gene M140 in Infected Macrophages. J. Virol. 2009, 83, 7449–7456. [Google Scholar] [CrossRef] [PubMed]
- Kattenhorn, L.M.; Mills, R.; Wagner, M.; Lomsadze, A.; Makeev, V.; Borodovsky, M.; Ploegh, H.L.; Kessler, B.M. Identification of Proteins Associated with Murine Cytomegalovirus Virions. J. Virol. 2004, 78, 11187–11197. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.A.; Carlson, M.E.; Henry, S.C.; Shanley, J.D. The Murine Cytomegalovirus M25 Open Reading Frame Encodes a Component of the Tegument. Virology 1999, 262, 265–276. [Google Scholar] [CrossRef]
- Kutle, I.; Sengstake, S.; Templin, C.; Glaß, M.; Kubsch, T.; Keyser, K.A.; Binz, A.; Bauerfeind, R.; Sodeik, B.; Čičin-Šain, L.; et al. The M25 Gene Products Are Critical for the Cytopathic Effect of Mouse Cytomegalovirus. Sci. Rep. 2017, 7, 15588. [Google Scholar] [CrossRef]
- Spaete, R.R.; Thayer, R.M.; Probert, W.S.; Masiarz, F.R.; Chamberlain, S.H.; Rasmussen, L.; Merigan, T.C.; Pachl, C. Human Cytomegalovirus Strain Towne Glycoprotein B Is Processed by Proteolytic Cleavage. Virology 1988, 167, 207–225. [Google Scholar] [CrossRef]
- Strive, T.; Borst, E.; Messerle, M.; Radsak, K. Proteolytic Processing of Human Cytomegalovirus Glycoprotein B Is Dispensable for Viral Growth in Culture. J. Virol. 2002, 76, 1252–1264. [Google Scholar] [CrossRef]
- Fraile-Ramos, A.; Cepeda, V.; Elstak, E.; Sluijs, P. van der Rab27a Is Required for Human Cytomegalovirus Assembly. PLoS ONE 2010, 5, e15318. [Google Scholar] [CrossRef]
