Shewanella algae and Morganella morganii Coinfection in Cobra-Bite Wounds: A Genomic Analysis
Abstract
1. Introduction
2. Case Presentation
3. Materials and Methods
3.1. Genome Sequencing and Assembly
3.2. Annotation of Protein-Coding Genes, Virulence Factors, and Antibiotic Resistance
3.3. Phylogeny Reconstruction
4. Results
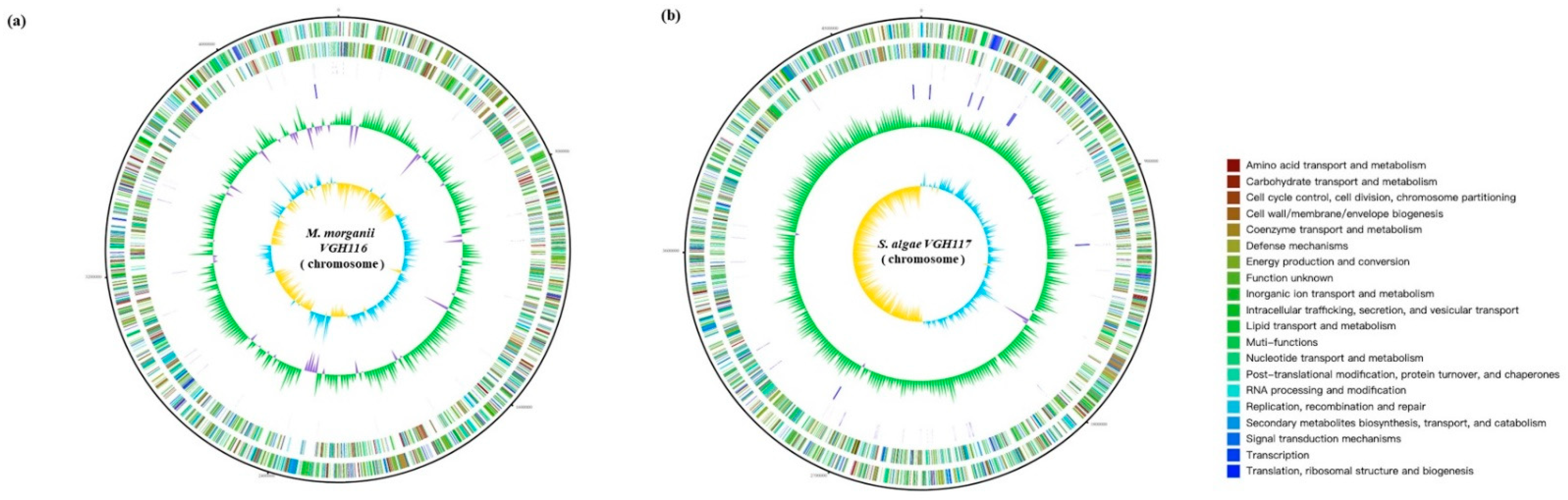
Genome Features of VGH116 and VGH117
5. Discussion
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- World Health Organization. Prevalence of Snakebite Envenoming. Available online: https://www.who.int/snakebites/epidemiology/en/ (accessed on 9 November 2020).
- Chippaux, J.P. Snake-bites: Appraisal of the global situation. Bull. World Health Organ. 1998, 76, 515–524. [Google Scholar] [PubMed]
- Statista. Countries with the Highest Population Density Worldwide in 2019. Available online: https://www.statista.com/statistics/264683/top-fifty-countries-with-the-highest-population-density/ (accessed on 20 November 2020).
- Hung, D.-Z. The Diagnosis and Epidemiological Study of Poisonous Snake Bite in Taiwan. 2010. Available online: https://www.cdc.gov.tw/En/Professional/ProgramResultInfo/ppxd4Xu5zcYwcLHniXKk6w?programResultId=L4ewBx_ufkiqDpeaz0_u5A (accessed on 10 April 2021).
