Hydrogen Bonding and Solvation of a Proline-Based Peptide Model in Salt Solutions
Abstract
:1. Introduction
2. Materials and Methods
3. Results
4. Discussion
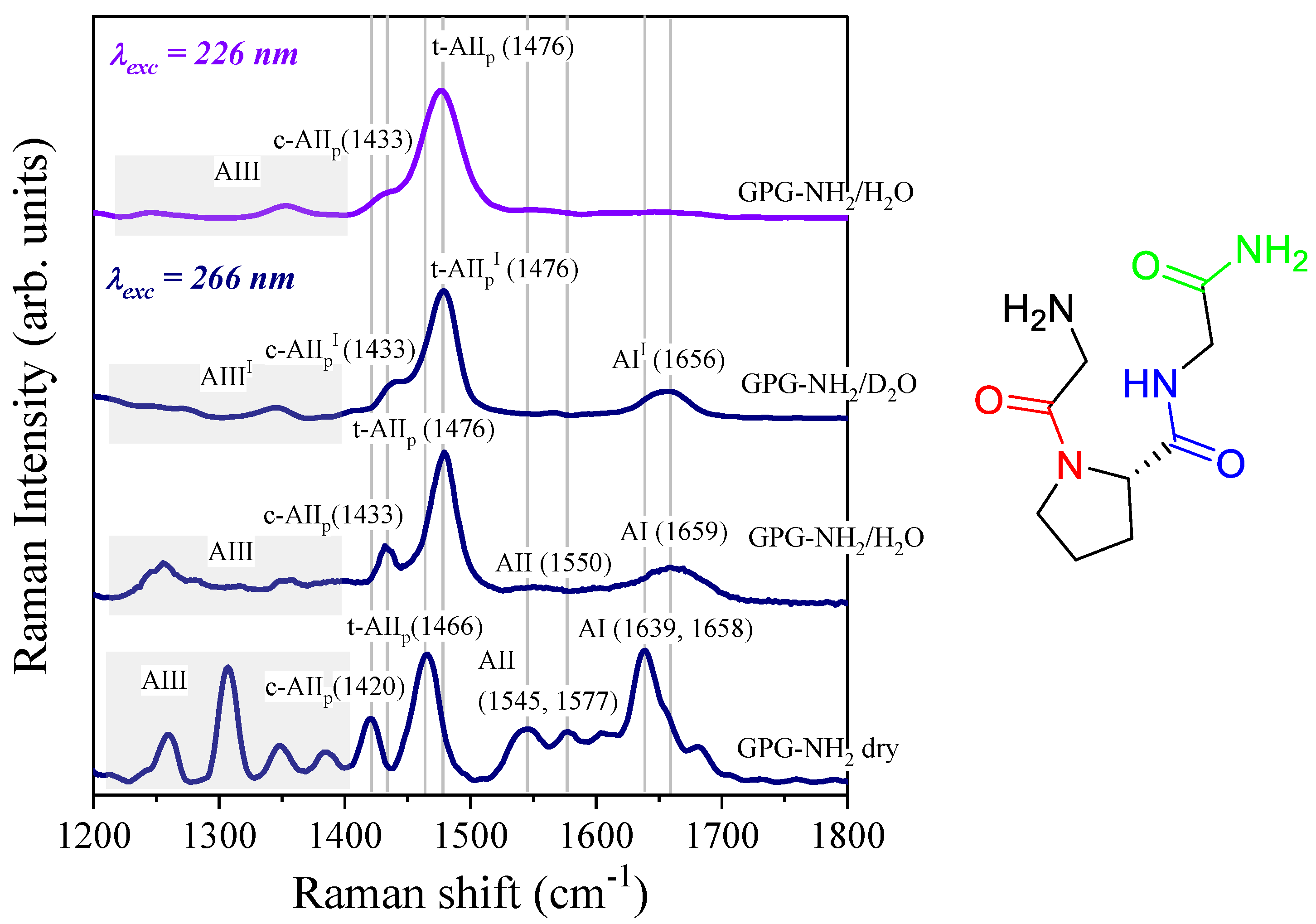
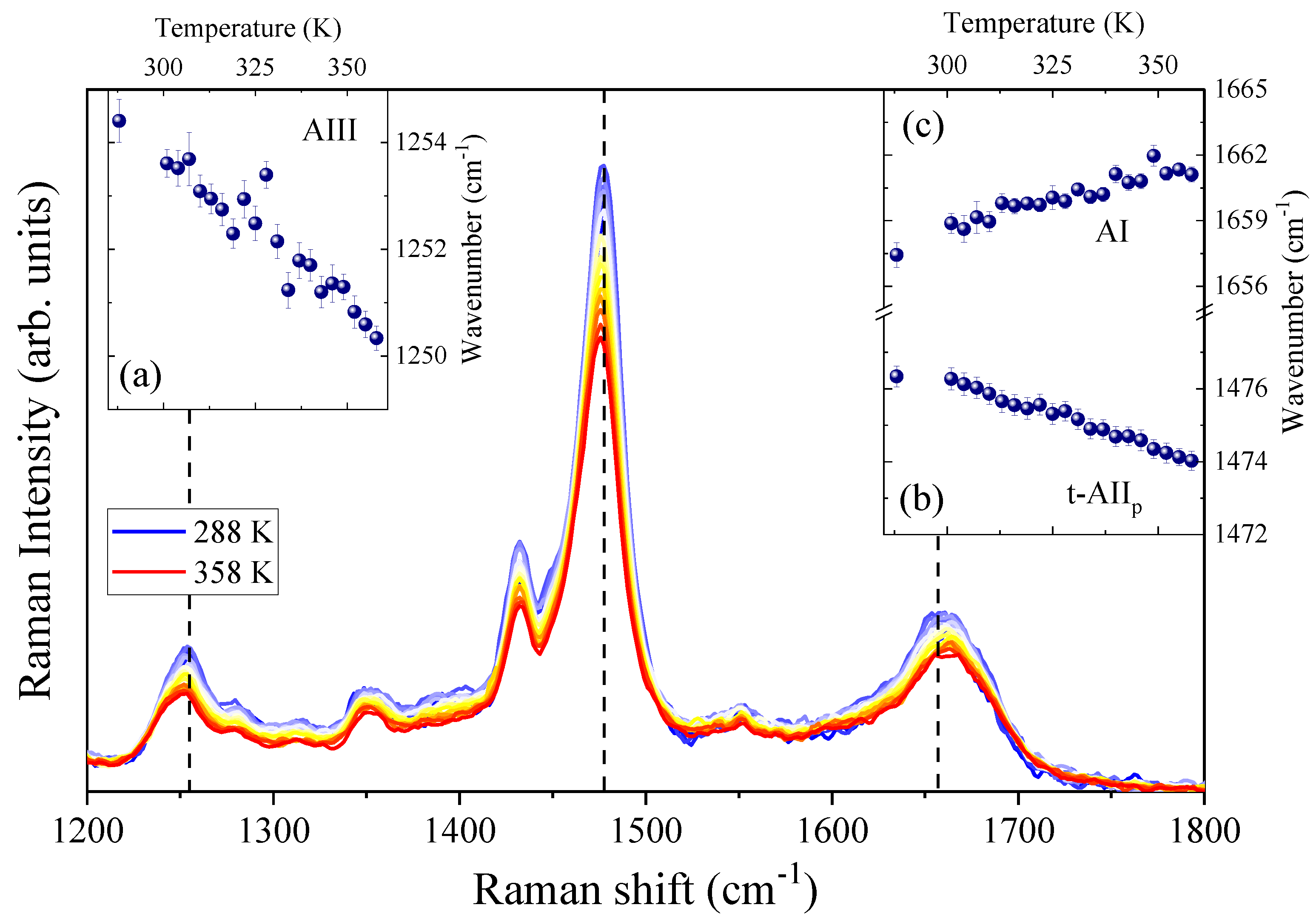
4.1. Temperature-Dependence of Hydrogen Bonding on the Amide Bonds of GPG-NH2
4.2. Effect of Salts on the Hydrogen Bonding at the Carbonyl Site of Pro
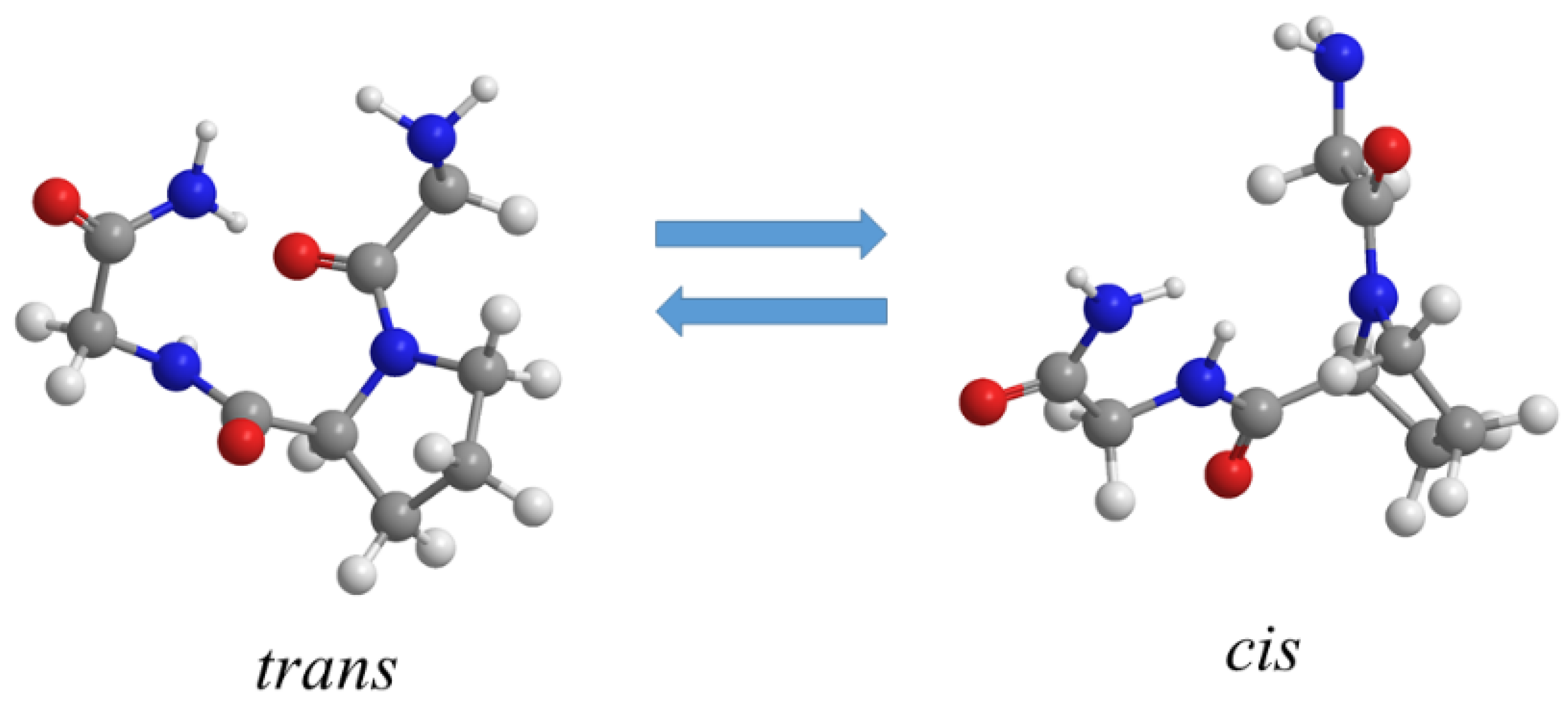
4.3. Influence of Salts on Trans/Cis Isomerization
4.4. Effect of Salts on the Structure of the Tripeptide GPG-NH2
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Maity, H.; Muttathukattil, A.N.; Reddy, G. Salt Effects on Protein Folding Thermodynamics. J. Phys. Chem. Lett. 2018, 9, 5063–5070. [Google Scholar] [CrossRef] [PubMed]
- Bröhl, A.; Albrecht, B.; Zhang, Y.; Maginn, E.; Giernoth, R. Influence of Hofmeister Ions on the Structure of Proline-Based Peptide Models: A Combined Experimental and Molecular Modeling Study. J. Phys. Chem. B 2017, 121, 2062–2072. [Google Scholar] [CrossRef]
- Nandi, P.K.; Robinson, D.R. The Effects of Salts on the Free Energy of the Peptide Group. J. Am. Chem. Soc. 1972, 94, 1308–1315. [Google Scholar] [CrossRef] [PubMed]
- Bosshard, H.R.; Marti, D.N.; Jelesarov, I. Protein stabilization by salt bridges: Concepts, experimental approaches and clarification of some misunderstandings. J. Mol. Recognit. 