- Hook, L.M.; Grey, F.; Grabski, R.; Tirabassi, R.; Doyle, T.; Hancock, M.; Landais, I.; Jeng, S.; McWeeney, S.; Britt, W.; et al. Cytomegalovirus MiRNAs Target Secretory Pathway Genes to Facilitate Formation of the Virion Assembly Compartment and Reduce Cytokine Secretion. Cell Host Microbe 2014, 15, 363–373. [Google Scholar] [CrossRef]
- Tomaš, M.I.; Kučić, N.; Mahmutefendić, H.; Blagojević, G.; Lučin, P. Murine Cytomegalovirus Perturbs Endosomal Trafficking of Major Histocompatibility Complex Class I Molecules in the Early Phase of Infection. J. Virol. 2010, 84, 11101–11112. [Google Scholar] [CrossRef] [PubMed]
- Hertel, L.; Chou, S.; Mocarski, E.S. Viral and Cell Cycle–Regulated Kinases in Cytomegalovirus-Induced Pseudomitosis and Replication. PLoS Pathog. 2007, 3, e6. [Google Scholar] [CrossRef] [PubMed]
- Derivery, E.; Sousa, C.; Gautier, J.J.; Lombard, B.; Loew, D.; Gautreau, A. The Arp2/3 Activator WASH Controls the Fission of Endosomes through a Large Multiprotein Complex. Dev. Cell 2009, 17, 712–723. [Google Scholar] [CrossRef]
- Nakajo, A.; Yoshimura, S.; Togawa, H.; Kunii, M.; Iwano, T.; Izumi, A.; Noguchi, Y.; Watanabe, A.; Goto, A.; Sato, T.; et al. EHBP1L1 Coordinates Rab8 and Bin1 to Regulate Apical-Directed Transport in Polarized Epithelial Cells. J. Cell Biol. 2016, 212, 297–306. [Google Scholar] [CrossRef]
