Comparative Analysis of Flavonoid Metabolites in Foxtail Millet (Setaria italica) with Different Eating Quality
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Photo Recording and Color Determination
2.3. Preparation and Extraction of Samples for Metabolomics Analysis
2.4. Ultra-High-Performance Liquid Chromatography (UPLC) Conditions
2.5. Electrospray-Triple Quadrupole-Linear Ion Trap Mass Spectrometry (ESI-Q TRAP-MS/MS) System
2.6. Qualitative and Quantitative Analysis of Metabolites
2.7. Quality Control Analysis of Samples
2.8. Statistical Analysis
3. Results
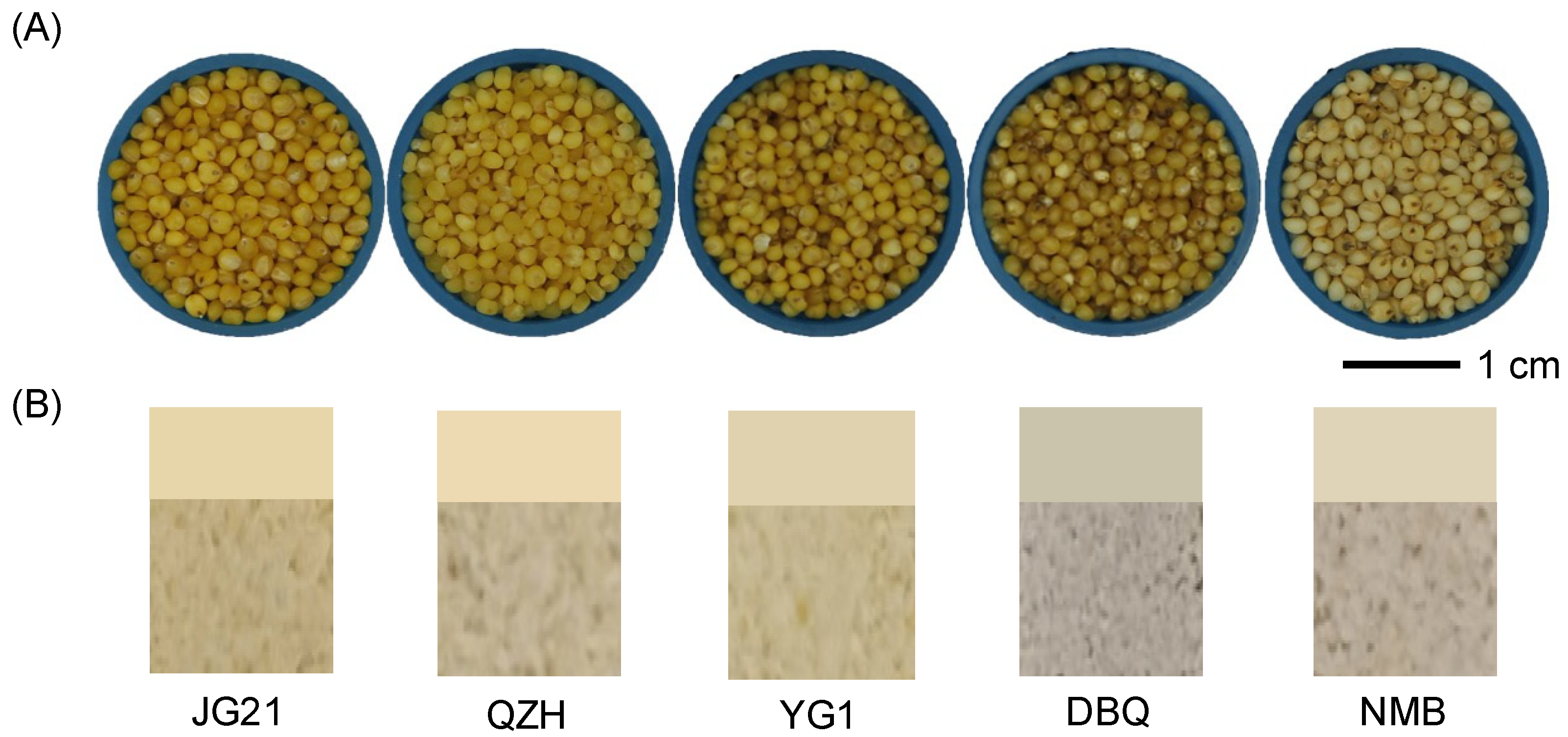
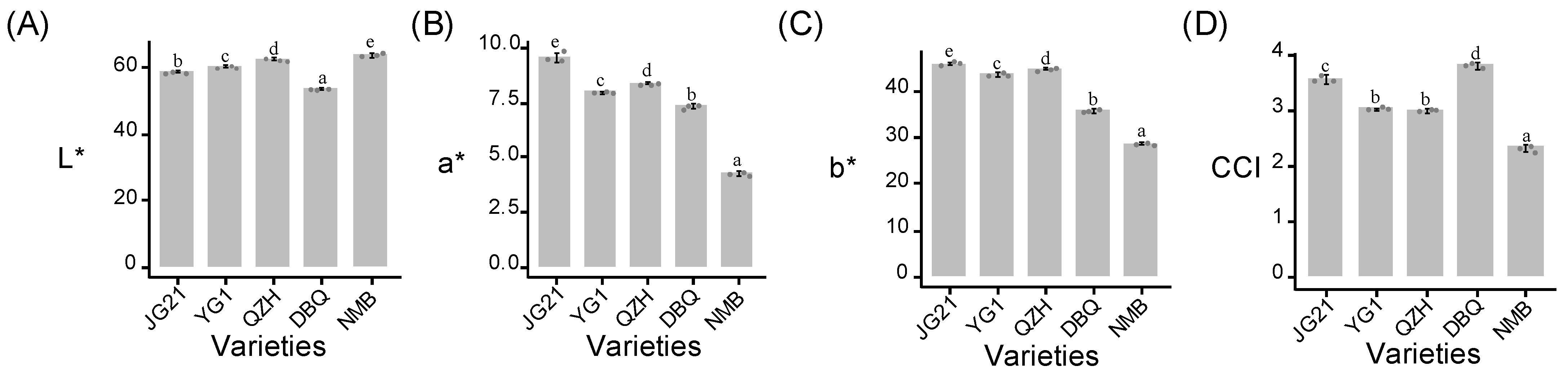
3.1. The Millet Color of Different Varieties
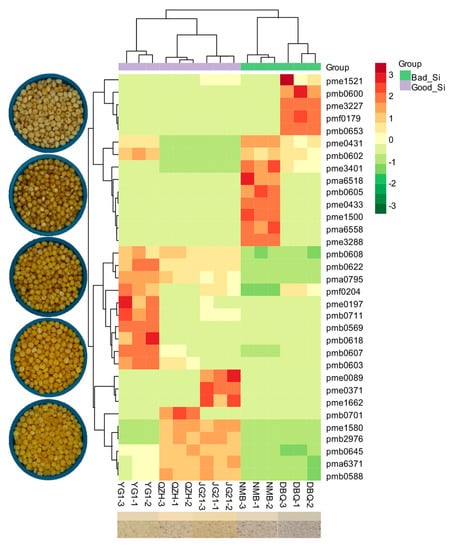
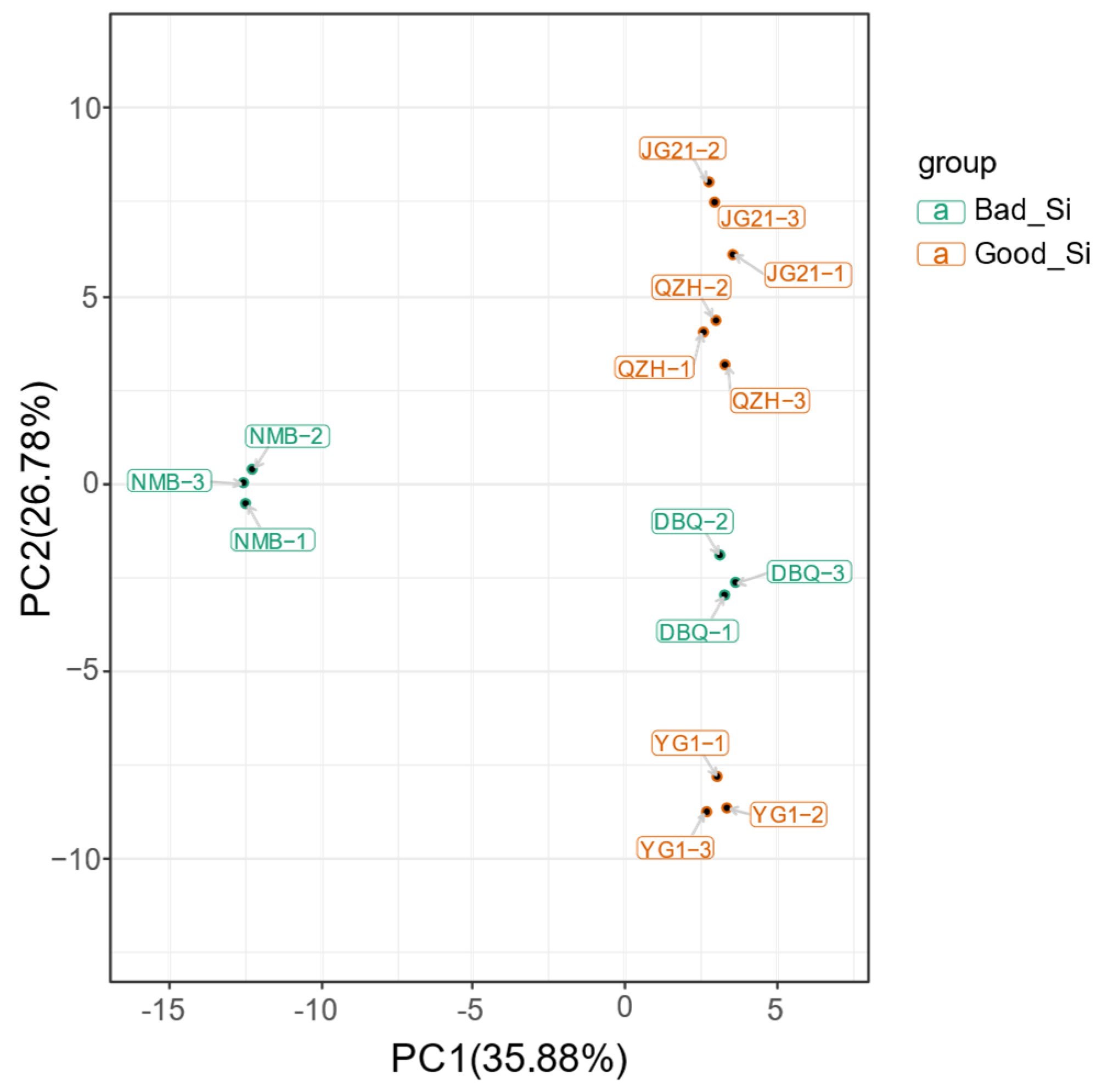
3.2. Flavonoid Metabolites Profiles in Millets of Different Varieties
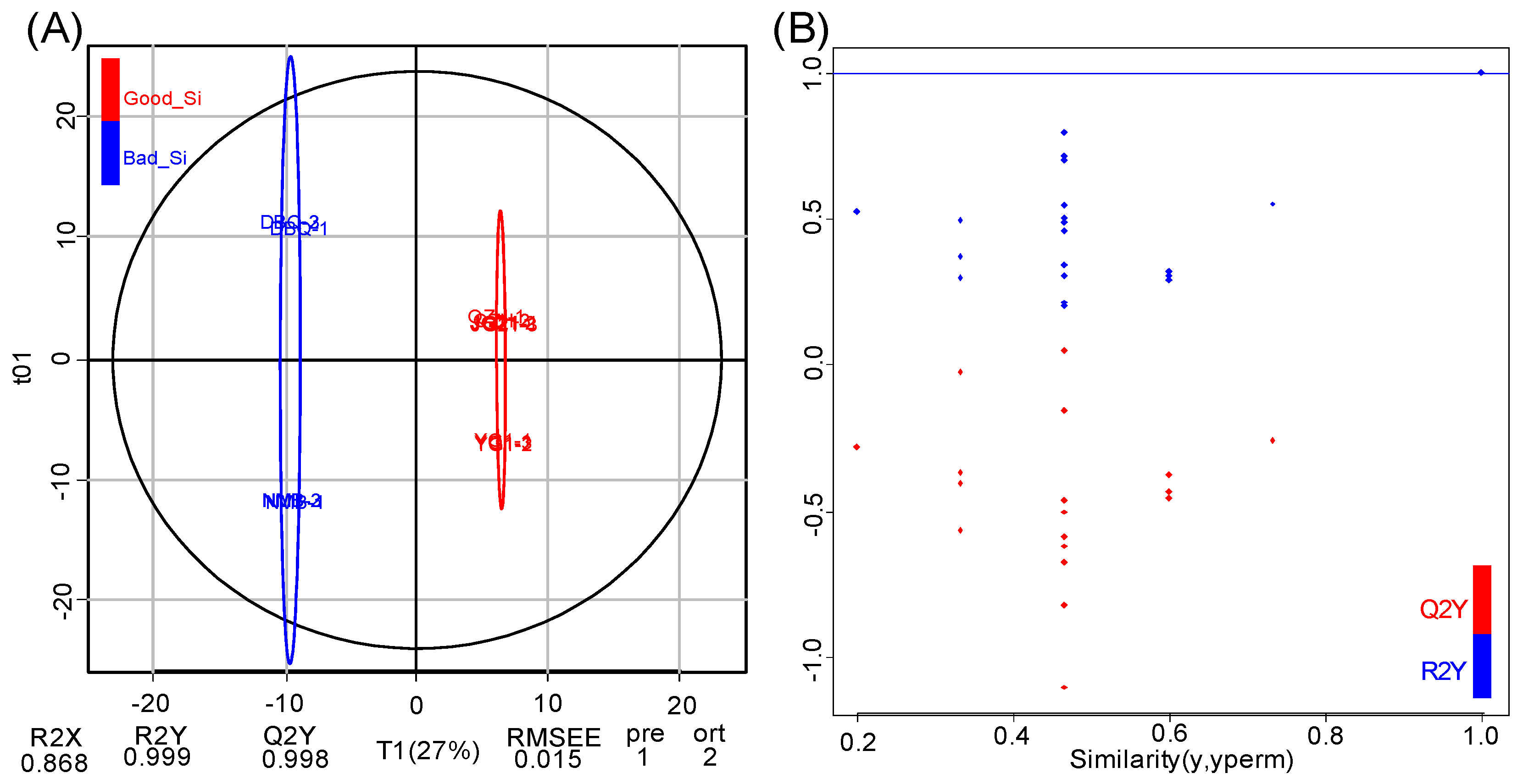
3.3. Pairwise Comparison of Flavonoid Metabolites Between High and Poor Eating Quality VarietiesBased on OPLS-DA Model
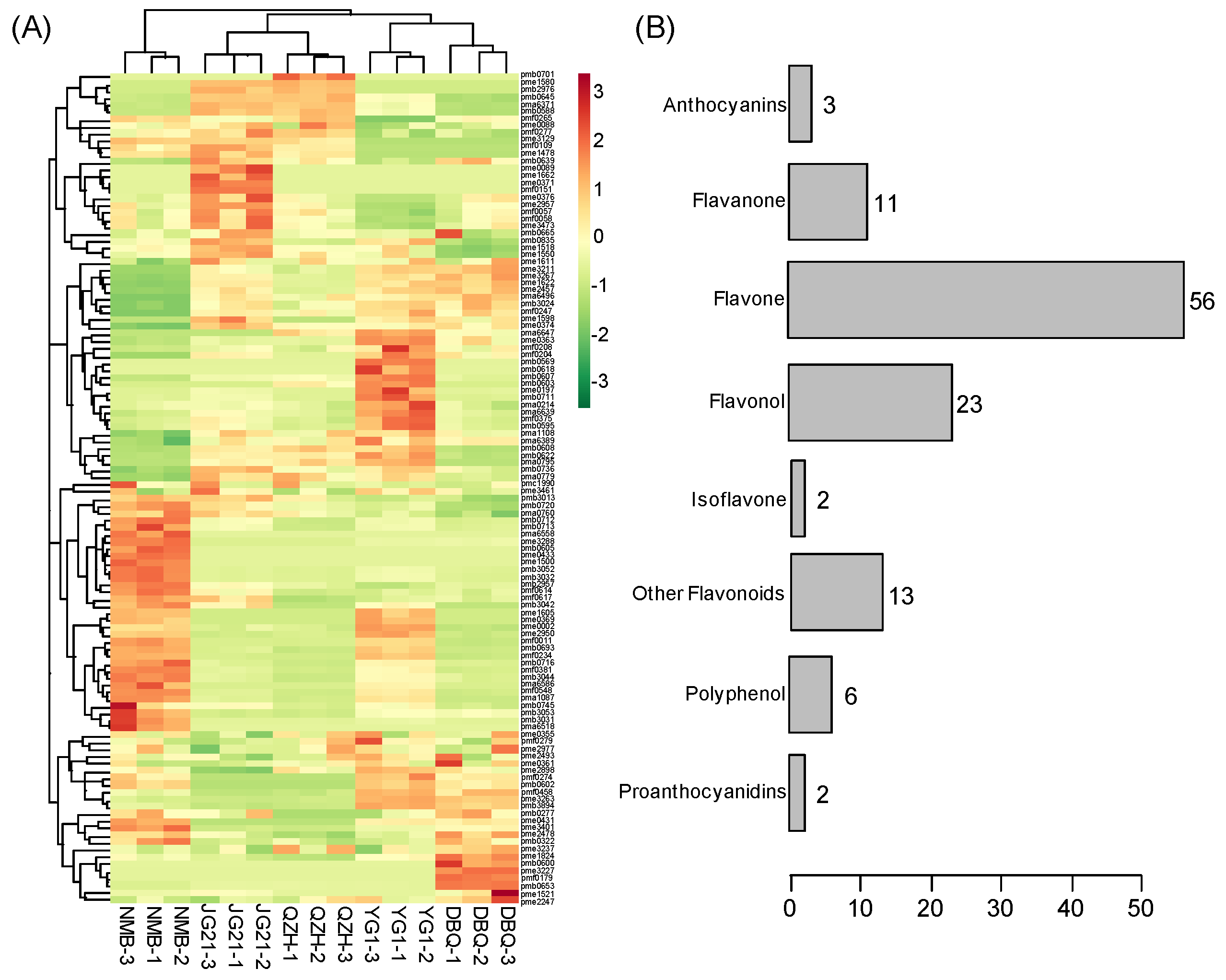
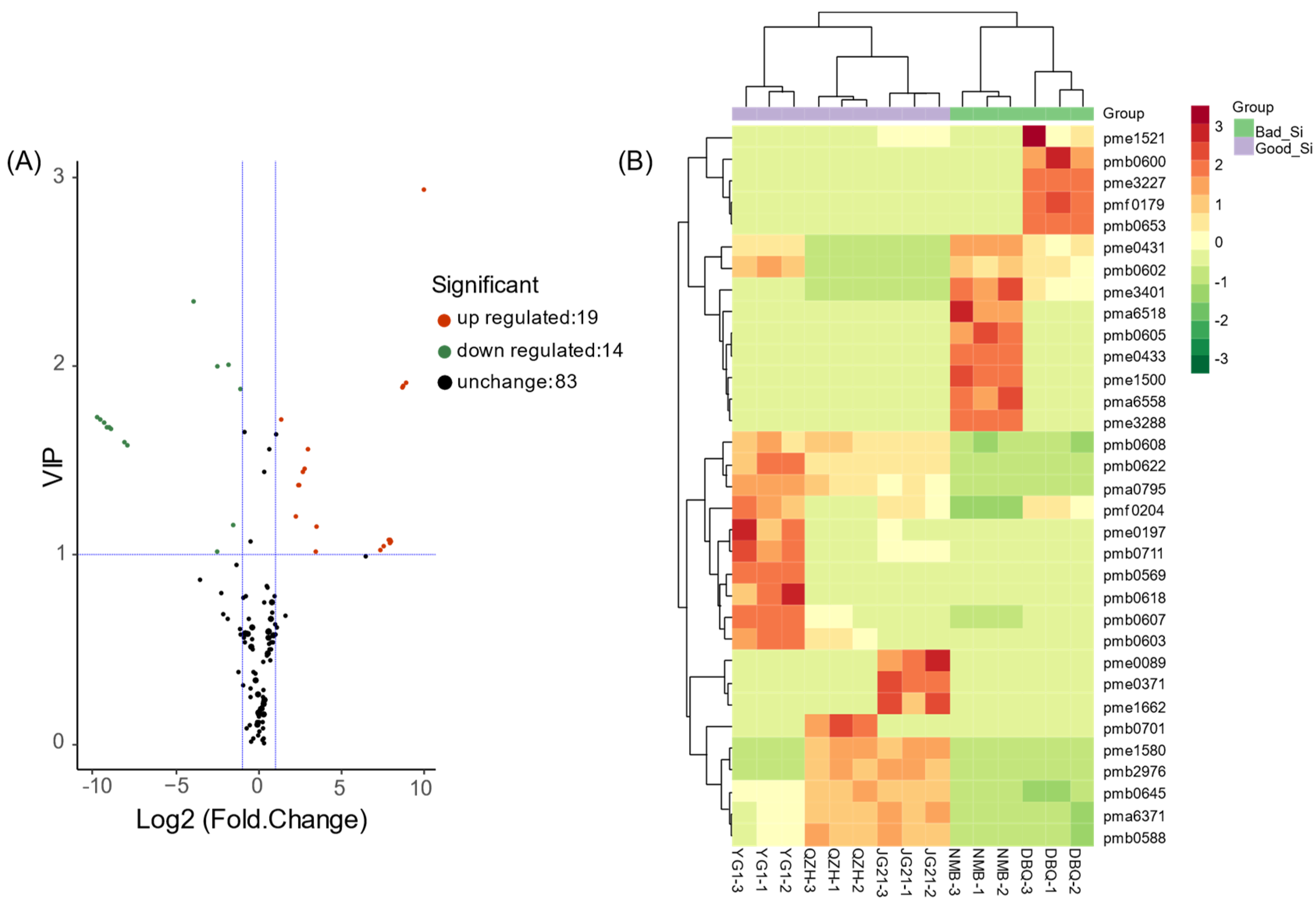
3.4. Identification of Differential Flavonoid Metabolites
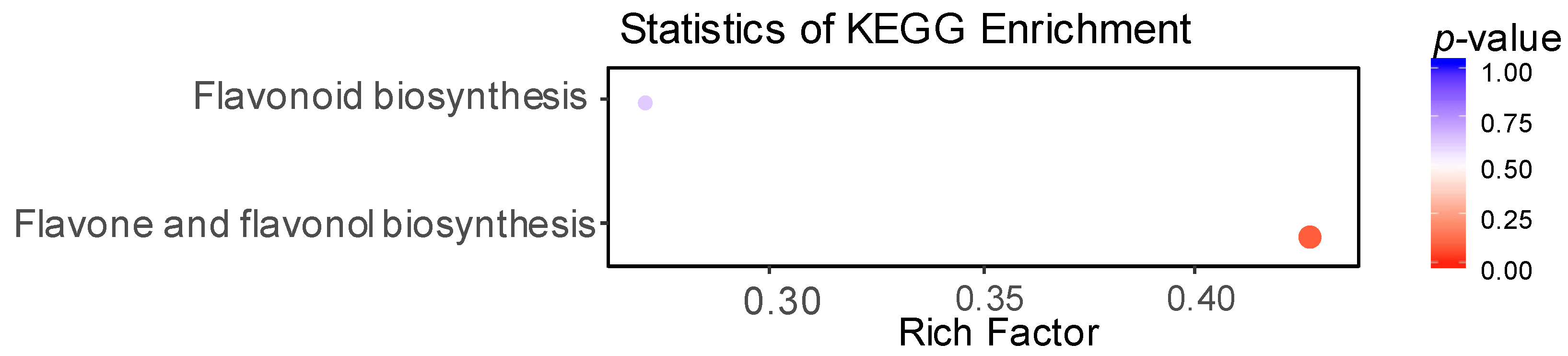
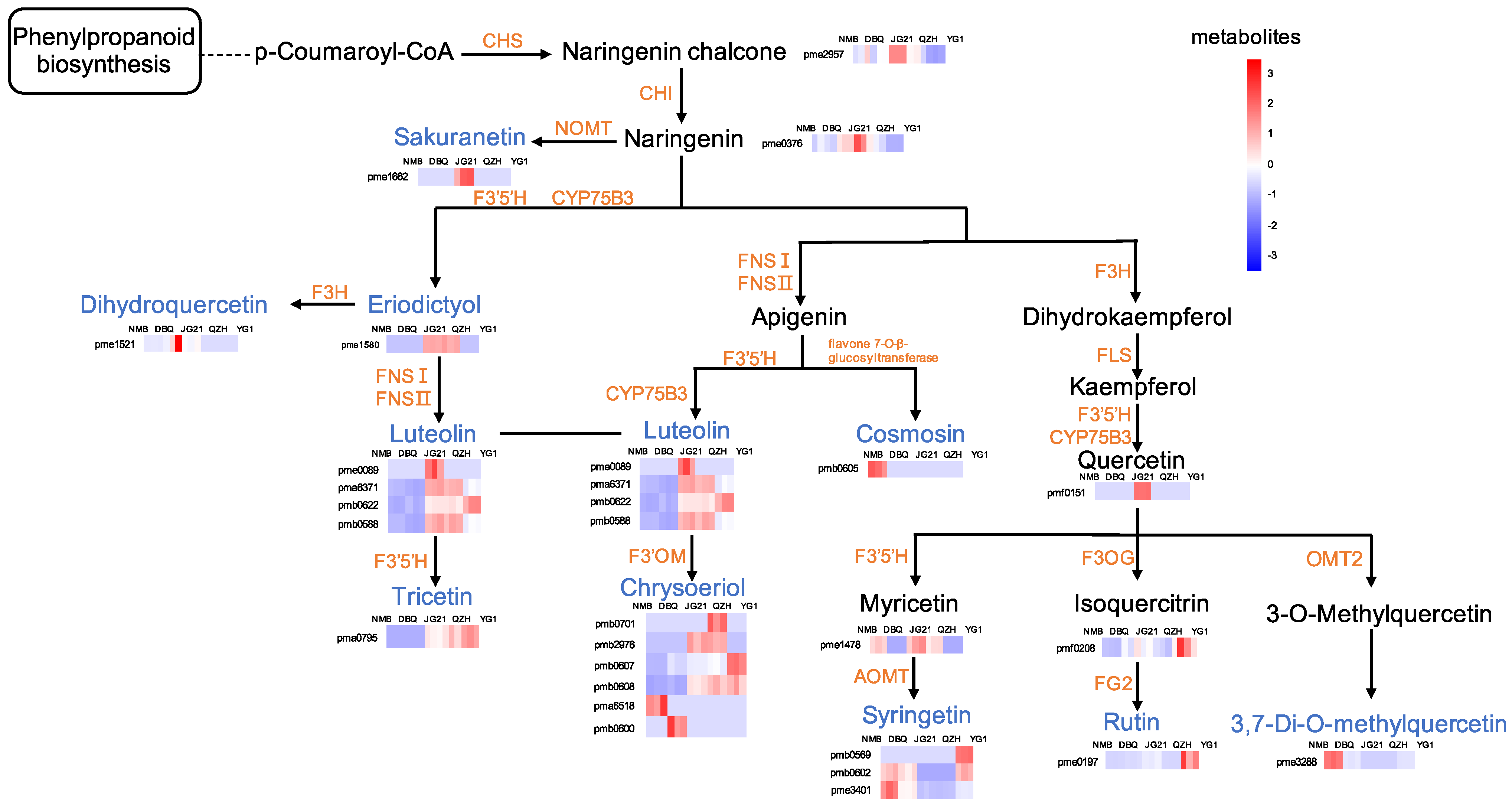
3.5. Enrichment Analysis Clarified the Metabolic Pathways of Differential Flavonoid Metabolites
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yang, X.Y.; Wan, Z.W.; Perry, L.; Lu, H.Y.; Wang, Q.; Zhao, C.H.; Li, J.; Xie, F.; Yu, J.C.; Cui, T.X.; et al. Early millet use in northern China. Proc. Natl. Acad. Sci. USA 2012, 109, 3726–3730. [Google Scholar] [CrossRef]
- Nithiyanantham, S.; Kalaiselvi, P.; Mahomoodally, M.F.; Zengin, G.; Abirami, A.; Srinivasan, G. Nutritional and functional roles of millets—A review. J. Food Biochem. 2019, 43, e12859. [Google Scholar] [CrossRef] [PubMed]
- Sachdev, N.; Goomer, S.; Singh, L.R. Foxtail millet: A potential crop to meet future demand scenario for alternative sustainable protein. J. Sci. Food Agric. 2020, 101, 831–842. [Google Scholar] [CrossRef] [PubMed]
