Comparative Physiological and Transcriptomic Profiling Offers Insight into the Sexual Dimorphism of Hepatic Metabolism in Size-Dimorphic Spotted Scat (Scatophagus argus)
Abstract
1. Introduction
2. Materials and Methods
2.1. Fish and Sample Preparation
2.2. Histological Procedures for Liver and Gonad Tissues
2.3. Measurement of Enzyme Activities and Serum Steroid Hormone Assay
2.4. RNA-Seq and Bioinformatics Analysis
2.5. Real-Time Quantitative PCR (RT-qPCR) Validation
2.6. Statistical Analysis
3. Results
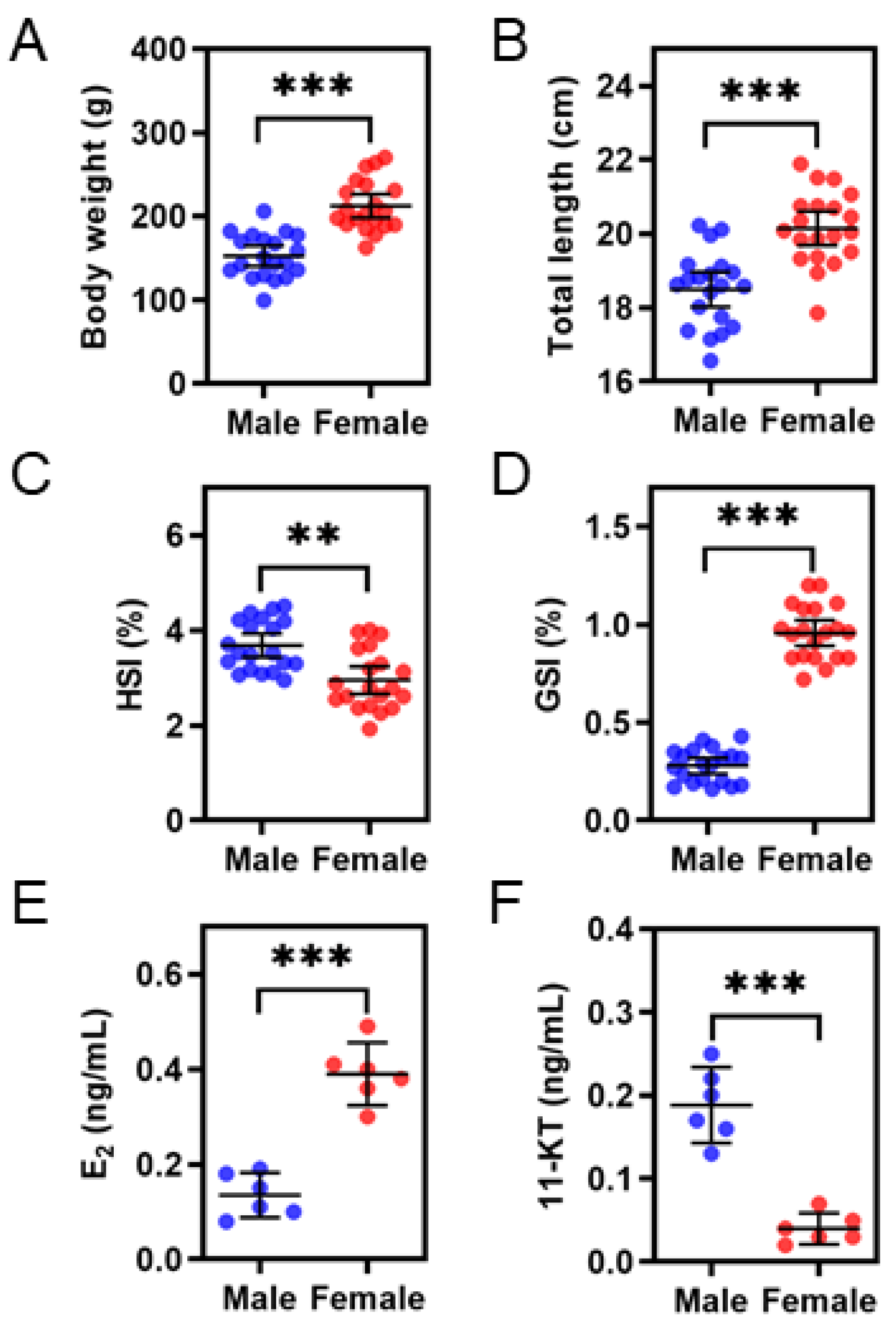
3.1. Comparisons of Growth, HSI, GSI, and Steroid Hormones Levels between the Sexes
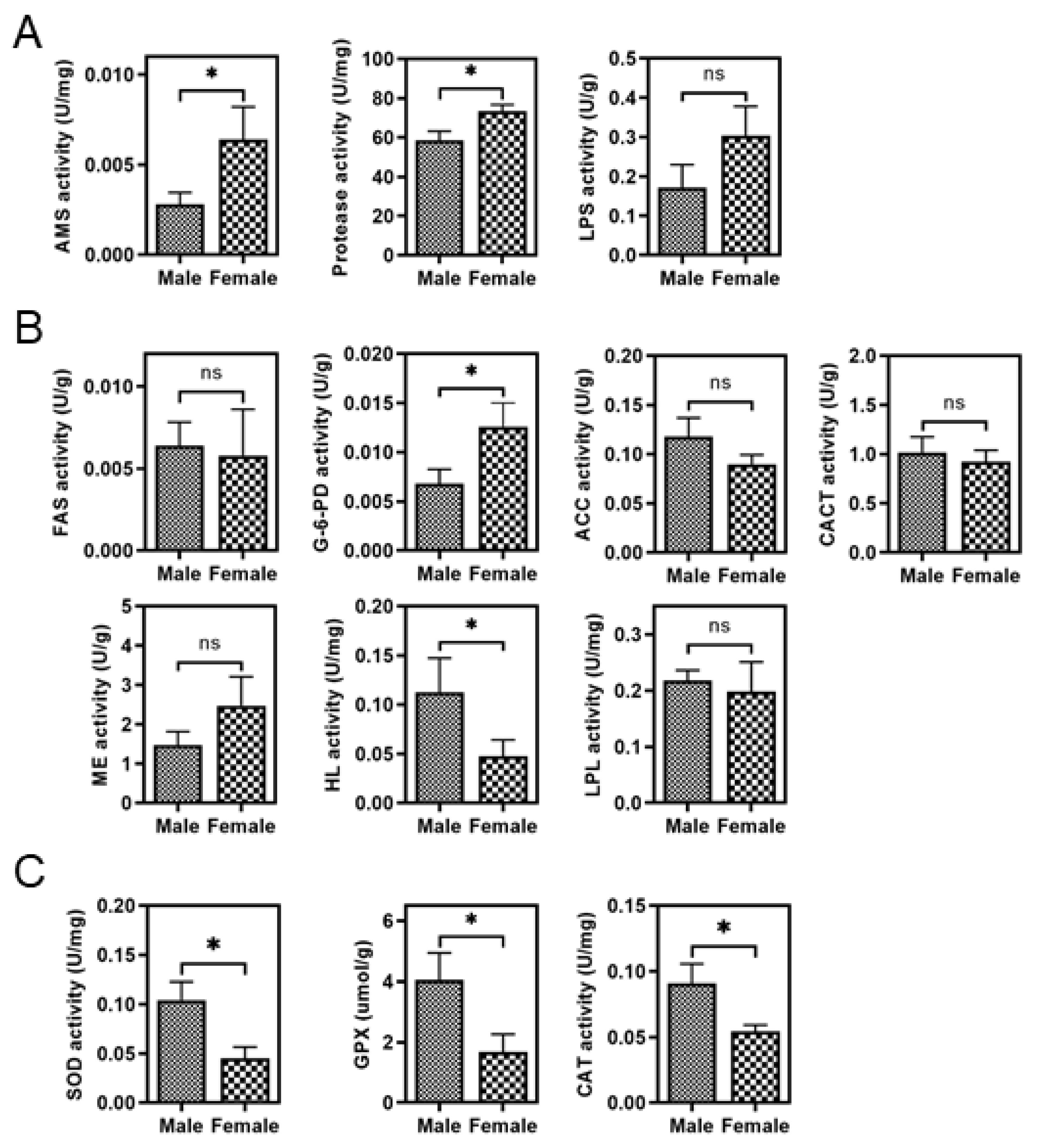
3.2. Sex Differences in the Activity of Liver Digestive and Metabolic Enzymes
3.3. RNA-Seq of the Liver Transcriptome
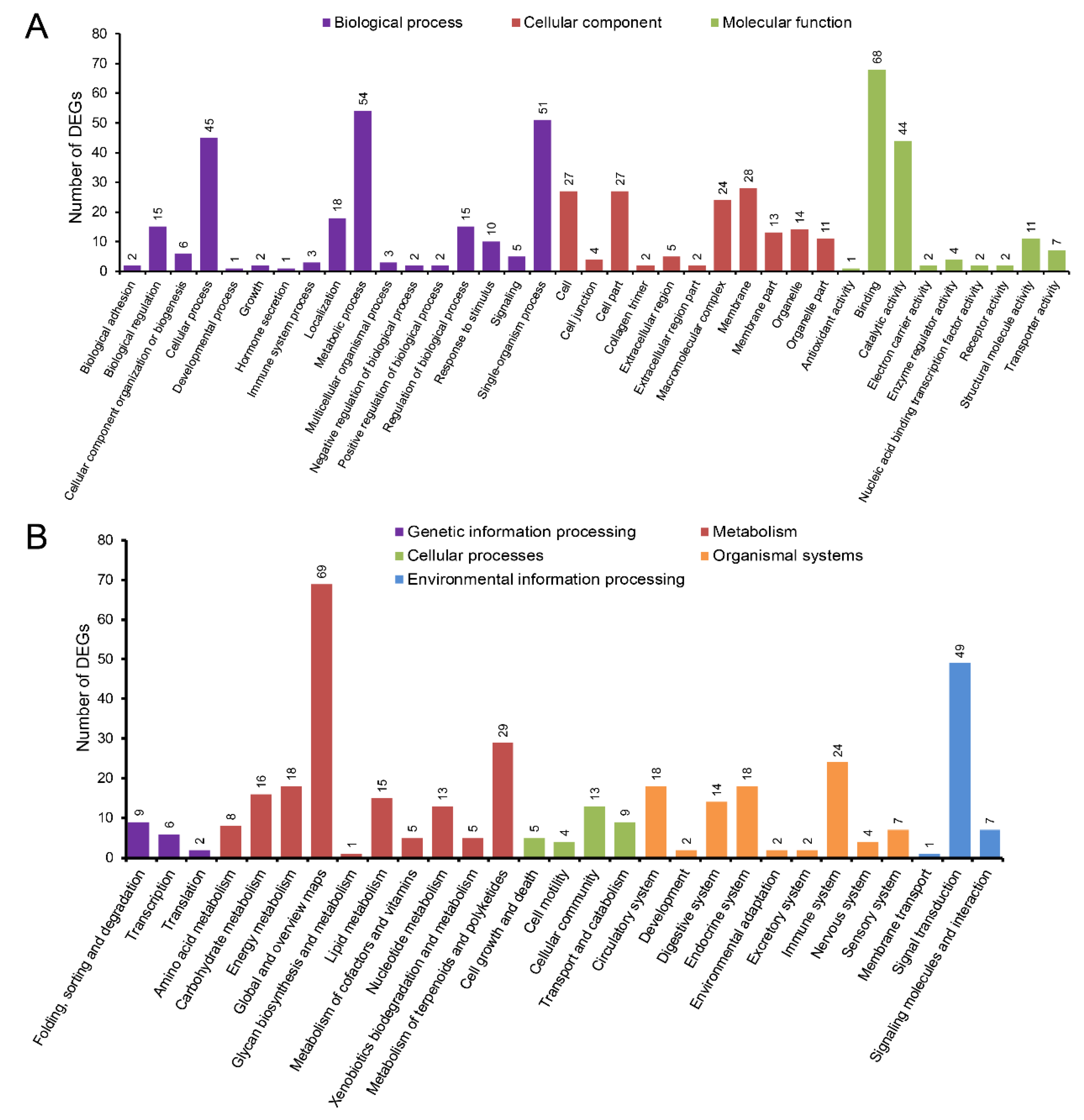
3.4. Differential Expression Analysis
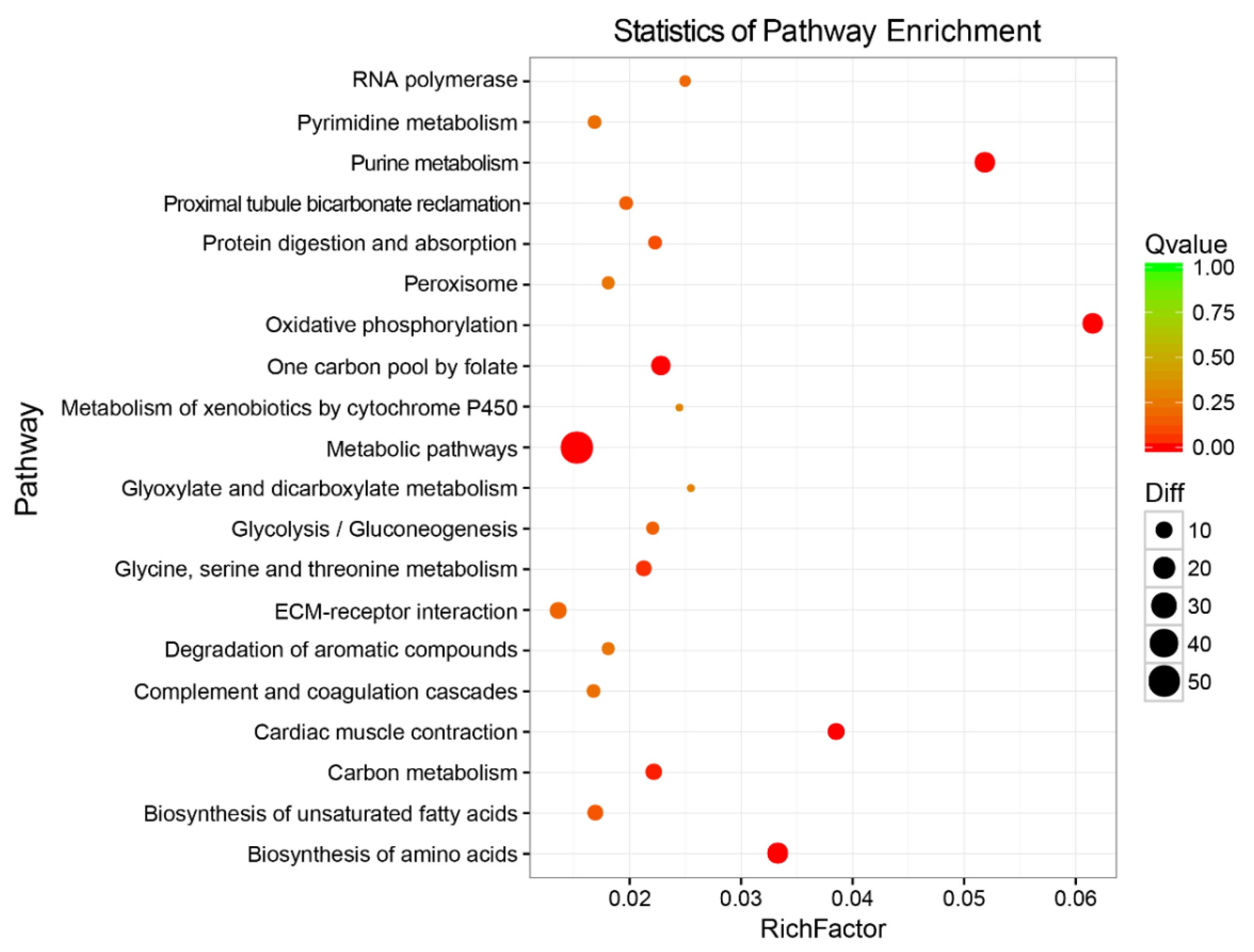
3.5. The Enriched GO Terms and KEGG Pathways
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Saillant, E.; Fostier, A.; Menu, B.; Haffray, P.; Chatain, B. Sexual growth dimorphism in sea bass Dicentrarchus labrax. Aquaculture 2001, 202, 371–387. [Google Scholar] [CrossRef]
- Wang, N.; Wang, R.; Wang, R.; Chen, S. Transcriptomics analysis revealing candidate networks and genes for the body size sexual dimorphism of Chinese tongue sole (Cynoglossus semilaevis). Funct. Integr. Genom. 2018, 18, 327–339. [Google Scholar] [CrossRef]
- Mei, J.; Gui, J.-F. Genetic basis and biotechnological manipulation of sexual dimorphism and sex determination in fish. Sci. China Life Sci. 2015, 58, 124–136. [Google Scholar] [CrossRef]
- Piferrer, F. Endocrine sex control strategies for the feminization of teleost fish. Aquaculture 2001, 197, 229–281. [Google Scholar] [CrossRef]
- Bower, N.; Li, X.; Taylor, R.; Johnston, I.A. Switching to fast growth: The insulin-like growth factor (IGF) system in skeletal muscle of Atlantic salmon. J. Exp. Biol. 2008, 211, 3859–3870. [Google Scholar] [CrossRef]
- Mandiki, S.; Houbart, M.; Babiak, I.; Vandeloise, E.; Gardeur, J.; Kestemont, P. Are sex steroids involved in the sexual growth dimorphism in Eurasian perch juveniles? Physiol. Behav. 2004, 80, 603–609. [Google Scholar] [CrossRef]
- Ji, X.-S.; Chen, S.-L.; Jiang, Y.-L.; Xu, T.-J.; Yang, J.-F.; Tian, Y.-S. Growth differences and differential expression analysis of pituitary adenylate cyclase activating polypeptide (PACAP) and growth hormone-releasing hormone (GHRH) between the sexes in half-smooth tongue sole Cynoglossus semilaevis. Gen. Comp. Endocrinol. 2011, 170, 99–109. [Google Scholar] [CrossRef]
- Jiang, D.; Shi, H.; Liu, Q.; Wang, T.; Huang, Y.; Huang, Y.; Deng, S.; Chen, H.; Tian, C.; Zhu, C.; et al. Isolation of Growth Hormone-Releasing Hormone and Its Receptor Genes from Scatophagus argus and Their Expression Analyses. J. Ocean Univ. China 2019, 18, 1486–1496. [Google Scholar] [CrossRef]