- Martin, C.; Leyton, L.; Hott, M.; Arancibia, Y.; Spichiger, C.; McNiven, M.A.; Court, F.A.; Concha, M.I.; Burgos, P.v.; Otth, C. Herpes Simplex Virus Type 1 Neuronal Infection Perturbs Golgi Apparatus Integrity through Activation of Src Tyrosine Kinase and Dyn-2 GTPase. Front. Cell. Infect. Microbiol. 2017, 7, 371. [Google Scholar] [CrossRef]
- Sanchez, V.; Greis, K.D.; Sztul, E.; Britt, W.J. Accumulation of Virion Tegument and Envelope Proteins in a Stable Cytoplasmic Compartment during Human Cytomegalovirus Replication: Characterization of a Potential Site of Virus Assembly. J. Virol. 2000, 74, 975–986. [Google Scholar] [CrossRef] [PubMed]
- Procter, D.J.; Banerjee, A.; Nukui, M.; Kruse, K.; Gaponenko, V.; Murphy, E.A.; Komarova, Y.; Walsh, D. The HCMV Assembly Compartment Is a Dynamic Golgi-Derived MTOC That Controls Nuclear Rotation and Virus Spread. Dev. Cell 2018, 45, 83–100.e7. [Google Scholar] [CrossRef] [PubMed]
- Raux, H.; Obiang, L.; Richard, N.; Harper, F.; Blondel, D.; Gaudin, Y. The Matrix Protein of Vesicular Stomatitis Virus Binds Dynamin for Efficient Viral Assembly. J. Virol. 2010, 84, 12609–12618. [Google Scholar] [CrossRef]
- Johns, H.L.; Gonzalez-Lopez, C.; Sayers, C.L.; Hollinshead, M.; Elliott, G. Rab6 Dependent Post-Golgi Trafficking of HSV1 Envelope Proteins to Sites of Virus Envelopment. Traffic 2014, 15, 157–178. [Google Scholar] [CrossRef]
- Maxfield, F.R.; McGraw, T.E. Endocytic Recycling. Nat. Rev. Mol. Cell Biol. 2004, 5, 121–132. [Google Scholar] [CrossRef]