2004, 17, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Pegrama, L.M.; Wendorff, T.; Erdmanna, R.; Shkela, I.; Bellissimo, D.; Felitskya, D.J.; Record, M.T., Jr. Why Hofmeister effects of many salts favor protein folding but not DNA helix formation. Proc. Natl. Acad. Sci. USA 2010, 107, 7716–7721. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Otzen, D.E.; Oliveberg, M. Salt-induced detour through compact regions of the protein folding landscape. Proc. Natl. Acad. Sci. USA 1999, 96, 11746–11751. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sedlák, E.; Stagg, L.; Wittung-Stafshede, P. Effect of Hofmeister ions on protein thermal stability: Roles of ion hydration and peptide groups? Arch. Biochem. Biophys. 2008, 479, 69–73. [Google Scholar] [CrossRef]
- Bellö, J.; Haas, D.; Bellö, H.R. On the Mechanism of Action of Neutral Salt on the Collagen-Fold. Biochemistry 1996, 5, 2539–2548. [Google Scholar] [CrossRef] [PubMed]
- Von Hippel, P.H.; Peticolas, V.; Schack, L.; Karlson, L. Ion effects on the solution structure of biological macromolecules. Biochemistry 1973, 12, 1256–1264. [Google Scholar] [CrossRef]
- Hamabata, A.; von Hippel, P.H. Model studies on the effects of neutral salts on the conformational stability of biological macromolecules. II. Effects of vicinal hydrophobic groups on the specificity of binding of ions to amide groups. Biochemistry 1973, 12, 1264–1271. [Google Scholar] [CrossRef]
- Baldwin, R.L. How Hofmeister ion interactions affect protein stability. Biophys. J. 1996, 71, 2056–2063. [Google Scholar] [CrossRef] [Green Version]
