Genomic Regions and Candidate Genes Linked to Capped Hock in Pig
Abstract
1. Introduction
2. Materials and Methods
2.1. Genotyping
2.2. Data Processing
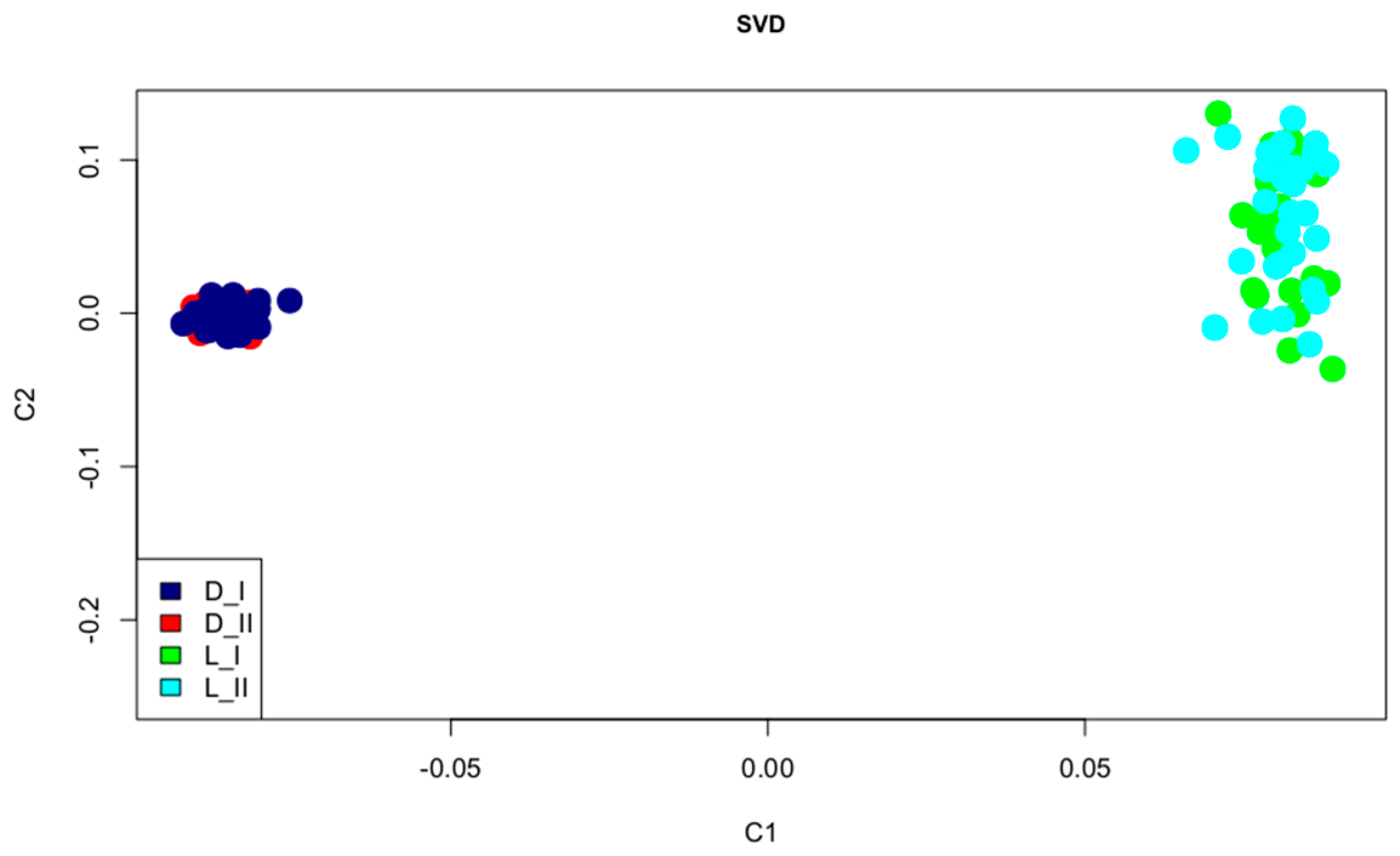
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ostner, F.; Hergt, T.; Klein, S.; Patzkéwitsch, D.; Reese, S.; Brühschwein, A.; Meyer-Lindenberg, A.; Schade, B.; Böhm, B.; Eisenreich, R.; et al. Technopathien der Gliedmaßen bei Mastschweinen: Ursachen, Entstehung, Tierschutzrelevanz [Technopathies of the limbs in finishing pigs: Risk factors, origin and impact on animal welfare—Study phase 1]. Tierarztl. Prax. Ausg. G Grosstiere Nutztiere 2018, 46, 307–315. [Google Scholar]
- Hergt, T.; Ostner, F.; Klein, S.; Zöls, S.; Erhard, M.; Reese, S.; Ritzmann, M.; Patzkéwitsch, D. Technopathien der Gliedmaßen bei Mastschweinen: Ursa chen, Entstehung und Tierschutzrelevanz [Technopathies of the limbs in finishing pigs: Risk factors, origin and impact on animal welfare—Study phase 2]. Tierarztl. Prax. Ausg. G Grosstiere Nutztiere. 2018, 46, 368–377. [Google Scholar]
- Bakoev, S.; Getmantseva, L.; Kolosova, M.; Kostyunina, O.; Chartier, D.R.; Tatarinova, T.V. PigLeg: Prediction of swine phenotype using machine learning. PeerJ 2020, 8, e8764. [Google Scholar] [CrossRef]
- Le, T.H.; Christensen, O.F.; Nielsen, B.; Sahana, G. Genome-wide association study for conformation traits in three Danish pig breeds. Genet. Sel. Evol. 2017, 49, 1–12. [Google Scholar] [CrossRef]
- Van Son, M.; Andersen-Ranberg, I.M.; Lopes, M.S.; Hamland, H.; Grindflek, E. Genome-wide association study and fine mapping of a QTL on SSC13 for Osteochondrosis in Duroc pigs. In Proceedings of the World Congress on Genetics Applied to Livestock Production, Electronic Poster Session: Aotea Centre, Auckland, New Zealand, 11–16 February 2018; Volume 1, p. 276. [Google Scholar]
- Gillman, C.E.; Kilbride, A.L.; Green, L.E. Prevalence of bursitis and capped hock in weaner to finisher pigs and risks associated with flooring factors: A cross-sectional study of 103 GB pig farms. In Proceedings of the ISVEE 11: 11th Symposium of the International Society for Veterinary Epidemiology and Economics, Theme 2—Disease Distribution & Determinants: Pigs Session, Cairns, Australia, 6–11 August 2006; p. 495. [Google Scholar]
- Meyer-Hamme, S.; Lambertz, C.; Gauly, M. Does group size have an impact on welfare indicators in fattening pigs? Animal 2016, 10, 142–149. [Google Scholar] [CrossRef]
- Teixeira, D.L.; Salazar, L.C.; Enriquez-Hidalgo, D.; Boyle, L.A. Assessment of Animal-Based Pig Welfare Outcomes on Farm and at the Abattoir: A Case Study. Front. Vet. Sci. 2020, 7, 576942. [Google Scholar] [CrossRef] [PubMed]