- Pavišić, V.; Mahmutefendić Lučin, H.; Blagojević Zagorac, G.; Lučin, P. Arf GTPases Are Required for the Establishment of the Pre-Assembly Compartment in the Early Phase of Cytomegalovirus Infection. Life 2021, 11, 867. [Google Scholar] [CrossRef]
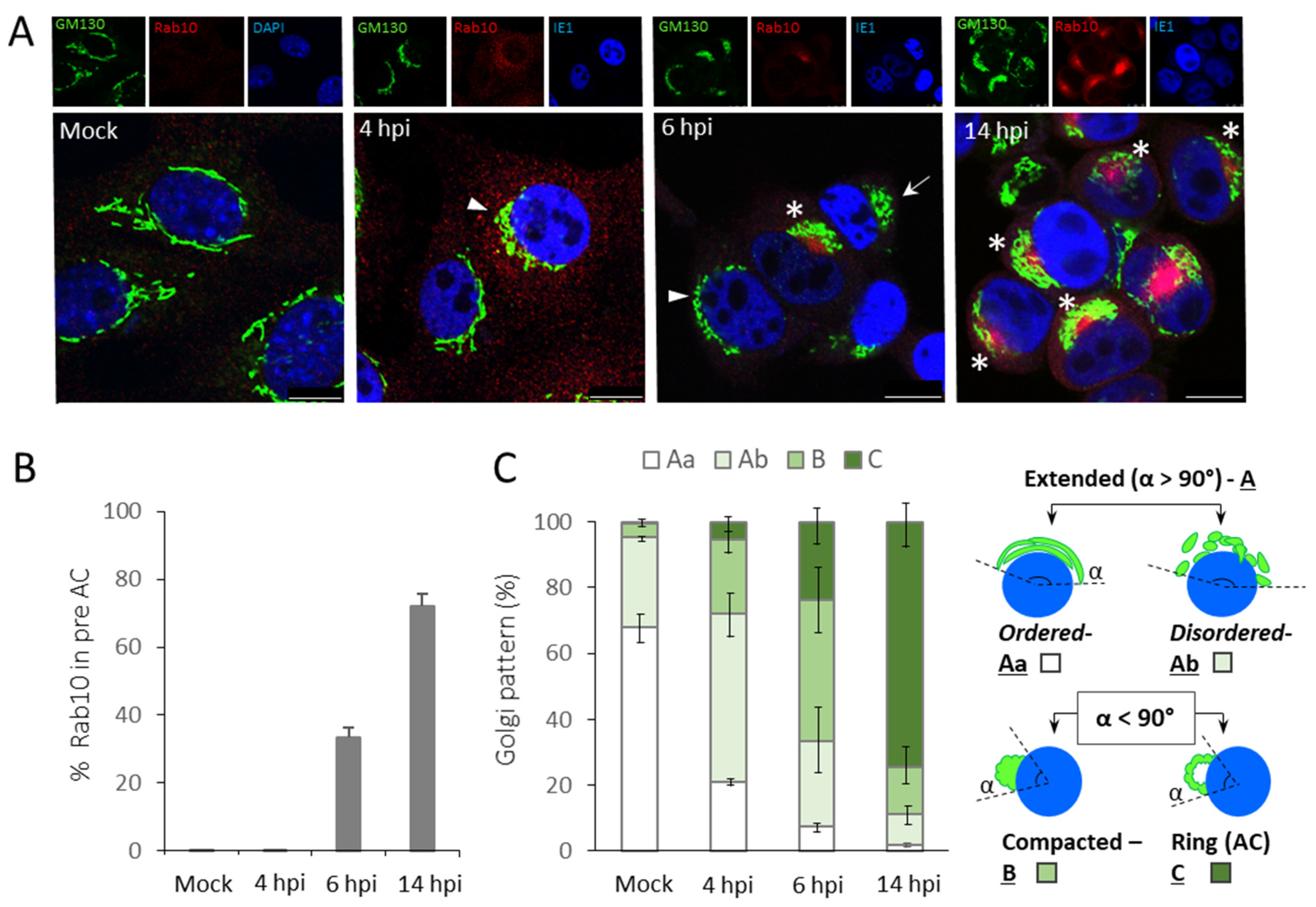
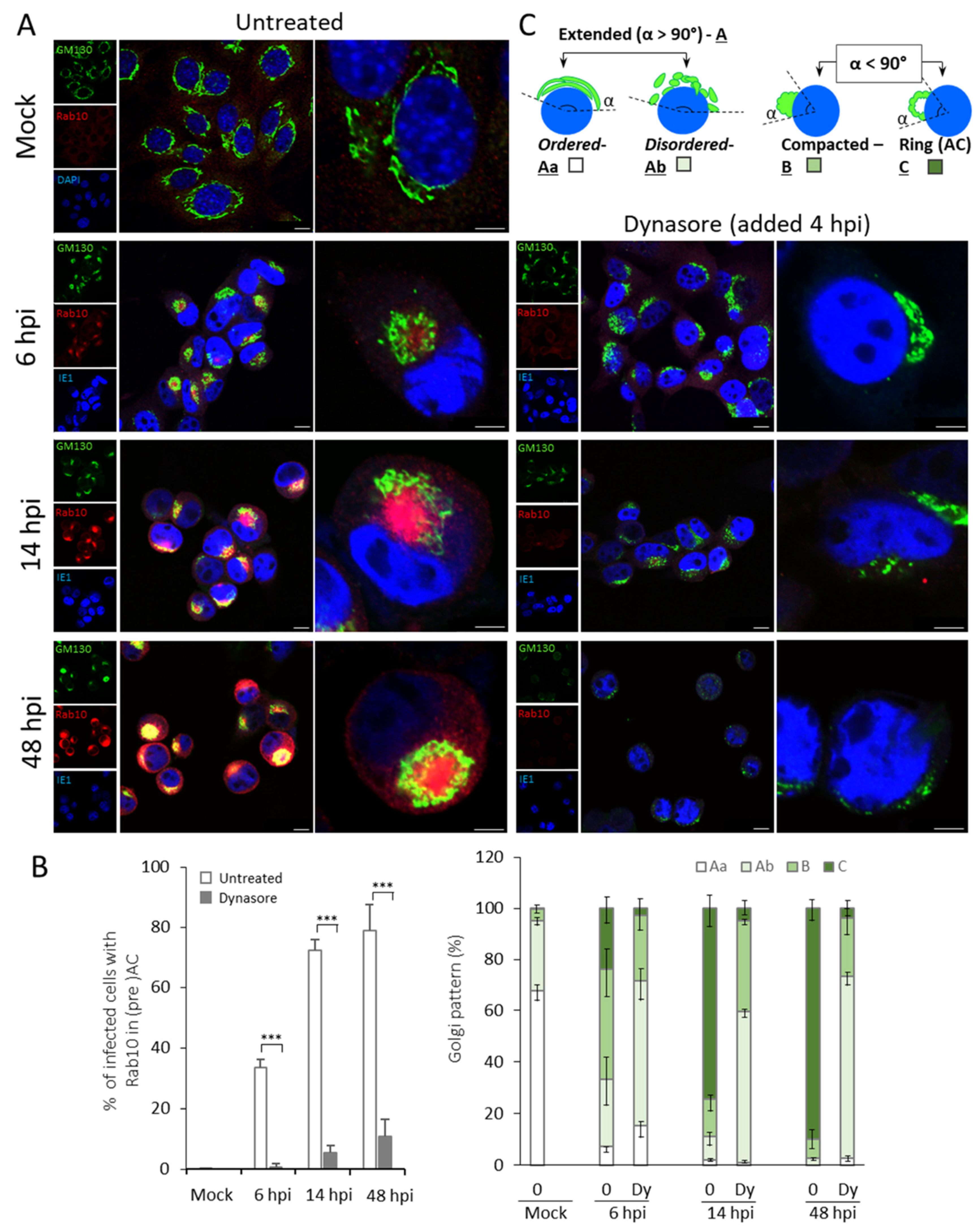
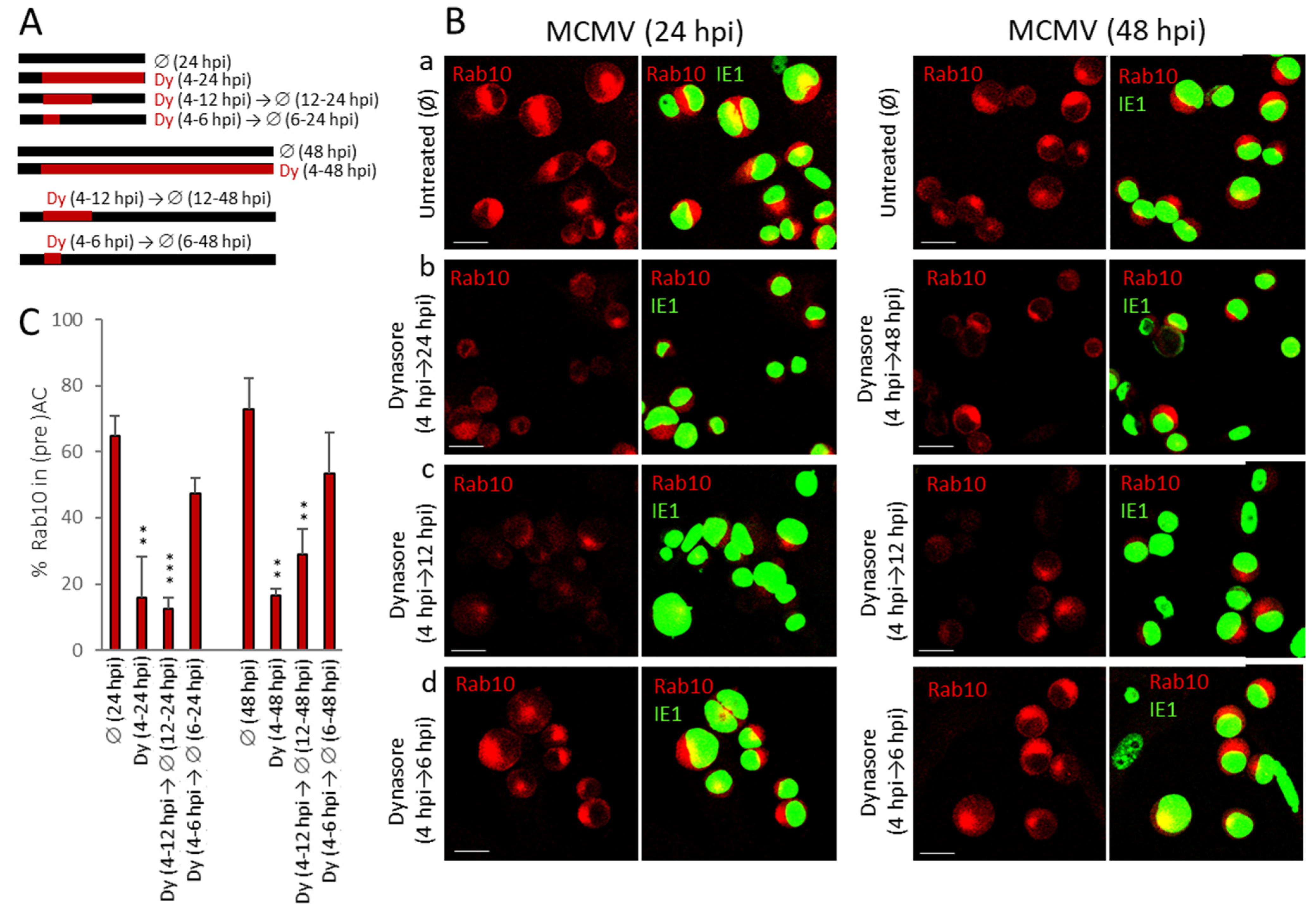
| Pericentriolar Rab10 (%) * | Golgi Patterns (%) * | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Aa | Ab | B | C | |||||||
| Ø | Dy | Ø | Dy | Ø | Dy | Ø | Dy | Ø | Dy | |
| Mock | 0 | - | 67.8 ± 4.2 | - | 27.2 ± 1.4 | - | 4.4 ± 2.3 | - | 0.4 ± 0.7 | - |
| 6 hpi | 33.3 ± 2.9 | 0.6 ± 1.1 | 7.3 ± 1.8 | 15.4 ± 4.9 | 25.9 ± 10.5 | 56.1 ± 7.2 | 42.7 ± 10.7 | 25.9 ± 9.0 | 23.9 ± 6.3 | 2.6 ± 2.9 |
| 14 hpi | 72.1 ± 3.6 | 5.4 ± 2.2 | 1.9 ± 0.7 | 0.9 ± 0.5 | 9.4 ± 3.9 | 58.7 ± 2.9 | 14.2 ± 4.2 | 35.3 ± 1.6 | 74.4 ± 7.5 | 5.0 ± 3.7 |
| 48 hpi | 78.9 ± 8.5 | 10.7 ± 5.8 | 0 | 2.7 ± 1.3 | 2.8 ± 0.7 | 70.5 ± 3.7 | 7.1 ± 4.2 | 22.9 ± 6.2 | 90.0 ± 5.0 | 3.8 ± 3.7 |
| Inhibitor | Number of Cells | Rab10 Accumulation (%) * | M55 in the oAC (%) * | Viability * | ||||
|---|---|---|---|---|---|---|---|---|
| 16 hpi | 40 hpi | 16 hpi | 40 hpi | 16 hpi | 40 hpi | 16 hpi | 40 hpi | |
| Untreated | 4209 | 2386 | 65.5 ± 11.5 | 81.0 ± 5.9 | 0 | 56.6 ± 8.6 | 96.2 ± 3.0 | 90.5 ± 4.7 |
| Dynasore (80 μM) | 3000 | 1757 | 2.2 ± 1.6 | 2.9 ± 2.5 | 0 | 0.1 ± 0.2 | 98.0 ± 2.8 | 92.5 ± 0.7 |
