Identification of Putative Biosynthetic Gene Clusters for Tolyporphins in Multiple Filamentous Cyanobacteria
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification of Tolyporphins BGCs Tol Functions and BGCs from Other Cyanobacteria
2.2. Phylogenetic Analysis
2.3. Cyanobacterial Strains—Identification and Procurement
2.4. Examination of Extracts from Cyanobacteria for Tolyporphins
3. Results
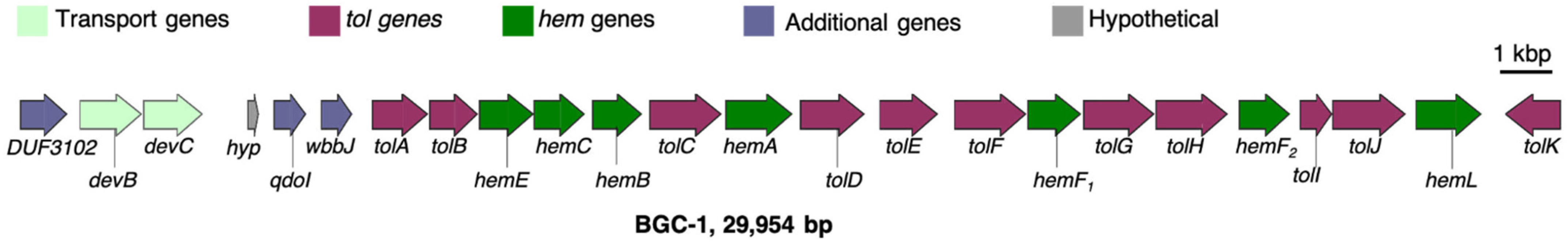
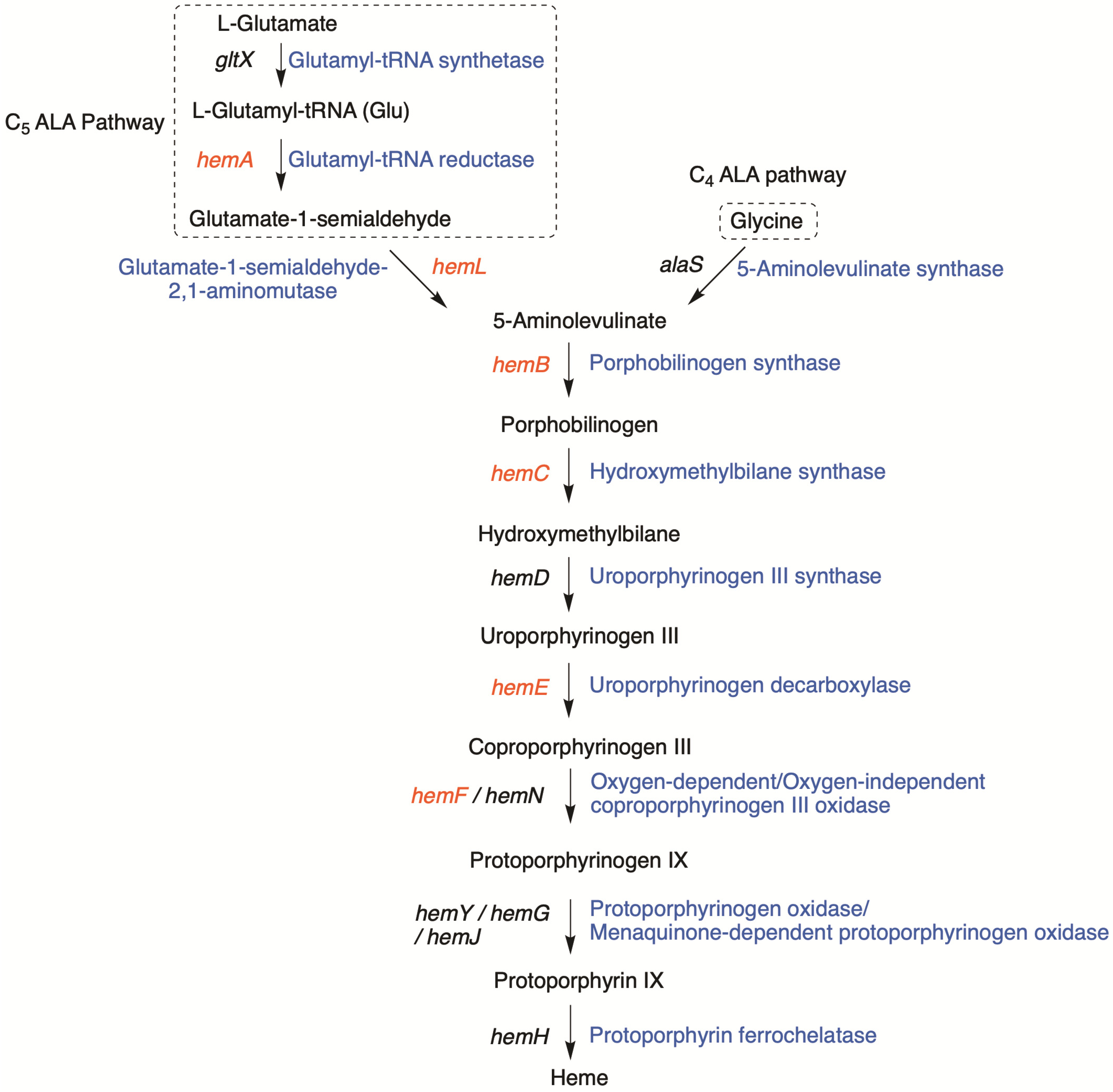
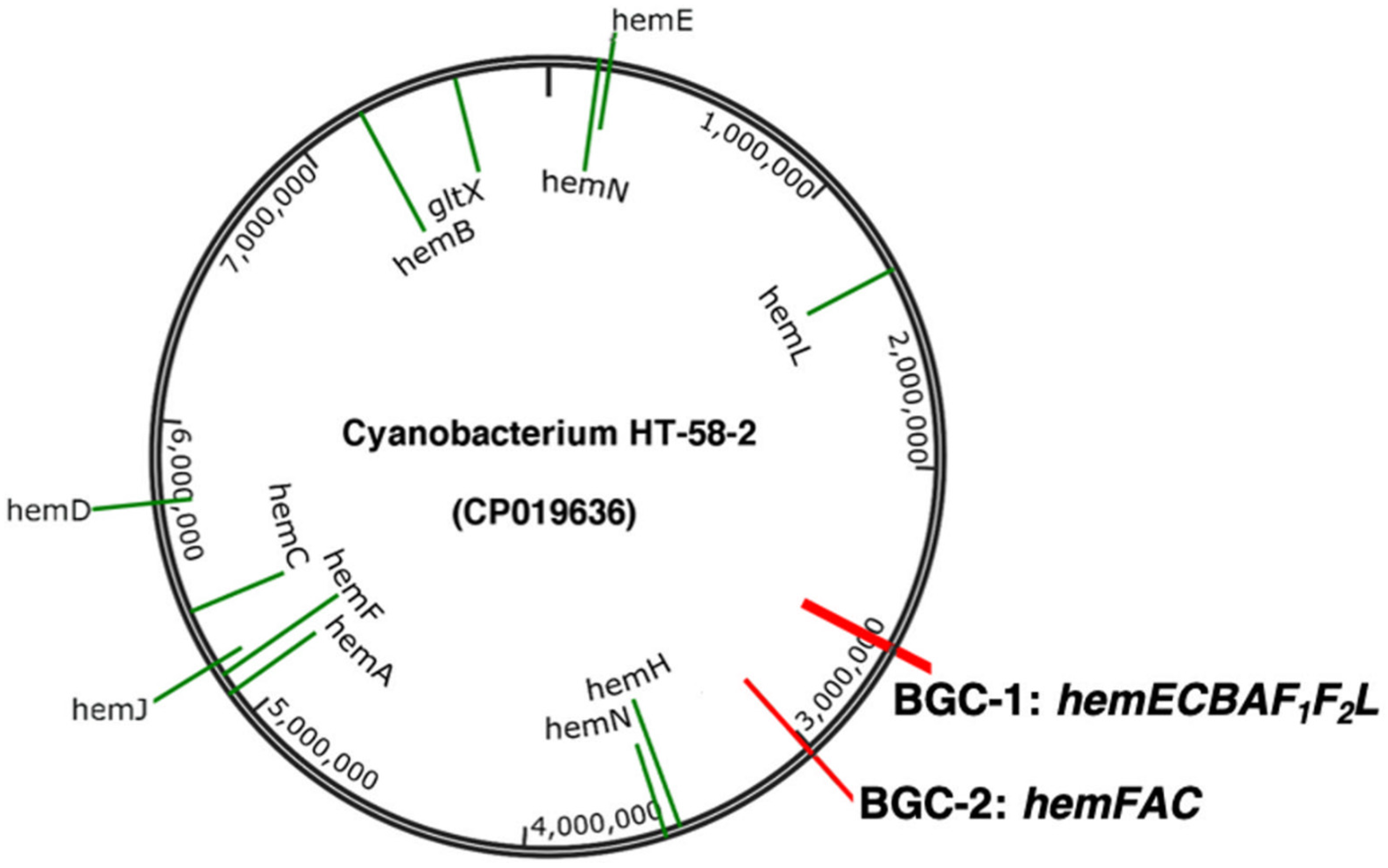
3.1. Significant Gene Features in BGC-1
3.2. Homology Searching for Additional Tolyporphins BGCs
- The genomic sequences of Nostoc sp.106C and Nostoc sp. RF31YmG were obtained from endophytic sub-communities from coralloid roots of Dioon merolae followed by metagenomic sequencing [5]. These Nostoc spp. cultures are nonaxenic, as is the case for the HT-58-2 culture. Indeed, both contain gene clusters of ~23 kbp that align with the tolyporphins BGC-1 in HT-58-2 (Figure 4). BLASTP alignments revealed that most Tol-like proteins from the two Nostoc strains align with relatively high identity to those from HT-58-2 BGC-1 (Table S1).