- Degani, G.; Tzchori, I.; Yom, S.; Goldberg, D.; Jackson, K. Growth differences and growth hormone expression in male and female European eels [Anguilla anguilla (L.)]. Gen. Comp. Endocrinol. 2003, 134, 88–93. [Google Scholar] [CrossRef]
- Din, S.Y.; Hurvitz, A.; Goldberg, D.; Jackson, K.; Levavi-Sivan, B.; Degani, G. Cloning of Russian sturgeon (Acipenser gueldenstaedtii) growth hormone and insulin-like growth factor I and their expression in male and female fish during the first period of growth. J. Endocrinol. Investig. 2008, 31, 201–210. [Google Scholar] [CrossRef] [PubMed]
- Deng, S.-P.; Wu, B.; Zhu, C.-H.; Li, G.-L. Molecular cloning and dimorphic expression of growth hormone (gh) in female and male spotted scat Scatophagus argus. Fish. Sci. 2014, 80, 715–723. [Google Scholar] [CrossRef]
- Ma, W.; Wu, J.; Zhang, J.; He, Y.; Gui, J.; Mei, J. Sex differences in the expression of GH/IGF axis genes underlie sexual size dimorphism in the yellow catfish (Pelteobagrus fulvidraco). Sci. China Life Sci. 2016, 59, 431–433. [Google Scholar] [CrossRef] [PubMed]
- Norbeck, L.A.; Sheridan, M.A. An in vitro model for evaluating peripheral regulation of growth in fish: Effects of 17β-estradiol and testosterone on the expression of growth hormone receptors, insulin-like growth factors, and insulin-like growth factor type 1 receptors in rainbow trout (Oncorhynchus mykiss). Gen. Comp. Endocrinol. 2011, 173, 270–280. [Google Scholar] [CrossRef]
- Davis, L.K.; Pierce, A.; Hiramatsu, N.; Sullivan, C.V.; Hirano, T.; Grau, E.G. Gender-specific expression of multiple estrogen receptors, growth hormone receptors, insulin-like growth factors and vitellogenins, and effects of 17β-estradiol in the male tilapia (Oreochromis mossambicus). Gen. Comp. Endocrinol. 2008, 156, 544–551. [Google Scholar] [CrossRef] [PubMed]
- Cleveland, B.M.; Weber, G. Effects of sex steroids on expression of genes regulating growth-related mechanisms in rainbow trout (Oncorhynchus mykiss). Gen. Comp. Endocrinol. 2015, 216, 103–115. [Google Scholar] [CrossRef]
- Zhang, K.-W.; Wu, T.-L.; Chen, H.-P.; Jiang, D.-N.; Zhu, C.-H.; Deng, S.-P.; Zhang, Y.; Li, G.-L. Estradiol-17β regulates the expression of insulin-like growth factors 1 and 2 via estradiol receptors in spotted scat (Scatophagus argus). Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2019, 237, 110328. [Google Scholar] [CrossRef] [PubMed]
- Yue, M.; Zhao, J.; Tang, S.; Zhao, Y. Effects of Estradiol and Testosterone on the Expression of Growth-related Genes in Female and Male Nile Tilapia, Oreochromis niloticus. J. World Aquac. Soc. 2017, 49, 216–228. [Google Scholar] [CrossRef]