- Gurgul, A.; Jasielczuk, I.; Ropka-Molik, K.; Semik-Gurgul, E.; Pawlina-Tyszko, K.; Szmatoła, T.; Szyndler-Nędza, M.; Bugno-Poniewierska, M.; Blicharski, T.; Szulc, K.; et al. A genome-wide detection of selection signatures in conserved and commercial pig breeds maintained in Poland. BMC Genet. 2018, 19, 1–17. [Google Scholar] [CrossRef] [PubMed]
- De Simoni Gouveia, J.J.; Da Silva, M.V.G.; Paiva, S.R.; De Oliveira, S.M.P. Identification of selection signatures in livestock species. Genet. Mol. Biol. 2014, 37, 330–342. [Google Scholar] [CrossRef] [PubMed]
- Kunhareang, S.; Zhou, H.; Hickford, J. Rapid DNA extraction of pig ear tissues. Meat Sci. 2010, 85, 589–590. [Google Scholar] [CrossRef]
- Barker, V.A.; Blackford, L.S.; Dongarra, J.; Du Croz, J.; Hammarling, S.; Marinova, M.; Waśniewski, J.; Yalamov, P. LAPACK95 Users’ Guide; Society for Industrial & Applied Mathematics (SIAM): Philadelphia, PA, USA, 2001. [Google Scholar]
- VanRaden, P.M. Efficient Methods to Compute Genomic Predictions. J. Dairy Sci. 2008, 91, 4414–4423. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, G.; Smith, A.V.; Jonsson, F.; Gustafsson, O.; Stefansson, K.; Donnelly, P. Assessing population differentiation and isolation from single-nucleotide polymorphism data. J. R. Stat. Soc. Ser. B Stat. Methodol. 2002, 64, 695–715. [Google Scholar] [CrossRef]
- Kilbride, A.; Gillman, C.; Ossent, P.; Green, L. A cross-sectional study of the prevalence and associated risk factors for capped hock and the associations with bursitis in weaner, grower and finisher pigs from 93 commercial farms in England. Prev. Vet. Med. 2008, 83, 272–284. [Google Scholar] [CrossRef]
- Ren, Z.; Wang, Y.; Ren, Y.; Zhang, Z.; Gu, W.; Wu, Z.; Chen, L.; Mou, L.; Li, R.; Yang, H.; et al. Enhancement of porcine intramuscular fat content by overexpression of the cytosolic form of phosphoenolpyruvate carboxykinase in skeletal muscle. Sci. Rep. 2017, 7, srep43746. [Google Scholar] [CrossRef]
- Piedrafita, J.; Christian, L.L.; Lonergan, S.M. Fatty acid profiles in three stress genotypes of swine and relationship with performance, carcass and meat quality traits. Meat Sci. 2001, 57, 71–77. [Google Scholar] [CrossRef]
- Getmantseva, L.; Kolosov, A.; Leonova, M.; Bakoev, S.; Klimenko, A.; Usatov, A.; Radyuk, A.; Vaselenko, V.; Makarenko, M.; Bakoev, N. Polymorphism in obesity—related leptin gene and its association with reproductive traits of sows. Bulg. J. Agric. Sci. 2017, 23, 848–855. [Google Scholar]
- Poklukar, K.; Čandek-Potokar, M.; Lukač, N.B.; Tomažin, U.; Škrlep, M. Lipid Deposition and Metabolism in Local and Modern Pig Breeds: A Review. Animal 2020, 10, 424. [Google Scholar] [CrossRef] [PubMed]