| Dyngo 4a (200 μM) | 1006 | 649 | 2.4 ± 1.5 | 1.3 ± 0.3 | 0 | 0.3 ± 0.5 | 90.0 ± 3.2 | 85.0 ± 2.1 |
| MiTMAB (20 μM) | 1547 | 935 | 30.0 ± 13.6 | 7.1 ± 3.0 | 0 | 0.3 ± 0.6 | 91.0 ± 4.0 | 81.0 ± 10.8 |
| Dynole 34-2 (10 μM) | 1078 | 623 | 41.9 ± 8.4 | 17.6 ± 5.0 | 0 | 4.9 ± 1.8 | 88.5 ± 6.4 | 70.5 ± 9.1 |
| Pitstop 2 (50 μM) | 1679 | 1652 | 45.8 ± 11.1 | 70.0 ± 9.4 | 0 | 10.5 ± 9.2 | 95.0 ± 2.4 | 89.0 ± 3.1 |
| Methyl-β-CD (7.5 mM) | 1985 | 1194 | 52.9 ± 19.2 | 67.5 ± 0.8 | 0 | 37.9 ± 15.9 | 97.0 ± 2.1 | 89.0 ± 1.5 |
| Marker of Cellular Compartment | The Ratio of Infected Cells (%) with the Accumulated Marker in Pericentriolar Membranes * | |||
|---|---|---|---|---|
| 6 hpi | 14 hpi | |||
| Untreated | Dynasore | Untreated | Dynasore | |
| Early Endosomes | ||||
| Rab5 | 50.1 ± 4.5 | 10.7 ± 1.9 | 71.5 ± 7.6 | 29.7 ± 1.6 |
| Vps34 | 28.0 ± 3.5 | 4.0 ± 0.2 | 63.8 ± 4.1 | 8.3 ± 3.2 |
| EEA1 | 20.7 ± 4.1 | 2.9 ± 0.6 | 62.4 ± 7.5 | 4.0 ± 1.3 |
| Hrs | 37.2 ± 1.8 | 4.0 ± 0.1 | 67.3 ± 3.6 | 5.6 ± 2.9 |
| Early Endosomes to Endosomal Recycling Compartment | ||||
| Rab10 | 33.3 ± 2.9 | 0.6 ± 1.1 | 72.1 ± 3.6 | 5.4 ± 2.2 |
| Endosomal Recycling Compartment | ||||
| Rab11 | 64.1 ± 4.3 | 15.9 ± 3.9 | 78.6 ± 3.7 | 51.3 ± 7.5 |
| Arf6 | 1.0 ± 0.5 | 0 | 67.81 ± 5.8 | 7.6 ± 2.7 |
| Golgi Compartments | ||||
| Grasp65 (ERGIC-cis-Golgi) | 28.0 ± 2.0 | 6.0 ± 0.5 | 71.8 ± 5.1 | 8.7 ± 2.8 |
| GM130 (cis-Golgi) | 23.9 ± 13.3 | 2.6 ± 2.9 | 74.4 ± 7.5 | 5.0 ± 3.7 |