- Nostoc sp. FACHB 892 was obtained from soil crusts in the Tengger Desert, China, for extracellular polysaccharide studies [37]. The total length of the BGC is ~30 kbp with an extended 8.5 kbp to include the nearby tolH gene.
- Oculatella sp. LEGE 06141 is from the Blue Biotechnology and Ecotoxicology Culture Collection in Portugal, where many of the LEGE strains are non-axenic [39].
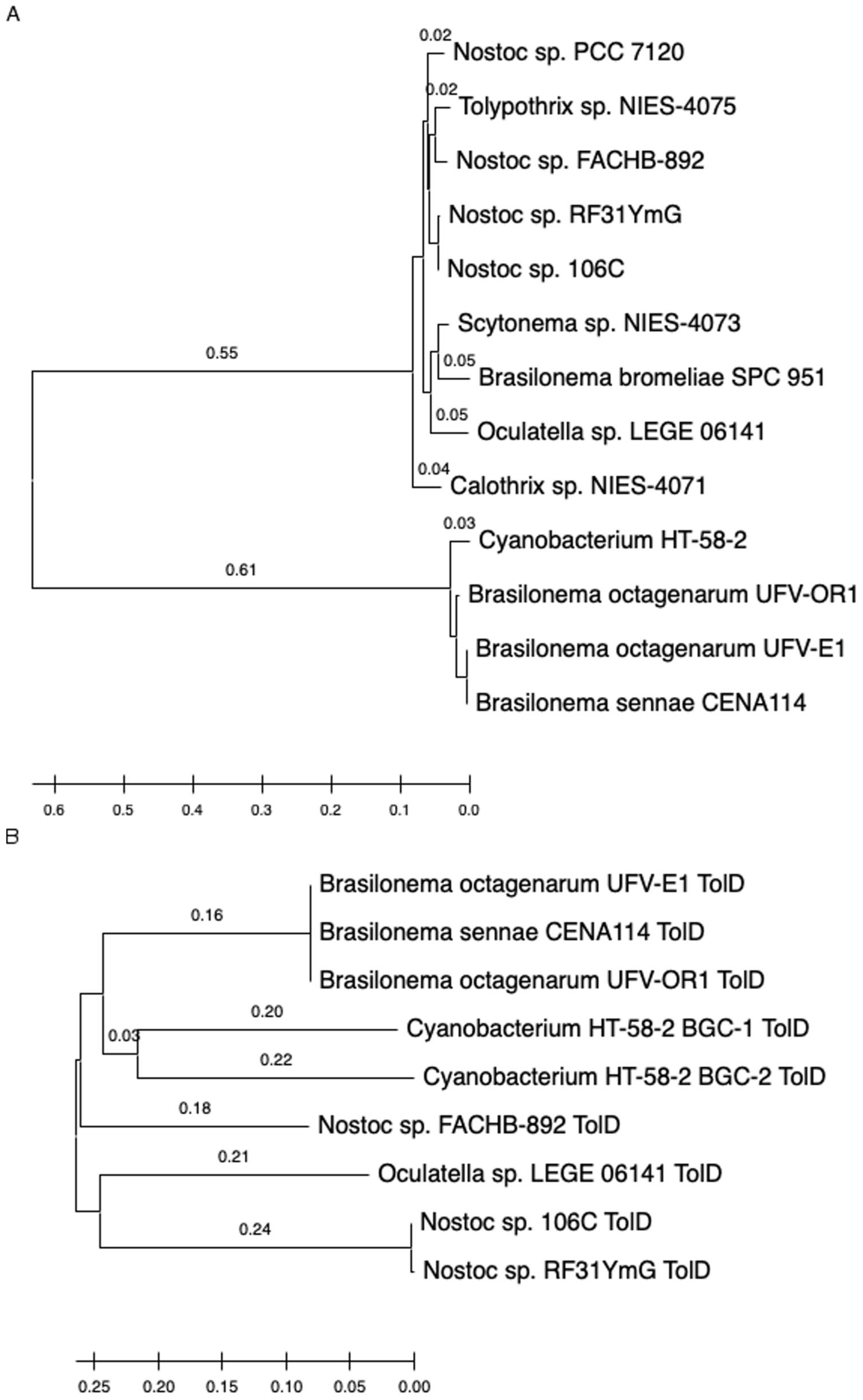
3.3. Phylogenetic Relationships among Cyanobacteria and Tolyporphins BGCs Genes
3.4. Examination of Samples from Cyanobacteria with Putative Tolyporphins BGC