- Vissers, L.E.L.M.; Bonetti, M.; Overman, J.P.; Nillesen, W.M.; Frints, S.G.M.; de Ligt, J.; Zampino, G.; Justino, A.; Machado, J.C.; Schepens, M.; et al. Heterozygous germline mutations in A2ML1 are associated with a disorder clinically related to Noonan syndrome. Eur. J. Hum. Genet. 2014, 23, 317–324. [Google Scholar] [CrossRef] [PubMed]
- Larson, E.D.; Magno, J.P.M.; Steritz, M.J.; Llanes, E.G.D.; Cardwell, J.; Pedro, M.; Roberts, T.B.; Einarsdottir, E.; Rosanes, R.A.Q.; Greenlee, C.; et al. A2ML1and otitis media: Novel variants, differential expression, and relevant pathways. Hum. Mutat. 2019, 40, 1156–1171. [Google Scholar] [CrossRef]
- Kidd, T.; Brose, K.; Mitchell, K.J.; Fetter, R.D.; Tessier-Lavigne, M.; Goodman, C.S.; Tear, G. Roundabout Controls Axon Crossing of the CNS Midline and Defines a Novel Subfamily of Evolutionarily Conserved Guidance Receptors. Cell 1998, 92, 205–215. [Google Scholar] [CrossRef]
- Long, H.; Sabatier, C.; Ma, L.; Plump, A.; Yuan, W.; Ornitz, D.M.; Tamada, A.; Murakami, F.; Goodman, C.S.; Tessier-Lavigne, M. Conserved Roles for Slit and Robo Proteins in Midline Commissural Axon Guidance. Neuron 2004, 42, 213–223. [Google Scholar] [CrossRef]
- Grieshammer, U.; Ma, L.; Plump, A.S.; Wang, F.; Tessier-Lavigne, M.; Martin, G.R. SLIT2-Mediated ROBO2 Signaling Restricts Kidney Induction to a Single Site. Dev. Cell 2004, 6, 709–717. [Google Scholar] [CrossRef]
- Greenberg, J.M.; Thompson, F.Y.; Brooks, S.K.; Shannon, J.M.; Akeson, A.L. Slit and robo expression in the developing mouse lung. Dev. Dyn. 2004, 230, 350–360. [Google Scholar] [CrossRef]
- Prasad, A.; Paruchuri, V.; Preet, A.; Latif, F.; Ganju, R.K. Slit-2 induces a tumor suppressive effect by regulating beta-catenin in breast cancer cells. J. Biol. Chem. 2008, 283, 26624–26633. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, J.; Li, B.H.; Qu, H.; Luo, C.L.; Shu, D.M. An association between genetic variation in the roundabout, axon guidance receptor, homolog 2 gene and immunity traits in chickens. Poult. Sci. 2014, 93, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Bovo, S.; Mazzoni, G.; Bertolini, F.; Schiavo, G.; Galimberti, G.; Gallo, M.; Dall’Olio, S.; Fontanesi, L. Genome-wide association studies for 30 haematological and blood clinical-biochemical traits in Large White pigs reveal genomic regions affecting intermediate phenotypes. Sci. Rep. 2019, 9, 1–17. [Google Scholar] [CrossRef]
- Sakakibara, S.-I.; Nakamura, Y.; Satoh, H.; Okano, H. RNA-Binding Protein Musashi2: Developmentally Regulated Expression in Neural Precursor Cells and Subpopulations of Neurons in Mammalian CNS. J. Neurosci. 2001, 21, 8091–8107. [Google Scholar] [CrossRef]