- Fu, B.; Wang, X.; Feng, X.; Yu, X.; Tong, J. Comparative transcriptomic analyses of two bighead carp (Hypophthalmichthys nobilis) groups with different growth rates. Comp. Biochem. Physiol. Part D Genom. Proteom. 2016, 20, 111–117. [Google Scholar] [CrossRef]
- Holloway, A.; Leatherland, J. Neuroendocrine Regulation of Growth Hormone Secretion in Teleost Fishes with Emphasis on the Involvement of Gonadal Sex Steroids. Rev. Fish Biol. Fish. 1998, 8, 409–429. [Google Scholar] [CrossRef]
- Dai, X.; Zhang, W.; Zhuo, Z.; He, J.; Yin, Z. Neuroendocrine regulation of somatic growth in fishes. Sci. China Life Sci. 2015, 58, 137–147. [Google Scholar] [CrossRef]
- Moriyama, S.; Ayson, F.G.; Kawauchi, H. Growth Regulation by Insulin-like Growth Factor-I in Fish. Biosci. Biotechnol. Biochem. 2000, 64, 1553–1562. [Google Scholar] [CrossRef]
- Very, N.; Sheridan, M. The Role of Somatostatins in the Regulation of Growth in Fish. Fish Physiol. Biochem. 2002, 27, 217–226. [Google Scholar] [CrossRef]
- Chatchaiphan, S.; Srisapoome, P.; Kim, J.-H.; Devlin, R.H.; Na-Nakorn, U. De Novo Transcriptome Characterization and Growth-Related Gene Expression Profiling of Diploid and Triploid Bighead Catfish (Clarias macrocephalus Günther, 1864). Mar. Biotechnol. 2017, 19, 36–48. [Google Scholar] [CrossRef]
- Fu, B.; Yu, X.; Tong, J.; Pang, M.; Zhou, Y.; Liu, Q.; Tao, W. Comparative transcriptomic analysis of hypothalamus-pituitary-liver axis in bighead carp (Hypophthalmichthys nobilis) with differential growth rate. BMC Genom. 2019, 20, 328. [Google Scholar] [CrossRef]
- Lin, G.; Thevasagayam, N.M.; Wan, Z.Y.; Ye, B.Q.; Yue, G.H. Transcriptome Analysis Identified Genes for Growth and Omega-3/-6 Ratio in Saline Tilapia. Front. Genet. 2019, 10, 244. [Google Scholar] [CrossRef]
- Zhang, J.; Shen, Y.; Xu, X.; Dai, Y.; Li, J. Transcriptome Analysis of the Liver and Muscle Tissues of Black Carp (Mylopharyngodon piceus) of Different Growth Rates. Mar. Biotechnol. 2020, 22, 1–11. [Google Scholar] [CrossRef]
- Escalante-Rojas, M.; Peña, E.; Hernández, C.; Llera-Herrera, R.; Garcia-Gasca, A. De novo transcriptome assembly for the rose spotted snapper Lutjanus guttatus and expression analysis of growth/atrophy-related genes. Aquac. Res. 2018, 49, 1709–1722. [Google Scholar] [CrossRef]
- Odei, D.K.; Hagen, Ø.; Peruzzi, S.; Falk-Petersen, I.-B.; Fernandes, J.M.O. Transcriptome sequencing and histology reveal dosage compensation in the liver of triploid pre-smolt Atlantic salmon. Sci. Rep. 2020, 10, 1–14. [Google Scholar] [CrossRef]
- Yang, W.; Chen, H.; Cui, X.; Zhang, K.; Jiang, D.; Deng, S.; Zhu, C.; Li, G. Sequencing, de novo assembly and characterization of the spotted scat Scatophagus argus (Linnaeus 1766) transcriptome for discovery of reproduction related genes and SSRs. J. Oceanol. Limnol. 2018, 36, 1329–1341. [Google Scholar] [CrossRef]