- Shek, K.C.; Tsui, K.L.; Lam, K.K.; Crow, P.; Ng, K.H.; Ades, G.; Yip, K.T.; Grioni, A.; Tan, K.S.; Lung, D.C.; et al. Oral bacterial flora of the Chinese cobra (Naja atra) and bamboo pit viper (Trimeresurus albolabris) in Hong Kong SAR, China. Hong Kong Med. J. 2009, 15, 183–190. [Google Scholar] [PubMed]
- To, K.K.; Wong, S.S.; Cheng, V.C.; Tang, B.S.; Li, I.W.; Chan, J.F.; Seto, W.K.; Tse, H.; Yuen, K.Y. Epidemiology and clinical features of Shewanella infection over an eight-year period. Scand. J. Infect. Dis. 2010, 42, 757–762. [Google Scholar] [CrossRef]
- WHO Regional Office for South-East Asia. Guidelines for the Management of Snakebites, 2nd ed.; WHO Regional Office for South-East Asia: New Delhi, India, 2016. [Google Scholar]
- Theakston, R.; Phillips, R.; Looareesuwan, S.; Echeverria, P.; Makin, T.; Warrell, D. Bacteriological studies of the venom and mouth cavities of wild Malayan pit vipers (Calloselasma rhodostoma) in southern Thailand. Trans. R. Soc. Trop. Med. Hyg. 1990, 84, 875–879. [Google Scholar] [CrossRef]
- Lam, K.K.; Crow, P.; Ng, K.H.; Shek, K.C.; Fung, H.T.; Ades, G.; Grioni, A.; Tan, K.S.; Yip, K.T.; Lung, D.C.; et al. A cross-sectional survey of snake oral bacterial flora from Hong Kong, SAR, China. Emerg. Med. J. EMJ 2011, 28, 107–114. [Google Scholar] [CrossRef] [PubMed]
- Goldstein, E.J.C.; Citron, D.M.; Gonzalez, H.; Russell, F.E.; Finegold, S.M. Bacteriology of Rattlesnake Venom and Implications for Therapy. J. Infect. Dis. 1979, 140, 818–821. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.-M.; Wu, K.-G.; Wang, C.-M. Bacterial infection in association with snakebite: A 10-year experience in a northern Taiwan medical center. J. Microbiol. Immunol. Infect. 2011, 44, 456–460. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.-W.; Wang, J.-D.; Huang, J.-A.; Hu, S.-Y.; Wang, L.-M.; Tsan, Y.-T. Wound infections secondary to snakebite in central Taiwan. J. Venom. Anim. Toxins Incl. Trop. Dis. 2012, 18, 272–276. [Google Scholar] [CrossRef]
- Mao, Y.-C.; Huang, S.-T.; Lai, W.-C.; Yang, C.-C.; Hung, D.-Z.; Liu, P.-Y.; Hung, Y.-M. Bacteriology of Naja atra Snakebite Wound and Its Implications for Antibiotic Therapy. Am. J. Trop. Med. Hyg. 2016, 94, 1129–1135. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.-Y.; Shi, Z.-Y.; Lin, C.-F.; Huang, J.-A.; Liu, J.-W.; Chan, K.-W.; Tung, K.-C. Shewanella infection of snake bites: A twelve-year retrospective study. Clinics 2012, 67, 431–435. [Google Scholar] [CrossRef]
- Holt, H.M.; Gahrn-Hansen, B.; Bruun, B. Shewanella algae and Shewanella putrefaciens: Clinical and microbiological characteristics. Clin. Microbiol. Infect. 2005, 11, 347–352. [Google Scholar] [CrossRef] [PubMed]
- Tsai, Y.-H.; Hsu, W.-H.; Huang, K.-C.; Yu, P.-A.; Chen, C.-L.; Kuo, L.T. Necrotizing fasciitis following venomous snakebites in a tertiary hospital of southwest Taiwan. Int. J. Infect. Dis. 2017, 63, 30–36. [Google Scholar] [CrossRef] [PubMed]