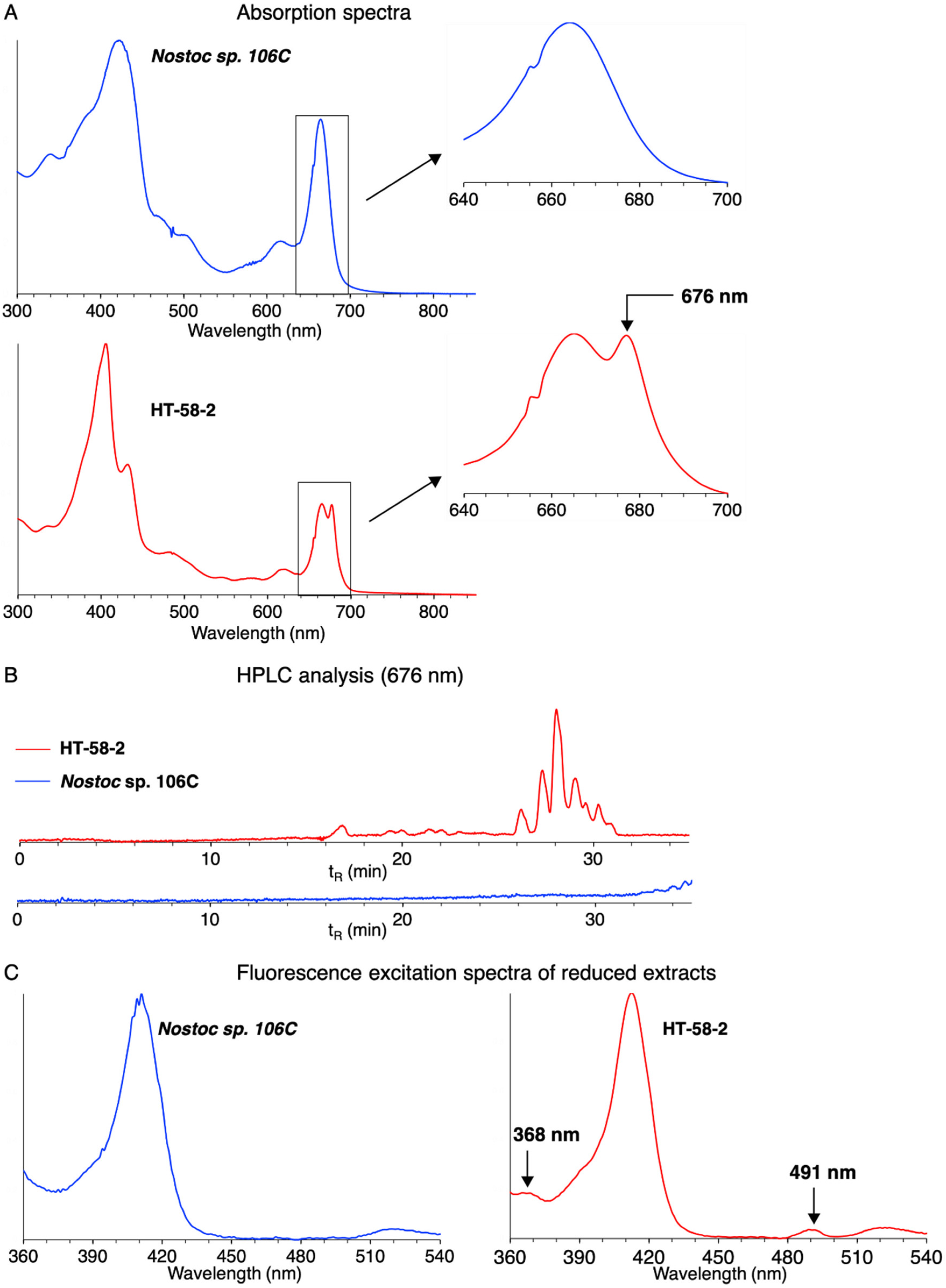
- The absorption spectral analysis relies on observation of the long-wavelength absorption band (~676 nm) of the dioxobacteriochlorin-type tolyporphins, which constitute the dominant members of the tolyporphins family [15,40]. The spectrum (panel A) of the extract from HT-58-2 indeed shows a peak at 676 nm; such a peak is absent for the extract from Nostoc sp. 106C. The peak at 676 nm is a shoulder on the long-wavelength absorption band of chlorophyll a (665 nm) [41].