- Yang, W.; Wang, Y.; Jiang, D.; Tian, C.; Zhu, C.; Li, G.; Chen, H. ddRADseq-assisted construction of a high-density SNP genetic map and QTL fine mapping for growth-related traits in the spotted scat (Scatophagus argus). BMC Genom. 2020, 21, 1–18. [Google Scholar] [CrossRef]
- Ru, X.-Y.; Shi, H.-J.; Wang, T.; Liu, Q.-Q.; Jiang, D.-N.; Peng, Y.-H.; Huang, Y.; Zhu, C.-H.; Li, S.-S.; Dong, R.-R.; et al. Effects of 17β-Estradiol on growth-related genes expression in female and male spotted scat (Scatophagus argus). Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2020, 250, 110492. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.-P.; Cui, X.-F.; Wang, Y.-R.; Li, Z.-Y.; Tian, C.-X.; Jiang, D.-N.; Zhu, C.-H.; Zhang, Y.; Li, S.-S.; Li, G.-L. Identification, functional characterization and estrogen regulation on gonadotropin-releasing hormone in the spotted scat, Scatophagus argus. Fish Physiol. Biochem. 2020, 46, 1743–1757. [Google Scholar] [CrossRef] [PubMed]
- Lowry, O.H.; Rosebrough, N.J.; Farr, A.L.; Randall, R.J. Protein measurement with the folin phenol reagent. J. Biol. Chem. 1951, 193, 265–275. [Google Scholar] [CrossRef]
- Cai, L.-S.; Wang, L.; Song, K.; Lu, K.-L.; Zhang, C.-X.; Rahimnejad, S. Evaluation of protein requirement of spotted seabass (Lateolabrax maculatus) under two temperatures, and the liver transcriptome response to thermal stress. Aquaculture 2020, 516, 734615. [Google Scholar] [CrossRef]
- He, F.-X.; Jiang, D.-N.; Huang, Y.-Q.; Mustapha, U.F.; Yang, W.; Cui, X.-F.; Tian, C.-X.; Chen, H.-P.; Shi, H.-J.; Deng, S.-P.; et al. Comparative transcriptome analysis of male and female gonads reveals sex-biased genes in spotted scat (Scatophagus argus). Fish Physiol. Biochem. 2019, 45, 1963–1980. [Google Scholar] [CrossRef]
- Chen, Y.; Chen, Y.; Shi, C.; Huang, Z.; Zhang, Y.; Li, S.; Li, Y.; Ye, J.; Yuxin, C.; Li, Z.; et al. SOAPnuke: A MapReduce acceleration-supported software for integrated quality control and preprocessing of high-throughput sequencing data. GigaScience 2018, 7, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; A Thompson, D.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef]
- Ye, J.; Fang, L.; Zheng, H.; Zhang, Y.; Chen, J.; Zhang, Z.; Wang, J.; Li, S.; Li, R.; Bolund, L. WEGO: A web tool for plotting GO annotations. Nucleic Acids Res. 2006, 34, W293–W297. [Google Scholar] [CrossRef]
- Xie, C.; Mao, X.; Huang, J.; Ding, Y.; Wu, J.; Dong, S.; Kong, L.; Gao, G.; Li, C.-Y.; Wei, L. KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011, 39, W316–W322. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Wang, F.; Dong, S.; Lu, Y. De novo assembly and transcriptome analysis of osmoregulation in Litopenaeus vannamei under three cultivated conditions with different salinities. Gene 2016, 578, 185–193. [Google Scholar] [CrossRef]