- Boström, M.; Tavares, F.W.; Finet, S.; Skouri-Panet, F.; Tardieu, A.; Ninham, B.W. Why forces between proteins follow different Hofmeister series for pH above and below pI. Biophys. Chem. 2005, 117, 217–224. [Google Scholar] [CrossRef]
- Rose, G.D.; Gierasch, L.M.; Smith, J.A. Turns in peptides and proteins. Adv. Protein. Chem. 1985, 37, 1–109. [Google Scholar]
- Wilmot, C.M.; Thornton, J.M. Analysis and Prediction of the Different Types of B-Turn in Proteins. J. Mol. Biol. 1988, 203, 221–232. [Google Scholar] [CrossRef]
- Richardson, J.S. The anatomy and taxonomy of protein structure. Adv. Protein. Chem. 1981, 34, 167–339. [Google Scholar] [PubMed]
- Schultz, G.D.; Schirmer, R.H. Principles of Protein Structure; Springer: New York, NY, USA, 1985. [Google Scholar]
- Kim, P.S.; Baldwin, R.L. Specific Intermediates in the Folding Reactions of Small Proteins and the Mechanism of Protein Folding. Annu. Rev. Biochem. 1982, 51, 459–489. [Google Scholar] [CrossRef] [PubMed]
- Kiefhaber, T.; Grunert, H.P.; Hahn, U.; Schmid, F.X. Replacement of a cis proline simplifies the mechanism of ribonuclease T1 folding. Biochemistry 1990, 29, 6475–6480. [Google Scholar] [CrossRef]
- Smyth, D.S.; Utsumi, S. Structure at the Hinge Region in Rabbit Immunoglobulin-G. Nature 1967, 216, 332–335. [Google Scholar] [CrossRef]
- Bretscher, L.E.; Jenkins, C.L.; Taylor, K.M.; Derider, M.L.; Raines, R.T. Conformational Stability of Collagen Relies on a Stereoelectronic Effect. J. Am. Chem. Soc. 2001, 123, 777–778. [Google Scholar] [CrossRef]