- The HPLC chromatogram with absorption detection at 676 nm (panel B) of the extract from HT-58-2 shows multiple bands with retention time tR ~26–32 min. Such bands are characteristic of the mixture of dioxobacteriochlorin-type tolyporphins. Chlorophyll a, which absorbs in the same wavelength region, elutes at longer time [15]. No such bands were observed for the chromatogram of the extract from Nostoc sp. 106C.
- The fluorescence assay relies on chemical reduction of the keto auxochromes of chlorophyll a and the appropriately substituted tolyporphins followed by fluorescence excitation spectroscopy [25]. The keto groups are present on the dioxobacteriochlorins and oxochlorins, but not the lone porphyrin member, of the tolyporphins family. The fluorescence assay (panel C) for the extract from HT-58-2 showed excitation peaks at 368 and 491 nm (λem 710 nm) characteristic of dioxobacteriochlorin-type tolyporphins [25], but no such peaks were observed for the extract from Nostoc sp. 106C.
4. Summary and Outlook
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tidgewell, K.; Clark, B.R.; Gerwick, W.H. The Natural Products Chemistry of Cyanobacteria. In Comprehensive Natural Products II Chemistry and Biology; Mander, L., Lui, H.-W., Eds.; Elsevier: Oxford, UK, 2010; Volume 2, pp. 141–188. ISBN 978-008-045-382-8. [Google Scholar]
- Singh, R.K.; Tiwari, S.P.; Rai, A.K.; Mohapatra, T.M. Cyanobacteria: An Emerging Source for Drug Discovery. J. Antibiot. 2011, 64, 401–412. [Google Scholar] [CrossRef]
- Leão, P.N.; Engene, N.; Antunes, A.; Gerwick, W.H.; Vasconcelos, V. The Chemical Ecology of Cyanobacteria. Nat. Prod. Rep. 2012, 29, 372–391. [Google Scholar] [CrossRef] [PubMed]
- Rague, A.L.; Parker, S.A.J.; Tidgewell, K.J. Evaluating Marine Cyanobacteria as a Source for CNS Receptor Ligands. Molecules 2018, 23, 2665. [Google Scholar] [CrossRef]
- Gutiérrez-García, K.; Bustos-Díaz, E.D.; Corona-Gómez, J.A.; Ramos-Aboites, H.E.; Sélem-Mojica, N.; Cruz-Morales, P.; Pérez-Farrera, M.A.; Barona-Gómez, F.; Cibrián-Jaramillo, A. Cycad Coralloid Roots Contain Bacterial Communities Including Cyanobacteria and Caulobacter spp. that Encode Niche-Specific Biosynthetic Gene Clusters. Genome Biol. Evol. 2019, 11, 319–334. [Google Scholar] [CrossRef]
- Genuário, D.B.; Vaz, M.G.M.V.; Santos, S.N.; Kavamura, V.N.; Melo, I.S. Cyanobacteria from Brazilian Extreme Environments: Toward Functional Exploitation. In Microbial Diversity in the Genomic Era; Elsevier: Amsterdam, The Netherlands, 2019; pp. 265–284. ISBN 978-0-12-814849-5. [Google Scholar]
- Gaysina, L.A.; Saraf, A.; Singh, P. Cyanobacteria in Diverse Habitats. In Cyanobacteria—From Basic Science to Applications; Mishra, A.K., Tiwari, D.N., Rai, A.N., Eds.; Academic Press: Cambridge, MA, USA, 2019; pp. 1–28. ISBN 978-012-814-667-5. [Google Scholar]
- Prinsep, M.R.; Caplan, F.R.; Moore, R.E.; Patterson, G.M.L.; Smith, C.D. Tolyporphin, a Novel Multidrug Resistance Reversing Agent from the Blue-Green Alga Tolypothrix nodosa. J. Am. Chem. Soc. 1992, 114, 385–387. [Google Scholar] [CrossRef]