- Cui, X.-F.; Zhao, Y.; Chen, H.-P.; Deng, S.-P.; Jiang, D.-N.; Wu, T.-L.; Zhu, C.-H.; Li, G.-L. Cloning, expression and functional characterization on vitellogenesis of estrogen receptors in Scatophagus argus. Gen. Comp. Endocrinol. 2017, 246, 37–45. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.; Ye, M.; Chen, L.; Deng, S.; Zhu, C.; Shi, S.; Li, G. Growth performance and digestive enzyme activity between male and female Scatophagus argus. J. Shanghai Ocean Univ. 2013, 22, 545–551. (In Chinese) [Google Scholar]
- Ijiri, S.; Kaneko, H.; Kobayashi, T.; Wang, D.-S.; Sakai, F.; Paul-Prasanth, B.; Nakamura, M.; Nagahama, Y. Sexual dimorphic expression of genes in gonads during early differentiation of a teleost fish, the Nile tilapia Oreochromis niloticus. Biol. Reprod. 2008, 78, 333–341. [Google Scholar] [CrossRef] [PubMed]
- Mandiki, S.N.; Babiak, I.; Bopopi, J.M.; Leprieur, F.; Kestemont, P. Effects of sex steroids and their inhibitors on endocrine parameters and gender growth differences in Eurasian perch (Perca fluviatilis) juveniles. Steroids 2005, 70, 85–94. [Google Scholar] [CrossRef]
- Riley, L.G.; Hirano, T.; Grau, E.G. Estradiol-17β and dihydrotestosterone differentially regulate vitellogenin and insulin-like growth factor-I production in primary hepatocytes of the tilapia Oreochromis mossambicus. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2004, 138, 177–186. [Google Scholar] [CrossRef]
- Rennie, M.D.; Purchase, C.F.; Lester, N.; Collins, N.C.; Shuter, B.J.; Abrams, P.A. Lazy males? Bioenergetic differences in energy acquisition and metabolism help to explain sexual size dimorphism in percids. J. Anim. Ecol. 2008, 77, 916–926. [Google Scholar] [CrossRef]
- Goetz, F.W.; Rise, M.; Goetz, G.W.; Binkowski, F.; Shepherd, B.S.; Rise, M.L. Stimulation of growth and changes in the hepatic transcriptome by 17β-estradiol in the yellow perch (Perca flavescens). Physiol. Genom. 2009, 38, 261–280. [Google Scholar] [CrossRef] [PubMed]
- Yue, M.M.; Zhao, J.L. Effects of estradiol and testosterone on digestive enzyme activity and expression of food intake genes between female and male Nile Tilapia Oreochromis niloticus. Fish. Sci. 2018, 37, 85–92. (In Chinese) [Google Scholar]
- Sangiao-Alvarellos, S.; Polakof, S.; Arjona, F.J.; García-López, A.; del Río, M.P.M.; Martínez-Rodríguez, G.; Míguez, J.M.; Mancera, J.M.; Soengas, J.L. Influence of testosterone administration on osmoregulation and energy metabolism of gilthead sea bream Sparus auratus. Gen. Comp. Endocrinol. 2006, 149, 30–41. [Google Scholar] [CrossRef]
- Thongprajukaew, K.; Kovitvadhi, U. Effects of sex on characteristics and expression levels of digestive enzymes in the adult guppy Poecilia reticulata. Zool. Stud. 2013, 52, 3. [Google Scholar] [CrossRef][Green Version]