- Wedemeyer, W.J.; Welker, E.; Scheraga, H.A. Proline Cis–Trans Isomerization and Protein Folding. Biochemistry 2002, 41, 14637–14644. [Google Scholar] [CrossRef]
- Eckert, B.; Martin, A.; Balbach, J.; Schmid, F.X. Prolyl Isomerization as a Molecular Timer in Phage Infection. Nat. Struct. Mol. Biol. 2005, 12, 619–623. [Google Scholar] [CrossRef]
- Schmid, F.; Baldwin, R.L. Detection of an early intermediate in the folding of ribonuclease A by protection of amide protons against exchange. J. Mol. Biol. 1979, 135, 199–215. [Google Scholar] [CrossRef]
- Mui, P.W.; Konishi, Y.; Scheraga, A. Kinetics and mechanism of the refolding of denatured ribonuclease A. Biochemistry 1985, 24, 4481–4489. [Google Scholar] [CrossRef] [PubMed]
- Brandts, J.F.; Halverson, H.R.; Brennan, M. Consideration of the Possibility that the slow step in protein denaturation reactions is due to cis-trans isomerism of proline residues. Biochemistry 1975, 14, 4953–4963. [Google Scholar] [CrossRef] [PubMed]
- Andreotti, A.H. Native State Proline Isomerization: An Intrinsic Molecular Switch. Biochemistry 2003, 42, 9515–9524. [Google Scholar] [CrossRef]
- Lummis, S.C.R.; Beene, D.L.; Lee, L.W.; Lester, H.; Broadhurst, R.W.; Dougherty, D.A. Cis-Trans Isomerization at a Proline Opens the Pore of a Neurotransmitter-Gated Ion Channel. Nature 2005, 438, 248–252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pandey, A.K.; Naduthambi, D.; Thomas, K.M.; Zondlo, N.J. Proline