- Smith, C.D.; Prinsep, M.R.; Caplan, F.R.; Moore, R.E.; Patterson, G.M.L. Reversal of Multiple Drug Resistance by Tolyporphin, a Novel Cyanobacterial Natural Product. Oncol. Res. 1994, 6, 211–218. [Google Scholar] [PubMed]
- Prinsep, M.R.; Patterson, G.M.L.; Larsen, L.K.; Smith, C.D. Further Tolyporphins from the Blue-Green Alga Tolypothrix nodosa. Tetrahedron 1995, 51, 10523–10530. [Google Scholar] [CrossRef]
- Prinsep, M.R.; Patterson, G.M.L.; Larsen, L.K.; Smith, C.D. Tolyporphins J and K, Two Further Porphinoid Metabolites from the Cyanobacterium Tolypothrix nodosa. J. Nat. Prod. 1998, 61, 1133–1136. [Google Scholar] [CrossRef]
- Gurr, J.R.; Dai, J.; Philbin, C.S.; Sartain, H.T.; O’Donnell, T.J.; Yoshida, W.Y.; Rheingold, A.L.; Williams, P.G. Tolyporphins L–R: Unusual Tetrapyrroles from a Brasilonema sp. of Cyanobacterium. J. Org. Chem. 2020, 85, 318–326. [Google Scholar] [CrossRef]
- Hughes, R.-A.; Zhang, Y.; Zhang, R.; Williams, P.G.; Lindsey, J.S.; Miller, E.S. Genome Sequence and Composition of a Tolyporphin-Producing Cyanobacterium-Microbial Community. Appl. Environ. Microbiol. 2017, 83, e01068-17. [Google Scholar] [CrossRef] [PubMed]
- Hughes, R.-A.; Jin, X.; Zhang, Y.; Zhang, R.; Tran, S.; Williams, P.G.; Lindsey, J.S.; Miller, E.S. Genome Sequence, Metabolic Properties and Cyanobacterial Attachment of Porphyrobacter sp. HT-58-2 Isolated from a Filamentous Cyanobacterium-Microbial Consortium. Microbiology 2018, 164, 1229–1239. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, R.; Hughes, R.-A.; Dai, J.; Gurr, J.R.; Williams, P.G.; Miller, E.S.; Lindsey, J.S. Quantitation of Tolyporphins, Diverse Tetrapyrrole Secondary Metabolites with Chlorophyll-Like Absorption, from a Filamentous Cyanobacterium–Microbial Community. Phytochem. Anal. 2018, 29, 205–216. [Google Scholar] [CrossRef]
- Barnhart-Dailey, M.; Zhang, Y.; Zhang, R.; Anthony, S.M.; Aaron, J.S.; Miller, E.S.; Lindsey, J.S.; Timlin, J.A. Cellular Localization of Tolyporphins, Unusual Tetrapyrroles, in a Microbial Photosynthetic Community Determined using Hyperspectral Confocal Fluorescence Microscopy. Photosynth. Res. 2019, 141, 259–271. [Google Scholar] [CrossRef]
- Jin, X.; Miller, E.S.; Lindsey, J.S. Natural Product Gene Clusters in the Filamentous Nostocales Cyanobacterium HT-58-2. Life 2021, 11, 356. [Google Scholar] [CrossRef]
- States, D.J.; Gish, W. Combined Use of Sequence Similarity and Codon Bias for Coding Region Identification. J. Comput. Biol. 1994, 1, 39–50. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The Neighbor-Joining Method: A New Method for Reconstructing Phylogenetic Trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Stecher, G.; Tamura, K.; Kumar, S. Molecular Evolutionary Genetics Analysis (MEGA) for macOS. Mol. Biol. Evol. 2020, 37, 1237–1239. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence Limits on Phylogenies: An Approach Using the Bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Nei, M.; Kumar, S. Prospects for Inferring Very Large Phylogenies by Using the Neighbor-Joining Method. Proc. Natl. Acad. Sci. USA 2004, 101, 11030–11035. [Google Scholar] [CrossRef]
- Zuckerkandl, E.; Pauling, L. Evolutionary Divergence and Convergence in Proteins. In Evolving Genes and Proteins, 1st ed.; Bryson, V., Vogel, H.J., Eds.; Academic Press: New York, NY, USA, 1965; pp. 97–165. ISBN 9781483266305. [Google Scholar]