- Holtby, L.B.; Healey, M.C. Sex-specific life history tactics and risk-taking in coho salmon. Ecology 1990, 71, 678–690. [Google Scholar] [CrossRef]
- A Henderson, B.; Collins, N.C.; E Morgan, G.; Vaillancourt, A. Sexual size dimorphism of walleye (Stizostedion vitreum vitreum). Can. J. Fish. Aquat. Sci. 2003, 60, 1345–1352. [Google Scholar] [CrossRef]
- Toguyeni, A.; Fauconneau, B.; Boujard, T.; Fostier, A.; Kuhn, E.R.; A Mol, K.; Baroiller, J.F. Feeding behaviour and food utilisation in tilapia, Oreochromis niloticus: Effect of Sex Ratio and Relationship with the Endocrine Status. Physiol. Behav. 1997, 62, 273–279. [Google Scholar] [CrossRef]
- Areekijseree, M.; Engkagul, A.; Kovitvadhi, U.; Thongpan, A.; Mingmuang, M.; Pakkong, P.; Rungruangsak-Torrissen, K. Temperature and pH characteristics of amylase and proteinase of adult freshwater pearl mussel, Hyriopsis (Hyriopsis) bialatus Simpson 1900. Aquaculture 2004, 234, 575–587. [Google Scholar] [CrossRef]
- Thongprajukaew, K.; Kovitvadhi, U.; Kovitvadhi, S.; Engkagul, A.; Rungruangsak-Torrissen, K. Evaluation of growth performance and nutritional quality of diets using enzymatic markers and in vitro digestibility in Siamese fighting fish (Betta splendens Regan, 1910). Afr. J. Biotechnol. 2013, 12, 1689–1702. [Google Scholar] [CrossRef]
- Leonard, J.; Iwata, M.; Ueda, H. Seasonal changes of hormones and muscle enzymes in adult lacustrine masu (Oncorhynchus masou) and sockeye salmon (O. nerka). Fish Physiol. Biochem. 2001, 25, 153–163. [Google Scholar] [CrossRef]
- Wang, L.Y.; Qi, P.P.; Chen, M.; Yuan, Y.C.; Shen, Z.G.; Fan, Q.X. Effects of sex steroid hormones on sexual size dimorphism in yellow catfish (Tachysurus fulvidraco). Acta Hydrobiol. Sin. 2020, 44, 379–388. [Google Scholar] [CrossRef]
- Figueiredo-Fernandes, A.; Fontaínhas-Fernandes, A.; Peixoto, F.; Rocha, E.; Reis-Henriques, M. Effects of gender and temperature on oxidative stress enzymes in Nile tilapia Oreochromis niloticus exposed to paraquat. Pestic. Biochem. Physiol. 2006, 85, 97–103. [Google Scholar] [CrossRef]
- Parolini, M.; Iacobuzio, R.; DE Felice, B.; Bassano, B.; Pennati, R.; Saino, N. Age- and sex-dependent variation in the activity of antioxidant enzymes in the brown trout (Salmo trutta). Fish Physiol. Biochem. 2018, 45, 145–154. [Google Scholar] [CrossRef]
- Zheng, W.; Xu, H.; Lam, S.H.; Luo, H.; Karuturi, R.K.M.; Gong, Z. Transcriptomic Analyses of Sexual Dimorphism of the Zebrafish Liver and the Effect of Sex Hormones. PLoS ONE 2013, 8, e53562. [Google Scholar] [CrossRef]
- Alonso-Alvarez, C.; Bertrand, S.; Faivre, B.; Sorci, G. Increased susceptibility to oxidative damage as a cost of accelerated somatic growth in zebra finches. Funct. Ecol. 2007, 21, 873–879. [Google Scholar] [CrossRef]