Editing: A General and Practical Approach to the Synthesis of Functionally and Structurally Diverse Peptides. J. Am. Chem. Soc. 2013, 135, 4333–4363. [Google Scholar] [CrossRef] [Green Version]
- Rossi, B.; Bottari, C.; Catalini, S.; Gessini, A.; D’Amico, F.; Masciovecchio, C. Synchrotron based UV Resonant Raman scattering for material science. In Molecular and Laser Spectroscopy, Volume 2, 1st ed.; Gupta, V.P., Ozaki, Y., Eds.; Elsevier: Amsterdam, The Netherlands, 2020; Chapter 13; pp. 447–478. [Google Scholar]
- Jordan, T.; Mukerji, I.; Wang, Y.; Spiro, T.G. UV resonance Raman spectroscopy and hydrogen bonding of the proline peptide bond. J. Mol. Struct. 1996, 379, 51–64. [Google Scholar] [CrossRef]
- Caswell, D.S.; Spiro, T.G. Proline Signals in Ultraviolet Resonance Raman Spectra of Proteins: Cis-Trans Isomerism in Polyproline and Ribonuclease A. J. Am. Chem. Soc. 1987, 109, 2796–2800. [Google Scholar] [CrossRef]
- Harhay, G.P.; Hudson, B.S. Ultraviolet Resonance Raman Study of Proline Isomerization. J. Chem. Phys. 1991, 95, 3511–3513. [Google Scholar] [CrossRef]
- Takeuchi, H.; Harada, I. Ultraviolet Resonance Raman Spectroscopy of X-Proline Bonds: A New Marker Band of Hydrogen Bonding at the hide C=O Site. J. Raman Spectrosc. 1990, 21, 509–515. [Google Scholar] [CrossRef]