- Nguyen, K.-U.; Zhang, R.; Taniguchi, M.; Lindsey, J.S. Fluorescence Assay for Tolyporphins amidst Abundant Chlorophyll in Crude Cyanobacterial Extracts. Photochem. Photobiol. 2021, 97. [Google Scholar] [CrossRef]
- Dailey, H.A.; Dailey, T.A.; Gerdes, S.; Jahn, D.; Jahn, M.; O’Brian, M.R.; Warren, M.J. Prokaryotic Heme Biosynthesis: Multiple Pathways to a Common Essential Product. Microbiol. Mol. Biol. Rev. 2017, 81, e00048-16. [Google Scholar] [CrossRef] [PubMed]
- Bryant, D.A.; Hunter, C.N.; Warren, M.J. Biosynthesis of the Modified Tetrapyrroles—The Pigments of Life. J. Biol. Chem. 2020, 295, 6888–6925. [Google Scholar] [CrossRef] [PubMed]
- Hansson, M.; Rutberg, L.; Schröder, I.; Hederstedt, L. The Bacillus subtilis hemAXCDBL Gene Cluster, which Encodes Enzymes of the Biosynthetic Pathway from Glutamate to Uroporphyrinogen III. J. Bacteriol. 1991, 173, 2590–2599. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Morita, H.; Toh, H.; Fukuda, S.; Horikawa, H.; Oshima, K.; Suzuki, T.; Murakami, M.; Hisamatsu, S.; Kato, Y.; Takizawa, T.; et al. Comparative Genome Analysis of Lactobacillus reuteri and Lactobacillus fermentum Reveal a Genomic Island for Reuterin and Cobalamin Production. DNA Res. 2008, 15, 151–161. [Google Scholar] [CrossRef]
- Kafala, B.; Sasarman, A. Isolation of the Staphylococcus aureus hemCDBL Gene Cluster Coding for Early Steps in Heme Biosynthesis. Gene 1997, 199, 231–239. [Google Scholar] [CrossRef]
- Echelard, Y.; Dymetryszyn, J.; Drolet, M.; Sasarman, A. Nucleotide Sequence of the hemB Gene of Escherichia coli K12. Mol. Gen. Genet. 1988, 214, 503–508. [Google Scholar] [CrossRef] [PubMed]
- Elliott, T. Cloning, Genetic Characterization, and Nucleotide Sequence of the hemA-prfA Operon of Salmonella typhimurium. J. Bacteriol. 1989, 171, 3948–3960. [Google Scholar] [CrossRef]
- Weber, T.; Blin, K.; Duddela, S.; Krug, D.; Kim, H.U.; Bruccoleri, R.; Lee, S.Y.; Fischbach, M.A.; Müller, R.; Wohlleben, W.; et al. AntiSMASH 3.0–A Comprehensive Resource for the Genome Mining of Biosynthetic Gene Clusters. Nucleic Acids Res. 2015, 43, W237–W243. [Google Scholar] [CrossRef] [PubMed]
- Medema, M.H.; Takano, E.; Breitling, R. Detecting Sequence Homology at the Gene Cluster Level with MultiGeneBlast. Mol. Biol. Evol. 2013, 30, 1218–1223. [Google Scholar] [CrossRef]
- Gilchrist, C.L.M.; Booth, T.J.; Chooi, Y.-H. Cblaster: A Remote Search Tool for Rapid Identification and Visualisation of Homologous Gene Clusters. bioRxiv 2020. [Google Scholar] [CrossRef]
- Díaz, E.; Ferrández, A.; García, J.L. Characterization of the hca Cluster Encoding the Dioxygenolytic Pathway for Initial Catabolism of 3-Phenylpropionic acid in Escherichia coli K-12. J. Bacteriol. 1998, 180, 2915–2923. [Google Scholar] [CrossRef]
- Hu, C.; Zhang, D.; Huang, Z.; Liu, Y. The Vertical Micro-Distribution of Cyanobacteria and Green Algae within Desert Crusts and the Development of the Algal Crusts. Plant Soil 2003, 257, 97–111. [Google Scholar] [CrossRef]