- Gopalan, C.; Ramasastri, B.V.; Balasubramanian, S.C. Nutritive Value of Indian Foods: National Institute of Nutrition; Indian Council of Medical Research: Hyderabad, India, 1989. [Google Scholar]
- Thathola, A.; Srivastava, S.; Singh, G. Effect of foxtail millet supplementation on serum glucose, serum lipids and glycosylated hemoglobin in type 2 diabetics. Diabetol. Croatica 2011, 40, 23–28. [Google Scholar]
- Zhang, Y.Y.; Lu, Y.; Zhang, B.; He, L.; Liu, L.L.; Wang, X.C.; Li, H.Y.; Han, Y.H. The relationship between the gene SiLCYB related to carotenoid synthesis and the colored formation of foxtail millet. Mol. Plant Breed. 2016, 14, 1341–1351. [Google Scholar] [CrossRef]
- Sun, J.; Luu, N.S.; Chen, Z.H.; Chen, B.; Cui, X.A.; Wu, J.X.; Zhang, Z.G.; Lu, T.G. Generation and characterization of a foxtail millet (Setaria italica) mutant library. Front. Plant Sci. 2019, 10, 369. [Google Scholar] [CrossRef] [PubMed]
- Pradeep, P.M.; Sreerama, Y.N. Phenolic antioxidants of foxtail and little millet cultivars and their inhibitory effects on alpha-amylase and alpha-glucosidase activities. Food Chem. 2018, 247, 46–55. [Google Scholar] [CrossRef]
- Xiang, J.L.; Zhang, M.; Apea-Bah, F.B.; Beta, T. Hydroxycinnamic acid amide (HCAA) derivatives, flavonoid C-glycosides, phenolic acids and antioxidant properties of foxtail millet. Food Chem. 2019, 295, 214–223. [Google Scholar] [CrossRef]
- Sun, Y.Q.; Qiao, L.P.; Shen, Y.; Jiang, P.; Chen, J.C.; Ye, X.Q. Phytochemical profile and antioxidant activity of physiological drop of citrus fruits. J. Food Sci. 2013, 78, 37–42. [Google Scholar] [CrossRef] [PubMed]
- Peng, M.; Shahzad, R.; Gul, A.; Subthain, H.; Shen, S.; Lei, L.; Zheng, Z.G.; Zhou, J.J.; Lu, D.D.; Wang, S.C.; et al. Differentially evolved glucosyltransferases determine natural variation of rice flavone accumulation and UV-tolerance. Nat. Commun. 2017, 8, 1975. [Google Scholar] [CrossRef]