- Busch, S.; Bruce, C.D.; Red, C.; Lorenz, C.D.; McLain, S.E. Water mediation is essential to nucleation of b-turn formation in peptide folding motifs. Angew. Chem. Int. Ed. 2013, 52, 13091–13095. [Google Scholar] [CrossRef] [Green Version]
- Di Gioacchino, M.; Bruni, F.; Ricci, M.A. GPG-NH2 solutions: A model system for b-turns formation. Possible role of trehalose against drought. J. Mol. Liq. 2021, 335, 116514. [Google Scholar] [CrossRef]
- Oladepo, S.A.; Xiong, K.; Hong, Z.; Asher, S.A.; Handen, J.; Lednev, I.K. UV Resonance Raman Investigations of Peptide and Protein Structure and Dynamics. Chem. Rev. 2012, 112, 2604–2628. [Google Scholar] [CrossRef] [Green Version]
- Catalini, S.; Rossi, B.; Foggi, P.; Masciovecchio, C.; Bruni, F. Aqueous solvation of Glutathione probed by UV Resonance Raman Spectroscopy. J. Mol. Liq. 2019, 283, 537–547. [Google Scholar] [CrossRef]
- Rossi, B.; Catalini, S.; Bottari, C.; Gessini, A.; Masciovecchio, C. Frontiers of UV resonant raman spectroscopy by using synchrotron radiation: The case of aqueous solvation of model peptides. In Proceedings Volume 11086, UV and Higher Energy Photonics: From Materials to Applications 2019; SPIE: San Diego, CA, USA, 2019. [Google Scholar]
- Frushour, B.G.; Koening, J.L. Advances in Infrared and Raman Spectroscopy; Clark, R.J.H., Hester, R.E., Eds.; Heyden: New York, NY, USA, 1975; Volume I, pp. 35–97. [Google Scholar]
- Mayne, L.; Hudson, B. Selective enhancement of proline Raman signals with ultraviolet excitation. J. Phys. Chem. 1987, 91, 4438–4440. [Google Scholar] [CrossRef]