- Zhang, X.; Han, Z.; Zhong, H.; Yin, Q.; Xiao, J.; Wang, F.; Zhou, Y.; Luo, Y. Regulation of triglyceride synthesis by estradiol in the livers of hybrid tilapia (Oreochromis niloticus ♀ × O. aureus ♂). Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2019, 238, 110335. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhong, H.; Han, Z.; Tang, Z.; Xiao, J.; Guo, Z.; Wang, F.; Luo, Y.; Zhou, Y. Effects of waterborne exposure to 17β-estradiol on hepatic lipid metabolism genes in tilapia (Oreochromis niloticus). Aquac. Rep. 2020, 17, 100382. [Google Scholar] [CrossRef]
- Cleveland, B.M.; Gao, G.; Leeds, T.D. Transcriptomic response to selective breeding for fast growth in rainbow trout (Oncorhynchus mykiss). Mar. Biotechnol. 2020, 22, 539–550. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, G.; Verma, P. Sex-specific metabolic changes in the annual reproductive cycle of a freshwater catfish. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2004, 137, 101–106. [Google Scholar] [CrossRef]
- Homma, H.; Kurachi, H.; Nishio, Y.; Takeda, T.; Yamamoto, T.; Adachi, K.; Morishige, K.; Ohmichi, M.; Matsuzawa, Y.; Murata, Y. Estrogen suppresses transcription of lipoprotein lipase gene: Existence of a unique estrogen response element on the lipoprotein lipase promoter. J. Biol. Chem. 2000, 275, 11404–11411. [Google Scholar] [CrossRef]
- Webster, T.M.U.; Shears, J.A.; Moore, K.; Santos, E.M. Identification of conserved hepatic transcriptomic responses to 17β-estradiol using high-throughput sequencing in brown trout. Physiol. Genom. 2015, 47, 420–431. [Google Scholar] [CrossRef]
- Weisgraber, K.H. Apolipoprotein E: Structure-function relationships. Adv. Protein Chem. 1994, 45, 249–302. [Google Scholar] [CrossRef]
- Poupard, G.; André, M.; Durliat, M.; Ballagny, C.; Boeuf, G.; Babin, P.J. Apolipoprotein E gene expression correlates with endogenous lipid nutrition and yolk syncytial layer lipoprotein synthesis during fish development. Cell Tissue Res. 2000, 300, 251–261. [Google Scholar] [CrossRef]
- Phumyu, N.; Boonanuntanasarn, S.; Jangprai, A.; Yoshizaki, G.; Na-Nakorn, U. Pubertal effects of 17α-methyltestosterone on GH–IGF-related genes of the hypothalamic–pituitary–liver–gonadal axis and other biological parameters in male, female and sex-reversed Nile tilapia. Gen. Comp. Endocrinol. 2012, 177, 278–292. [Google Scholar] [CrossRef]
- Duan, C.; Xu, Q. Roles of insulin-like growth factor (IGF) binding proteins in regulating IGF actions. Gen. Comp. Endocrinol. 2005, 142, 44–52. [Google Scholar] [CrossRef]
- Safian, D.; Fuentes, E.N.; Valdés, J.A.; Molina, A. Dynamic transcriptional regulation of autocrine/paracrine igfbp1, 2, 3, 4, 5, and 6 in the skeletal muscle of the fine flounder during different nutritional statuses. J. Endocrinol. 2012, 214, 95–108. [Google Scholar] [CrossRef]