- Chen, J.J.; Yuan, Z.Y.; Zhang, H.P.; Li, W.Y.; Shi, M.Y.; Peng, Z.X.; Li, M.Y.; Tian, J.; Deng, X.X.; Cheng, Y.J.; et al. Cit1,2RhaT and two novel CitdGlcTs participate in flavor-related flavonoid metabolism during citrus fruit development. J. Exp. Bot. 2019, 70, 2759–2771. [Google Scholar] [CrossRef]
- Zhang, J.; Qiu, X.; Tan, Q.; Xiao, Q.; Mei, S. A comparative metabolomics study of flavonoids in radish with different skin and flesh colors (Raphanus sativus L.). J. Agric. Food Chem. 2020, 68, 14463–14470. [Google Scholar] [CrossRef]
- Ni, J.B.; Zhao, Y.; Tao, R.Y.; Yin, L.; Gao, L.; Strid, A.; Qian, M.J.; Li, J.C.; Li, Y.J.; Shen, J.Q.; et al. Ethylene mediates the branching of the jasmonate-induced flavonoid biosynthesis pathway by suppressing anthocyanin biosynthesis in red Chinese pear fruits. Plant Biotechnol. J. 2020, 18, 1223–1240. [Google Scholar] [CrossRef]
- George, V.C.; Dellaire, G.; Rupasinghe, H.P.V. Plant flavonoids in cancer chemoprevention: Role in genome stability. J. Nutr. Biochem. 2017, 45, 1–14. [Google Scholar] [CrossRef]
- Perez-Vizcaino, F.; Fraga, C.G. Research trends in flavonoids and health. Arch. Biochem. Biophys. 2018, 646, 107–112. [Google Scholar] [CrossRef]
- Ali, S.S.; Ahsan, H.; Zia, M.K.; Siddiqui, T.; Khan, F.H. Understanding oxidants and antioxidants: Classical team with new players. J. Food Biochem. 2020, 44, e13145. [Google Scholar] [CrossRef] [PubMed]
- Koes, R.; Verweij, W.; Quattrocchio, F. Flavonoids: A colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci. 2005, 10, 236–242. [Google Scholar] [CrossRef] [PubMed]
- Winkel-Shirley, B. Biosynthesis of flavonoids and effects of stress. Curr. Opin. Plant Biol. 2002, 5, 218–223. [Google Scholar] [CrossRef]
- Dunn, W.B.; Ellis, D.I. Metabolomics: Current analytical platforms and methodologies. Trac-Trend. Anal. Chem. 2005, 24, 285–294. [Google Scholar]
- Chan, C.O.; Xie, X.J.; Wan, S.W.; Zhou, G.L.; Yuen, C.A.; Mok, D.K.; Chen, S.B. Qualitative and quantitative analysis of sesquiterpene lactones in Centipeda minima by UPLC–Orbitrap–MS & UPLC-QQQ-MS. J. Pharm. Biomed. Anal. 2019, 174, 360–366. [Google Scholar] [CrossRef] [PubMed]