- Poirel, L.; Guibert, M.; Girlich, D.; Naas, T.; Nordmann, P. Cloning, Sequence Analyses, Expression, and Distribution of ampC-ampR from Morganella morganii Clinical Isolates. Antimicrob. Agents Chemother. 1999, 43, 769–776. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Héritier, C.; Poirel, L.; Nordmann, P. Genetic and Biochemical Characterization of a Chromosome-Encoded Carbapenem-Hydrolyzing Ambler Class D β-Lactamase from Shewanella algae. Antimicrob. Agents Chemother. 2004, 48, 1670–1675. [Google Scholar] [CrossRef] [PubMed]
- Tseng, C.-H.; Cheng, J.-F.; Chen, S.-Y.; Chen, W.-H.; Shi, Z.-Y.; Lin, Y.-C.; Tsai, C.-A.; Lin, S.-P.; Chen, Y.-C.; Huang, Y.-T.; et al. Detection of S83V GyrA mutation in quinolone-resistant Shewanella algae using comparative genomics. J. Microbiol. Immunol. Infect. 2020. [Google Scholar] [CrossRef] [PubMed]
- Arakawa, K. Measures of Compositional Strand Bias Related to Replication Machinery and its Applications. Curr. Genom. 2012, 13, 4–15. [Google Scholar] [CrossRef] [PubMed]
- Saravia-Otten, P.; Gutierrez, J.M.; Arvidson, S.; Thelestam, M.; Flock, J.I. Increased infectivity of Staphylococcus aureus in an experimental model of snake venom-induced tissue damage. J. Infect. Dis. 2007, 196, 748–754. [Google Scholar] [CrossRef] [PubMed]
- Lambrecht, E.; Van Coillie, E.; Boon, N.; Heyndrickx, M.; Van de Wiele, T. Transfer of Antibiotic Resistance Plasmid from Commensal E. coli towards Human Intestinal Microbiota in the M-SHIME: Effect of E. coli dosis, Human Individual and Antibiotic Use. Life 2021, 11, 192. [Google Scholar] [CrossRef] [PubMed]

| M. morganii VGH 116 | S. algae VGH 117 | |
|---|---|---|
| MIC (mg/L); Interpretation | ||
| Cefazolin | 64; R | NA |
| Ceftazidime | ≤ 0.12; S | ≤ 0.12; S |
| Ceftriaxone | ≤ 0.25; S | ≤ 0.25; S |
| Cefepime | ≤ 0.12; S | ≤ 0.12; S |
| Piperacillin–tazobactam | ≤ 0.25; S | ≤ 4; S |
| Imipenem | 1; S | 2; S |
| Gentamicin | ≤ 1; S | ≤ 1; S |
| Trimethoprim–sulfamethoxazole | ≤ 20; S | ≤ 20; S |
| Organism | Strain | Genome Size | GC Content | Genes (Coding) | Genes (RNA) |
|---|---|---|---|---|---|
| Morganella morganii | VGH116 | 3.8 Mbp | 50.89% | 4141 | 64 |
| Shewanella algae | VGH117 | 4.7 Mbp | 53.44% | 4102 | 64 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, W.-H.; Kao, C.-C.; Mao, Y.-C.; Lai, C.-S.; Lai, K.-L.; Lai, C.-H.; Tseng, C.-H.; Huang, Y.-T.; Liu, P.-Y. Shewanella algae and Morganella morganii Coinfection in Cobra-Bite Wounds: A Genomic Analysis. Life 2021, 11, 329. https://doi.org/10.3390/life11040329
Huang W-H, Kao C-C, Mao Y-C, Lai C-S, Lai K-L, Lai C-H, Tseng C-H, Huang Y-T, Liu P-Y. Shewanella algae and Morganella morganii Coinfection in Cobra-Bite Wounds: A Genomic Analysis. Life. 2021; 11(4):329. https://doi.org/10.3390/life11040329
Chicago/Turabian StyleHuang, Wei-Hsuan, Chin-Chuan Kao, Yan-Chiao Mao, Chih-Sheng Lai, Kuo-Lung Lai, Chung-Hsu Lai, Chien-Hao Tseng, Yao-Ting Huang, and Po-Yu Liu. 2021. "Shewanella algae and Morganella morganii Coinfection in Cobra-Bite Wounds: A Genomic Analysis" Life 11, no. 4: 329. https://doi.org/10.3390/life11040329
APA StyleHuang, W.-H., Kao, C.-C., Mao, Y.-C., Lai, C.-S., Lai, K.-L., Lai, C.-H., Tseng, C.-H., Huang, Y.-T., & Liu, P.-Y. (2021). Shewanella algae and Morganella morganii Coinfection in Cobra-Bite Wounds: A Genomic Analysis. Life, 11(4), 329. https://doi.org/10.3390/life11040329