- Kelley, K.M.; E Schmidt, K.; Berg, L.; Sak, K.; Galima, M.M.; Gillespie, C.; Balogh, L.; Hawayek, A.; A Reyes, J.; Jamison, M. Comparative endocrinology of the insulin-like growth factor-binding protein. J. Endocrinol. 2002, 175, 3–18. [Google Scholar] [CrossRef]
- Martyniuk, C.J.; Gerrie, E.R.; Popesku, J.T.; Ekker, M.; Trudeau, V.L. Microarray analysis in the zebrafish (Danio rerio) liver and telencephalon after exposure to low concentration of 17alpha-ethinylestradiol. Aquat. Toxicol. 2007, 84, 38–49. [Google Scholar] [CrossRef]

| Sample | Raw Reads (×106) | Raw Bases (Gb) | Clean Reads (×106) | Clean Bases (Gb) | % ≥ Q20 | Raito of Clean Read (%) |
|---|---|---|---|---|---|---|
| Female_1 | 45.53 | 6.83 | 45.12 | 6.77 | 92.25 | 99.10 |
| Female_2 | 44.84 | 6.73 | 44.31 | 6.65 | 91.51 | 98.81 |
| Female_3 | 46.97 | 7.04 | 46.49 | 6.97 | 92.46 | 98.98 |
| Male_1 | 48.40 | 7.26 | 47.96 | 7.19 | 95.91 | 99.10 |
| Male_2 | 45.40 | 6.81 | 44.84 | 6.73 | 95.90 | 98.77 |
| Male_3 | 44.44 | 6.67 | 43.92 | 6.59 | 96.06 | 98.84 |
| Mean | 45.93 | 6.89 | 45.44 | 6.82 | 94.01 | 98.93 |
| Total | 275.58 | 41.34 | 272.65 | 40.90 | - | - |
| Category | Value |
|---|---|
| Unigene number | 79,115 |
| Min length (bp) | 200 |
| Max length (bp) | 17,724 |
| Mean length (bp) | 1258 |
| N50 (bp) | 2462 |
| N90 (bp) | 471 |
| GC (%) | 46.86 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, H.; Jiang, D.; Li, Z.; Wang, Y.; Yang, X.; Li, S.; Li, S.; Yang, W.; Li, G. Comparative Physiological and Transcriptomic Profiling Offers Insight into the Sexual Dimorphism of Hepatic Metabolism in Size-Dimorphic Spotted Scat (Scatophagus argus). Life 2021, 11, 589. https://doi.org/10.3390/life11060589
Chen H, Jiang D, Li Z, Wang Y, Yang X, Li S, Li S, Yang W, Li G. Comparative Physiological and Transcriptomic Profiling Offers Insight into the Sexual Dimorphism of Hepatic Metabolism in Size-Dimorphic Spotted Scat (Scatophagus argus). Life. 2021; 11(6):589. https://doi.org/10.3390/life11060589
Chicago/Turabian StyleChen, Huapu, Dongneng Jiang, Zhiyuan Li, Yaorong Wang, Xuewei Yang, Shuangfei Li, Shuisheng Li, Wei Yang, and Guangli Li. 2021. "Comparative Physiological and Transcriptomic Profiling Offers Insight into the Sexual Dimorphism of Hepatic Metabolism in Size-Dimorphic Spotted Scat (Scatophagus argus)" Life 11, no. 6: 589. https://doi.org/10.3390/life11060589
APA StyleChen, H., Jiang, D., Li, Z., Wang, Y., Yang, X., Li, S., Li, S., Yang, W., & Li, G. (2021). Comparative Physiological and Transcriptomic Profiling Offers Insight into the Sexual Dimorphism of Hepatic Metabolism in Size-Dimorphic Spotted Scat (Scatophagus argus). Life, 11(6), 589. https://doi.org/10.3390/life11060589