- Keurentjes, J.J.B.; Fu, J.Y.; de Vos, C.H.R.; Lommen, A.; Hall, R.D.; Bino, R.J.; van der Plas, L.H.W.; Jansen, R.C.; Vreugdenhil, D.; Koornneef, M. The genetics of plant metabolism. Nat. Genet. 2006, 38, 842–849. [Google Scholar] [CrossRef] [PubMed]
- Brazier-Hicks, M.; Evans, K.M.; Gershater, M.C.; Puschmann, H.; Steel, P.G.; Edwards, R. The C-glycosylation of flavonoids in cereals. J. Biol. Chem. 2009, 284, 17926–17934. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.F.; Xiao, K.; Wang, Z.; Wang, S.H.; Xu, F.X. Comparison of metabolism substances in Cordyceps sinensis and Cordyceps militaris cultivated with tussah pupa based on LC-MS. J. Food Biochem. 2021, 45, e13735. [Google Scholar] [CrossRef]
- Bennetzen, J.L.; Schmutz, J.; Wang, H.; Percifield, R.; Hawkins, J.; Pontaroli, A.C.; Estep, M.; Feng, L.; Vaughn, J.N.; Grimwood, J.; et al. Reference genome sequence of the model plant Setaria. Nat. Biotechnol. 2012, 30, 555–561. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.Y.; Sun, C.D.; Zhang, L.L.; Dai, X.; Xu, C.J.; Chen, K.S. Preferential accumulation of orange-colored carotenoids in Ponkan (Citrus reticulata) fruit peel following postharvest application of ethylene or ethephon. Sci. Hortic. 2010, 126, 229–235. [Google Scholar] [CrossRef]
- Dong, X.K.; Chen, W.; Wang, W.S.; Zhang, H.Y.; Liu, X.Q.; Luo, J. Comprehensive profiling and natural variation of flavonoids in rice. J. Integr. Plant Biol. 2014, 56, 876–886. [Google Scholar] [CrossRef]
- Wang, A.M.; Li, R.S.; Ren, L.; Gao, X.L.; Zhang, Y.G.; Ma, Z.M.; Ma, D.F.; Luo, Y.H. A comparative metabolomics study of flavonoids in sweet potato with different flesh colors (Ipomoea batatas (L.) Lam). Food Chem. 2018, 260, 124–134. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Gong, L.; Guo, Z.L.; Wang, W.S.; Zhang, H.Y.; Liu, X.Q.; Yu, S.B.; Xiong, L.Z.; Luo, J. A novel integrated method for large-scale detection, identification, and quantification of widely targeted metabolites: Application in the study of rice metabolomics. Mol. Plant 2013, 6, 1769–1780. [Google Scholar] [CrossRef]