- Myshakina, N.S.; Ahmed, Z.; Asher, S.A. Dependence of amide vibrations on hydrogen bonding. J. Phys. Chem. B 2008, 112, 11873–11877. [Google Scholar] [CrossRef] [Green Version]
- Asher, S.A.; Mikhonin, A.V.; Bykov, S. UV Raman demonstrates that α-helical polyalanine peptides melt to polyproline II conformations. J. Am. Chem. Soc. 2004, 126, 8433–8440. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Wang, J. Direct anionic effect on water structure and indirect anionic effect on peptide back-bone hydration state revealed by thin-layer infrared spectroscopy. J. Phys. Chem. B 2018, 122, 68–76. [Google Scholar] [CrossRef]
- Kim, H.; Lee, H.; Lee, G.; Kim, H.; Cho, M. Hofmeister anionic effects on hydration electric fields around water and peptide. J. Chem. Phys. 2012, 136, 03B612. [Google Scholar] [CrossRef]
- Li, P.; Chen, X.G.; Shulin, E.; Asher, S.A. UV resonance Raman ground and excited state studies of amide and peptide isomerization dynamics. J. Am. Chem. Soc. 1997, 119, 1116–1120. [Google Scholar] [CrossRef]
- Holtz, J.S.; Li, P.; Asher, S.A. UV resonance Raman studies of cis-to-trans isomerization of glycyglycine derivatives. J. Am. Chem. Soc. 1999, 121, 3762–3766. [Google Scholar] [CrossRef]
- Imperiali, B.; Fisher, S.L.; Moats, R.A.; Prins, T.J. A Conformational Study of Peptides with the General Structure Ac-l-Xaa-Pro-d-Xaa-l-Xaa-NH2: Spectroscopic Evidence for a Peptide with Significant/3-Turn Character in Water and in Dimethyl Sulfoxide. J. Am. Chem. Soc. 1992, 114, 3182–3188. [Google Scholar] [CrossRef]