- Ito, T.; Kwon, H.Y.; Zimdahl, B.; Congdon, K.L.; Blum, J.; Lento, W.E.; Zhao, C.; Lagoo, A.; Gerrard, G.; Foroni, L.; et al. Regulation of myeloid leukaemia by the cell-fate determinant Musashi. Nature 2010, 466, 765–768. [Google Scholar] [CrossRef]
- Wang, S.; Li, N.; Yousefi, M.; Nakauka-Ddamba, A.; Li, F.; Parada, K.; Rao, S.; Minuesa, G.; Katz, Y.; Gregory, B.D.; et al. Faculty Opinions recommendation of Transformation of the intestinal epithelium by the MSI2 RNA-binding protein. Nat. Commun. 2015, 6, 6517. [Google Scholar] [CrossRef] [PubMed]
- Bennett, C.G.; Riemondy, K.; Chapnick, U.A.; Bunker, E.; Liu, X.; Kuersten, S.; Yi, R. Genome-wide analysis of Musashi-2 targets reveals novel functions in governing epithelial cell migration. Nucleic Acids Res. 2016, 44, 3788–3800. [Google Scholar] [CrossRef]
- Okano, H. Stem cell biology of the central nervous system. J. Neurosci. Res. 2002, 69, 698–707. [Google Scholar] [CrossRef]
- Battelli, C.; Nikopoulos, G.N.; Mitchell, J.G.; Verdi, J.M. The RNA-binding protein Musashi-1 regulates neural development through the translational repression of p21WAF-1. Mol. Cell Neurosci. 2006, 31, 85–96. [Google Scholar] [CrossRef] [PubMed]
- Forouzanfar, M.; Lachinani, L.; Dormiani, K.; Nasr-Esfahani, M.H.; Gure, A.O.; Ghaedi, K. Intracellular functions of RNA-binding protein, Musashi1, in stem and cancer cells. Stem Cell Res. Ther. 2020, 11, 1–10. [Google Scholar] [CrossRef]
- Becker, S.F.; Langhe, R.; Huang, C.; Wedlich, R.; Kashef, J. Giving the right tug for migration: Cadherins in tissue movements. Arch. Biochem. Biophys. 2012, 524, 30–42. [Google Scholar] [CrossRef]
- Langhe, R.P.; Gudzenko, T.; Bachmann, M.; Becker, S.F.; Gonnermann, C.; Winter, C.; Abbruzzese, G.; Alfandari, D.; Kratzer, M.-C.; Franz, C.M.; et al. Cadherin-11 localizes to focal adhesions and promotes cell–substrate adhesion. Nat. Commun. 2016, 7, 10909. [Google Scholar] [CrossRef] [PubMed]
- Ding, X.; Zhang, Y.; Huang, Y.; Liu, S.; Lü, H.; Sun, T. Cadherin-11 involves in synovitis and increases the migratory and invasive capacity of fibroblast-like synoviocytes of osteoarthritis. Int. Immunopharmacol. 2015, 26, 153–161. [Google Scholar] [CrossRef]
- Okano, H.; Kawahara, H.; Toriya, M.; Nakao, K.; Shibata, S.; Imai, T. Function of RNA-binding protein Musashi-1 in stem cells. Exp. Cell Res. 2005, 306, 349–356. [Google Scholar] [CrossRef] [PubMed]
| Symbol | Full Name | SSC | Location |
|---|---|---|---|
| SASH1 | SAM and SH3 domain containing 1 | 1 | 17306364..17663117 |
| NAV2 | Neuron navigator 2 | 2 | 39263939..40083539 |
| DTWD2 | DTW domain containing 2 | 2 | 122680254..122937002 |
| SIL1 | SIL1 nucleotide exchange factor | 2 | 140884170..141118201 |
| A2ML1 | Alpha-2-macroglobulin like 1 | 5 | 62603260..62648170 |
| HS6ST3 | Heparan sulfate 6-O-sulfotransferase 3 | 11 | 65516180..66193410 |