- Fraga, C.G.; Clowers, B.H.; Moore, R.J.; Zink, E.M. Signature-Discovery approach for sample matching of a nerve-agent precursor using liquid chromatography-mass spectrometry, XCMS, and chemometrics. Anal. Chem. 2010, 82, 4165–4173. [Google Scholar] [CrossRef]
- Hadley, W. Ggplot2: Elegant graphics for Data Analysis; Springer: New York, NY, USA, 2016. [Google Scholar]
- Gaude, E.; Chignola, F.; Spiliotopoulos, D.; Spitaleri, A.; Ghitti, M.; Garcìa-Manteiga, J.M.; Mari, S.; Musco, G. muma, An R package for metabolomics univariate and multivariate statistical analysis. Curr. Metabolomics 2013, 1, 180–189. [Google Scholar] [CrossRef]
- Kolde., R. pheatmap: Pretty Heatmaps. Available online: https://CRAN.R-project.org/package=pheatmap (accessed on 2019).
- Wang, Y.; Li, H.; Sun, M.; Shi, Q.; Guo, E. Relationship between cooked millet palatability and both visual quality and RVA profile character of starch. J. Shanxi Agric. Sci. 2008, 36, 34–39. [Google Scholar]
- Bylesjö, M.; Rantalainen, M.; Cloarec, O.; Nicholson, J.K.; Holmes, E.; Trygg, J. OPLS discriminant analysis: Combining the strengths of PLS-DA and SIMCA classification. J. Chemometr. 2006, 20, 341–351. [Google Scholar] [CrossRef]
- Paull, R.; Seymour, G.; Taylor, J.; Tucker, G. Biochemistry of Fruit Ripening; Chapman & Hall: London, UK, 1993. [Google Scholar]
- Smolikova, G.N. and Medvedev, S.S. Seed carotenoids: Synthesis, diversity, and functions. Russ. J. Plant Physiol. 2015, 62, 3–16. [Google Scholar] [CrossRef]
- Carazo, A.; Macáková, K.; Matoušová, K.; Krčmová, L.; Protti, M.; Mladěnka, P. Vitamin A update: Forms, sources, kinetics, detection, function, deficiency, therapeutic use and toxicity. Nutrients 2021, 13, 1703. [Google Scholar] [CrossRef]
- Cheng, L.; Zhang, B.; He, L.; Ma, F.F.; Wang, X.C.; Li, H.Y.; Han, Y.H. Constitutive down-regulation of SiSGR gene is related to green millet in Setaria italica. Russ. J. Plant Physiol. 2017, 64, 608–615. [Google Scholar] [CrossRef]