- Fiore, M.F.; Sant’Anna, C.L.; Azevedo, M.T.P.; Komárek, J.; Kaštovský, J.; Sulek, J.; Lorenzi, A.S. The Cyanobacterial Genus Brasilonema, gen. nov., a Molecular and Phenotypic Evaluation. J. Phycol. 2007, 43, 789–798. [Google Scholar] [CrossRef]
- Eusebio, N.; Rego, A.; Glasser, N.R.; Castelo-Branco, R.; Balskus, E.P.; Leão, P.N. Distribution and Diversity of Dimetal-Carboxylate Halogenases in Cyanobacteria. bioRxiv 2021. [Google Scholar] [CrossRef]
- O’Donnell, T.J.; Gurr, J.R.; Dai, J.; Taniguchi, M.; Williams, P.G.; Lindsey, J.S. Tolyporphins A–R, Unusual Tetrapyrrole Macrocycles in a Cyanobacterium from Micronesia, Assessed Quantitatively from the Culture HT-58-2. New J. Chem. 2021, 45, 11481–11494. [Google Scholar] [CrossRef]
- Taniguchi, M.; Lindsey, J.S. Absorption and Fluorescence Spectral Database of Chlorophylls and Analogues. Photochem. Photobiol. 2021, 97, 136–165. [Google Scholar] [CrossRef]
- Lindsey, J.S. Considerations of the Biosynthesis and Molecular Diversity of Tolyporphins. New J. Chem. 2021, 45, 12097–12107. [Google Scholar] [CrossRef]
- Sokolovskaya, O.M.; Shelton, A.N.; Taga, M.E. Sharing Vitamins: Cobamides Unveil Microbial Interactions. Science 2020, 369, eaba0165. [Google Scholar] [CrossRef] [PubMed]

| Scheme | Location | Sample Origin | BGC Composition |
|---|---|---|---|
| HT-58-2 BGC-1 a | Pohnpei, Micronesia | Soil | 7 hem genes, 11 tol genes |
| HT-58-2 BGC-2 | Pohnpei, Micronesia | Soil | 3 hem genes, 7 tol genes |
| Nostoc sp.106C | Chiapas, Mexico | Coralloid roots | 6 hem genes, 7 tol genes |
| Nostoc sp. RF31YmG | Chiapas, Mexico | Coralloid roots | 6 hem genes, 7 tol genes |
| Nostoc sp. FACHB-892 | Tengger Desert, China | Algal crusts | 6 hem genes, 6 tol genes |
| Brasilonema octagenarum UFV-OR1 | Minas Gerais, Brazil | Orchid leaves | 8 hem genes, 6 tol genes |
| Brasilonema octagenarum UFV-E1 | Minas Gerais, Brazil | Eucalyptus grandis leaves | 8 hem genes, 6 tol genes (assembly gap) |
| Brasilonema sennae CENA114 | São Paulo, Brazil | Iron water pipe | 8 hem genes, 6 tol genes |
| Oculatella sp. LEGE 06141 | Lagos, Portugal | Green macroalgae | 6 hem genes, 9 tol genes |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jin, X.; Zhang, Y.; Zhang, R.; Nguyen, K.-U.; Lindsey, J.S.; Miller, E.S. Identification of Putative Biosynthetic Gene Clusters for Tolyporphins in Multiple Filamentous Cyanobacteria. Life 2021, 11, 758. https://doi.org/10.3390/life11080758
Jin X, Zhang Y, Zhang R, Nguyen K-U, Lindsey JS, Miller ES. Identification of Putative Biosynthetic Gene Clusters for Tolyporphins in Multiple Filamentous Cyanobacteria. Life. 2021; 11(8):758. https://doi.org/10.3390/life11080758
Chicago/Turabian StyleJin, Xiaohe, Yunlong Zhang, Ran Zhang, Kathy-Uyen Nguyen, Jonathan S. Lindsey, and Eric S. Miller. 2021. "Identification of Putative Biosynthetic Gene Clusters for Tolyporphins in Multiple Filamentous Cyanobacteria" Life 11, no. 8: 758. https://doi.org/10.3390/life11080758
APA StyleJin, X., Zhang, Y., Zhang, R., Nguyen, K.-U., Lindsey, J. S., & Miller, E. S. (2021). Identification of Putative Biosynthetic Gene Clusters for Tolyporphins in Multiple Filamentous Cyanobacteria. Life, 11(8), 758. https://doi.org/10.3390/life11080758