| MBNL2 | muscle blind-like splicing regulator 2 | 11 | 66481923..66641653 |
| MSI2 | Musashi RNA binding protein 2 | 12 | 33537163..33974845 |
| ROBO2 | Roundabout guidance receptor 2 | 13 | 177365712..179040027 |
| RIMBP2 | RIMS binding protein 2 | 14 | 24394793..24722237 |
| MSI1 | Musashi RNA binding protein 1 | 14 | 40331371..40356793 |
| SVOP | SV2 related protein | 14 | 41829649..41931359 |
| SART3 | Spliceosome associated factor 3, U4/U6 recycling protein | 14 | 42223752..42259992 |
| SGSM1 | Small G protein signaling modulator 1 | 14 | 42882101..42974755 |
| CDH23 | Cadherin related 23 | 14 | 74267547..74734623 |
| PID1 | Phosphotyrosine interaction domain containing 1 | 15 | 130080821..130215548 |
| DNER | Delta/notch-like EGF repeat containing | 15 | 130414159..130754043 |
| Symbol | Full Name | SSC | Location |
|---|---|---|---|
| RBFOX1 | RNA binding fox-1 homolog 1 | 3 | 34938703..37209772 |
| A2ML1 | Alpha-2-macroglobulin like 1 | 5 | 62603260..62648170 |
| NUMB | NUMB endocytic adaptor protein | 7 | 96624270..96798690 |
| KCNK2 | Potassium two pore domain channel subfamily K member 2 | 9 | 128429232..128659662 |
| DTL | Denticleless E3 ubiquitin protein ligase homolog | 9 | 131225840..131275426 |
| FLT1 | FMs-related receptor tyrosine kinase 1 | 11 | 5620698..5797095 |
| MSI2 | Musashi RNA binding protein 2 | 12 | 33537163..33974845 |
| ZNF385D | Zinc finger protein 385D | 13 | 8234734..9214628 |
| SCN10A | Sodium voltage-gated channel alpha subunit 10 | 13 | 23481537..23570454 |
| SCN11A | Sodium voltage-gated channel alpha subunit 11 | 13 | 23602378..23692281 |
| WDR48 | WD repeat domain 48 | 13 | 23735566..23786312 |
| TTC21A | Tetratricopeptide repeat domain 21A | 13 | 23797518..23830334 |
| ROBO2 | Roundabout guidance receptor 2 | 13 | 177365712..179040027 |
| MYO10 | Myosin X | 16 | 5907111..6145485 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Getmantseva, L.; Kolosova, M.; Bakoev, F.; Zimina, A.; Bakoev, S. Genomic Regions and Candidate Genes Linked to Capped Hock in Pig. Life 2021, 11, 510. https://doi.org/10.3390/life11060510
Getmantseva L, Kolosova M, Bakoev F, Zimina A, Bakoev S. Genomic Regions and Candidate Genes Linked to Capped Hock in Pig. Life. 2021; 11(6):510. https://doi.org/10.3390/life11060510
Chicago/Turabian StyleGetmantseva, Lyubov, Maria Kolosova, Faridun Bakoev, Anna Zimina, and Siroj Bakoev. 2021. "Genomic Regions and Candidate Genes Linked to Capped Hock in Pig" Life 11, no. 6: 510. https://doi.org/10.3390/life11060510
APA StyleGetmantseva, L., Kolosova, M., Bakoev, F., Zimina, A., & Bakoev, S. (2021). Genomic Regions and Candidate Genes Linked to Capped Hock in Pig. Life, 11(6), 510. https://doi.org/10.3390/life11060510