- Daglia, M.; Di Lorenzo, A.; Nabavi, S.F.; Talas, Z.S.; Nabavi, S.M. Polyphenols: Well beyond the antioxidant capacity: Gallic acid and related compounds as neuroprotective agents: You are what you eat! Curr. Pharm. Biotechnol. 2014, 15, 362–372. [Google Scholar] [CrossRef]
- Harborne, J.B.; Williams, C.A. Advances in flavonoid research since 1992. Phytochemistry 2000, 55, 481–504. [Google Scholar] [CrossRef]
- Yonekura-sakakibara, K.; Higashi, Y. and Nakabayashi, R. The origin and evolution of plant flavonoid metabolism. Front. Plant Sci. 2019, 10, 943. [Google Scholar] [CrossRef] [PubMed]
- Mou, J.L.; Zhang, Z.H.; Qiu, H.J.; Lu, Y.; Zhu, X.; Fan, Z.Q.; Zhang, Q.H.; Ye, J.L.; Fernie, A.R.; Cheng, Y.J.; et al. Multiomics-based dissection of citrus flavonoid metabolism using a Citrus reticulata × Poncirus trifoliata population. Hortic. Res. 2021, 8, 56. [Google Scholar] [CrossRef]
- Sharma, M.; Sandhir, R.; Singh, A.; Kumar, P.; Mishra, A.; Jachak, S.; Singh, S.P.; Singh, J.; Roy, J. Comparative analysis of phenolic compound characterization and their biosynthesis genes between two diverse bread wheat (Triticum aestivum) varieties differing for chapatti (Unleavened Flat Bread) quality. Front. Plant Sci. 2016, 7, 1870. [Google Scholar] [CrossRef]
- Li, J.; Yang, P.; Yang, Q.H.; Gong, X.W.; Ma, H.C.; Dang, K.; Chen, G.H.; Gao, X.L.; Feng, B.L. Analysis of flavonoid metabolites in buckwheat leaves using UPLC-ESI-MS/MS. Molecules 2019, 24, 1310. [Google Scholar] [CrossRef]
- Böttner, L.; Grabe, V.; Gablenz, S.; Böhme, N.; Appenroth, K.J.; Gershenzon, J.; Huber, M. Differential localization of flavonoid glucosides in an aquatic plant implicates different functions under abiotic stress. Plant Cell Environ. 2021, 44, 900–914. [Google Scholar] [CrossRef]
- Li, S.D.; Dong, X.K.; Fan, G.Y.; Yang, Q.F.; Shi, J.; Wei, W.; Zhao, F.; Li, N.; Wang, X.M.; Wang, F.; et al. Comprehensive profiling and inheritance patterns of metabolites in foxtail millet. Front. Plant Sci. 2018, 9, 1716. [Google Scholar] [CrossRef]