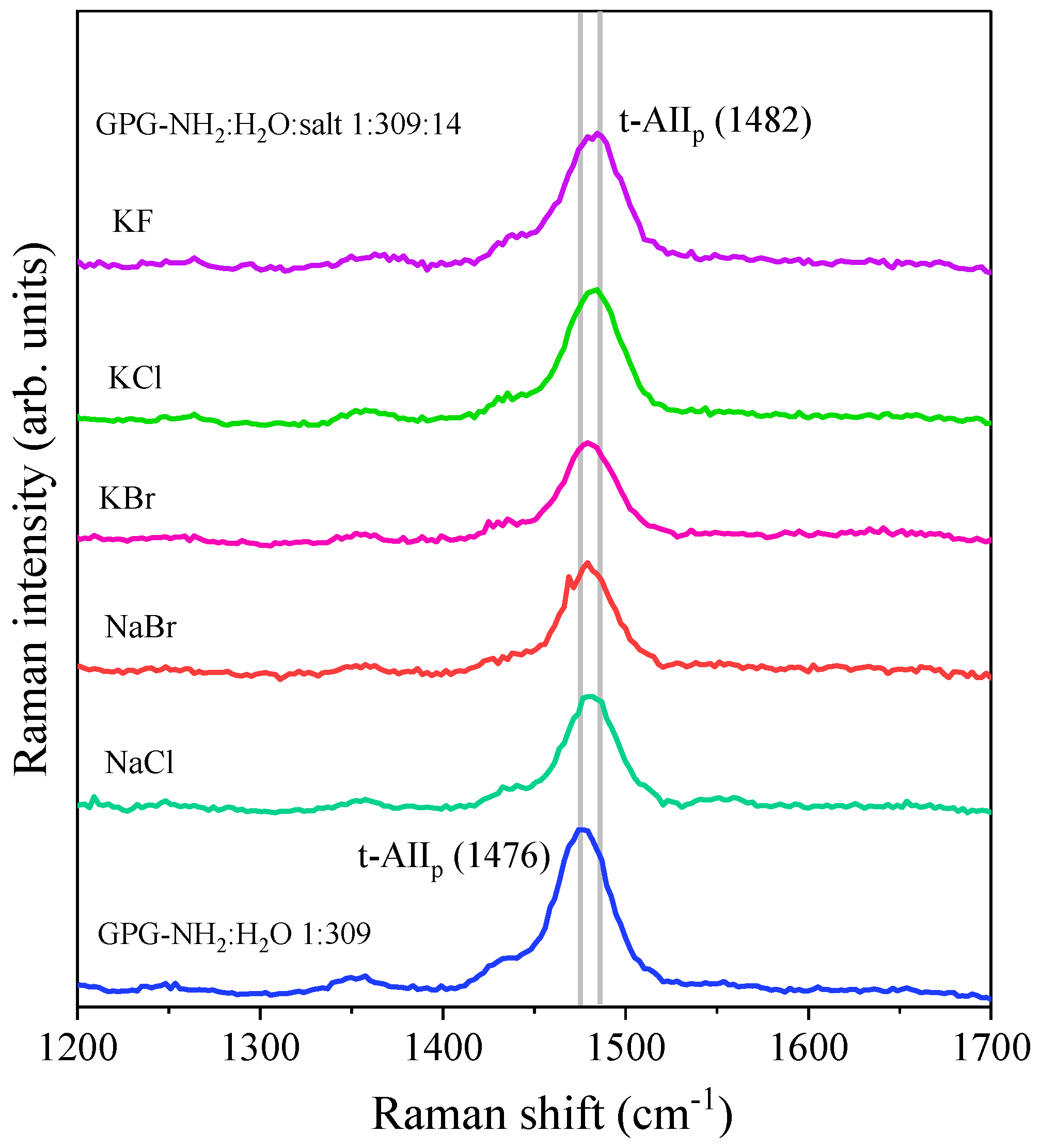
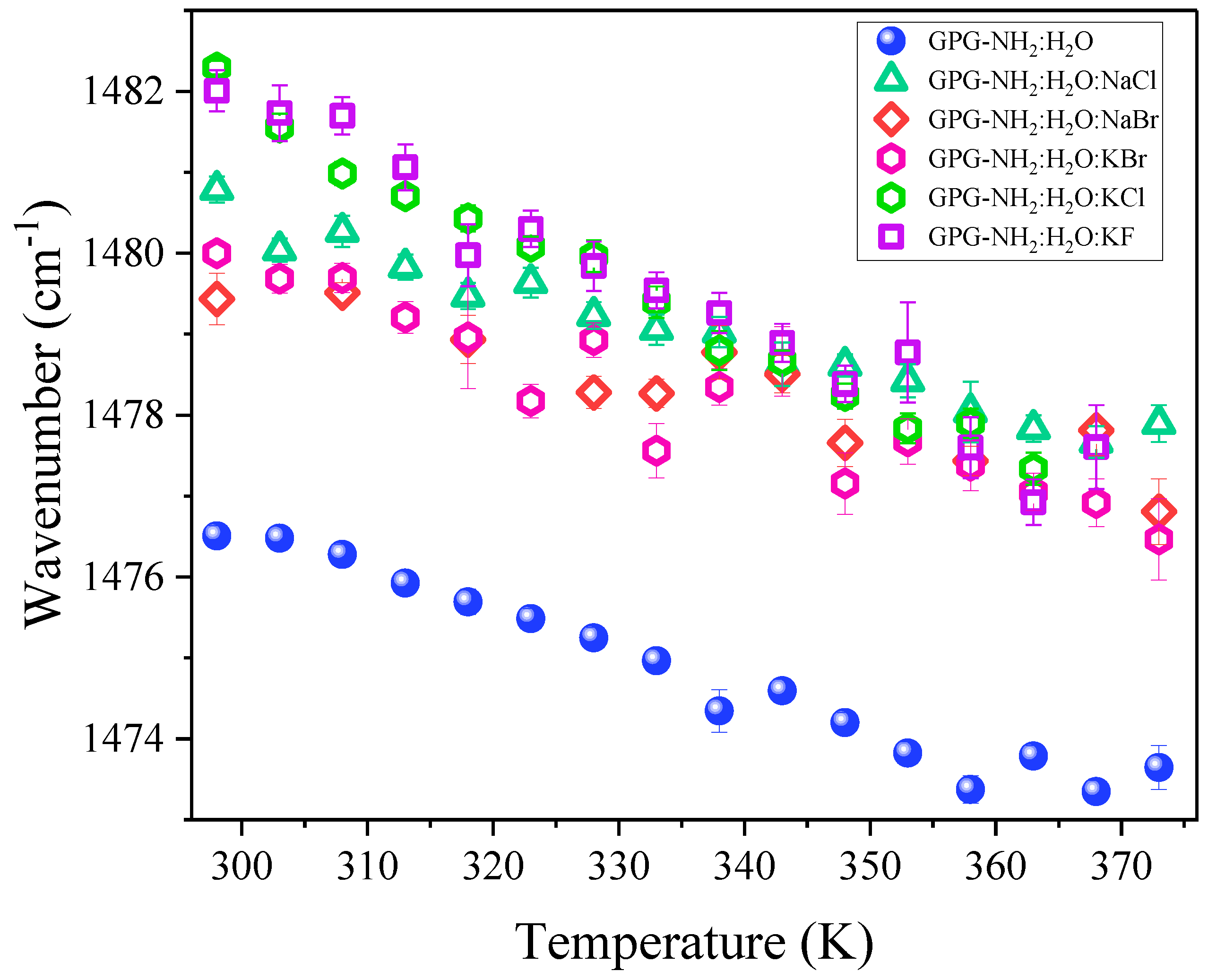
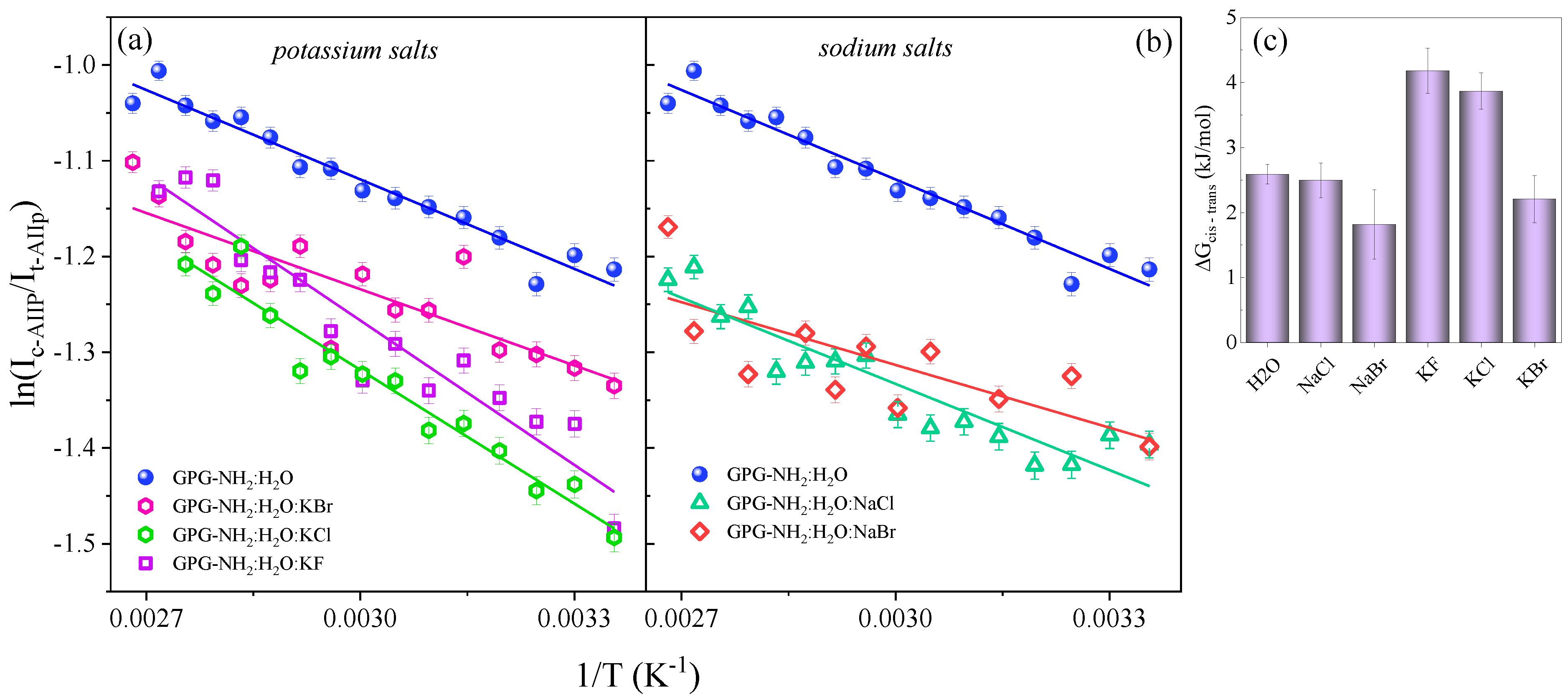
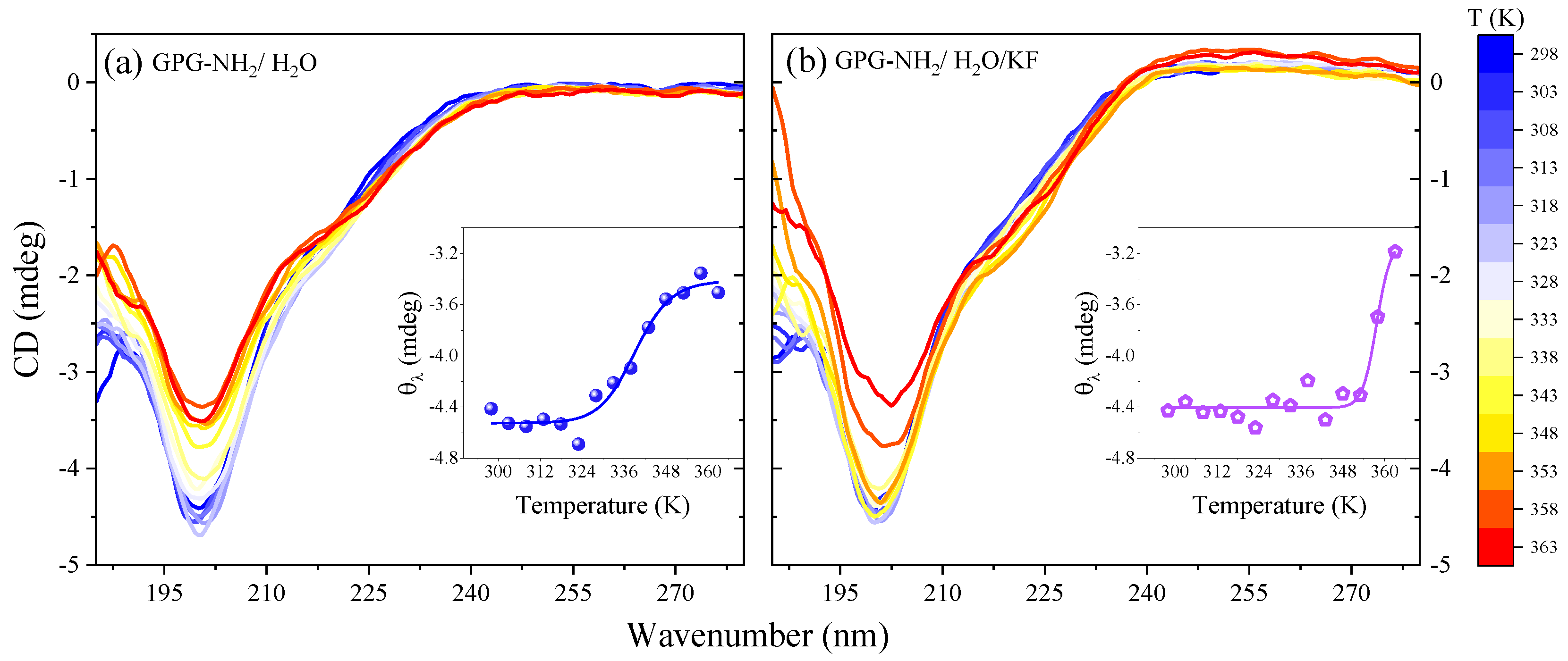
| Co-Solvent | Position of t-AIIp (cm−1) | Δν (cm−1) |
|---|---|---|
| NaCl | 1480.8 ± 0.2 | 4.8 |
| NaBr | 1479.4 ± 0.3 | 3.4 |
| KCl | 1482.3 ± 0.1 | 6.3 |
| KBr | 1480.0 ± 0.1 | 4.0 |
| KF | 1482.0 ± 0.3 | 6.2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Catalini, S.; Rossi, B.; Tortora, M.; Foggi, P.; Gessini, A.; Masciovecchio, C.; Bruni, F. Hydrogen Bonding and Solvation of a Proline-Based Peptide Model in Salt Solutions. Life 2021, 11, 824. https://doi.org/10.3390/life11080824
Catalini S, Rossi B, Tortora M, Foggi P, Gessini A, Masciovecchio C, Bruni F. Hydrogen Bonding and Solvation of a Proline-Based Peptide Model in Salt Solutions. Life. 2021; 11(8):824. https://doi.org/10.3390/life11080824
Chicago/Turabian StyleCatalini, Sara, Barbara Rossi, Mariagrazia Tortora, Paolo Foggi, Alessandro Gessini, Claudio Masciovecchio, and Fabio Bruni. 2021. "Hydrogen Bonding and Solvation of a Proline-Based Peptide Model in Salt Solutions" Life 11, no. 8: 824. https://doi.org/10.3390/life11080824
APA StyleCatalini, S., Rossi, B., Tortora, M., Foggi, P., Gessini, A., Masciovecchio, C., & Bruni, F. (2021). Hydrogen Bonding and Solvation of a Proline-Based Peptide Model in Salt Solutions. Life, 11(8), 824. https://doi.org/10.3390/life11080824