| Golgin97 (trans-Golgi) | 29.1 ± 2.0 | 5.4 ± 3.1 | 69.2 ± 6.5 | 7.0 ± 0.6 |
| Grasp65 (ERGIC-cis-Golgi) | 28.0 ± 2.0 | 6.0 ± 0.5 | 71.8 ± 5.1 | 8.7 ± 2.8 |
| Late Endosomes | ||||
| Lamp1 | 0 | 0 | 0 | 0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Štimac, I.; Jug Vučko, N.; Blagojević Zagorac, G.; Marcelić, M.; Mahmutefendić Lučin, H.; Lučin, P. Dynamin Inhibitors Prevent the Establishment of the Cytomegalovirus Assembly Compartment in the Early Phase of Infection. Life 2021, 11, 876. https://doi.org/10.3390/life11090876
Štimac I, Jug Vučko N, Blagojević Zagorac G, Marcelić M, Mahmutefendić Lučin H, Lučin P. Dynamin Inhibitors Prevent the Establishment of the Cytomegalovirus Assembly Compartment in the Early Phase of Infection. Life. 2021; 11(9):876. https://doi.org/10.3390/life11090876
Chicago/Turabian StyleŠtimac, Igor, Natalia Jug Vučko, Gordana Blagojević Zagorac, Marina Marcelić, Hana Mahmutefendić Lučin, and Pero Lučin. 2021. "Dynamin Inhibitors Prevent the Establishment of the Cytomegalovirus Assembly Compartment in the Early Phase of Infection" Life 11, no. 9: 876. https://doi.org/10.3390/life11090876
APA StyleŠtimac, I., Jug Vučko, N., Blagojević Zagorac, G., Marcelić, M., Mahmutefendić Lučin, H., & Lučin, P. (2021). Dynamin Inhibitors Prevent the Establishment of the Cytomegalovirus Assembly Compartment in the Early Phase of Infection. Life, 11(9), 876. https://doi.org/10.3390/life11090876