- Nabavi, S.F.; Braidy, N.; Gortzi, O.; Sobarzo-Sanchez, E.; Daglia, M.; Skalicka-Woźniak, K.; Nabavi, S.M. Luteolin as an anti-inflammatory and neuroprotective agent: A brief review. Brain Res. Bull. 2015, 119, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Gendrisch, F.; Esser, P.R.; Schempp, C.M.; Wölfle, U. Luteolin as a modulator of skin aging and inflammation. Biofactors 2021, 47, 170–180. [Google Scholar] [CrossRef] [PubMed]
- López-Lázaro, M. Distribution and biological activities of the flavonoid luteolin. Mini Rev. Med. Chem. 2009, 9, 31–59. [Google Scholar] [CrossRef]
- Tang, S.M.; Deng, X.T.; Zhou, J.; Li, Q.P.; Ge, X.X.; Miao, L. Pharmacological basis and new insights of quercetin action in respect to its anti-cancer effects. Biomed. Pharmacother. 2020, 121, 109604. [Google Scholar] [CrossRef]
- Wong, S.K.; Chin, K.Y.; Ima-Nirwana, S. Quercetin as an agent for protecting the bone: A review of the current evidence. Int. J. Mol. Sci. 2020, 21, 6448. [Google Scholar] [CrossRef] [PubMed]
- David, A.V.A.; Arulmoli, R.; Parasuraman, S. Overviews of biological importance of quercetin: A bioactive flavonoid. Pharmacogn. Rev. 2016, 10, 84–89. [Google Scholar] [CrossRef]
- Shabbir, U.; Rubab, M.; Daliri, E.B.; Chelliah, R.; Javed, A.; Oh, D.H. Curcumin, quercetin, catechins and metabolic diseases: The role of gut microbiota. Nutrients 2021, 13, 206. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Micaelo, N.; Gonzalez-Abuin, N.; Ardevol, A.; Pinent, M.; Blay, M.T. Procyanidins and inflammation: Molecular targets and health implications. Biofactors 2012, 38, 257–265. [Google Scholar] [CrossRef]
- Sasaki, Y.; Ito, S.; Zhang, Z.; Lyu, X.; Takizawa, S.Y.; Kubota, R.; Minami, T. Supramolecular sensor for astringent procyanidin C1: Fluorescent artificial tongue for wine components. Chemistry 2020, 26, 16236–16240. [Google Scholar] [CrossRef]
- Cave, J.R.; Waterhouse, A.L. Combinatorics of proanthocyanidins in wine. Analyst 2019, 144, 4395–4399. [Google Scholar] [CrossRef] [PubMed]
- Tang, D.; Chen, K.; Huang, L.; Li, J. Pharmacokinetic properties and drug interactions of apigenin, a natural flavone. Expert Opin. Drug Metab. Toxicol. 2017, 13, 323–330. [Google Scholar] [CrossRef] [PubMed]
- Madunic, J.; Madunic, I.V.; Gajski, G.; Popic, J.; Garaj-Vrhovac, V. Apigenin: A dietary flavonoid with diverse anticancer properties. Cancer Lett. 2018, 413, 11–22. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Hu, X.; Shi, T.T.; Yin, H.Y.; Sun, D.F.; Hao, Y.F.; Xia, X.C.; Luo, J.; Fernie, A.R.; He, Z.H.; et al. Metabolite-based genome-wide association study enables dissection of the flavonoid decoration pathway of wheat kernels. Plant Biotechnol. J. 2020, 18, 1722–1735. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.R.; Zhang, H.S.; Li, X.K.; Shen, H.M.; Gao, J.H.; Hou, S.Y.; Zhang, B.; Mayes, S.; Bennett, M.; Ma, J.X.; et al. A mini foxtail millet with an Arabidopsis-like life cycle as a C4 model system. Nat. Plants 2020, 6, 1167–1178. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Gao, J.; Qie, Q.; Yang, Y.; Hou, S.; Wang, X.; Li, X.; Han, Y. Comparative Analysis of Flavonoid Metabolites in Foxtail Millet (Setaria italica) with Different Eating Quality. Life 2021, 11, 578. https://doi.org/10.3390/life11060578
Zhang Y, Gao J, Qie Q, Yang Y, Hou S, Wang X, Li X, Han Y. Comparative Analysis of Flavonoid Metabolites in Foxtail Millet (Setaria italica) with Different Eating Quality. Life. 2021; 11(6):578. https://doi.org/10.3390/life11060578
Chicago/Turabian StyleZhang, Yakun, Jianhua Gao, Qianru Qie, Yulu Yang, Siyu Hou, Xingchun Wang, Xukai Li, and Yuanhuai Han. 2021. "Comparative Analysis of Flavonoid Metabolites in Foxtail Millet (Setaria italica) with Different Eating Quality" Life 11, no. 6: 578. https://doi.org/10.3390/life11060578
APA StyleZhang, Y., Gao, J., Qie, Q., Yang, Y., Hou, S., Wang, X., Li, X., & Han, Y. (2021). Comparative Analysis of Flavonoid Metabolites in Foxtail Millet (Setaria italica) with Different Eating Quality. Life, 11(6), 578. https://doi.org/10.3390/life11060578