The Effect of Continuous Light on Growth and Muscle-Specific Gene Expression in Atlantic Salmon (Salmo salar L.) Yearlings
Abstract
1. Introduction
2. Results
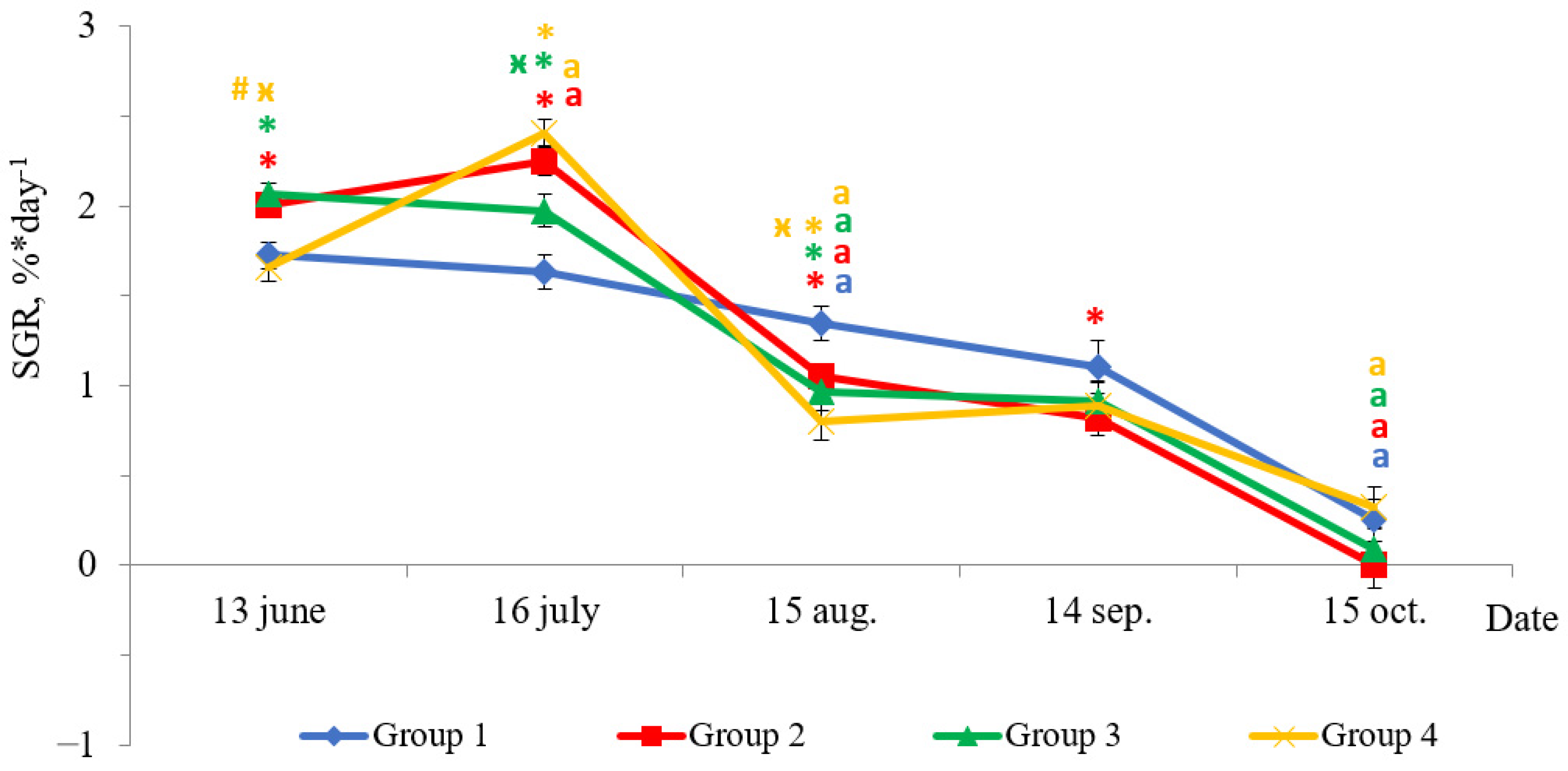
2.1. Weight Gain and Growth Rate
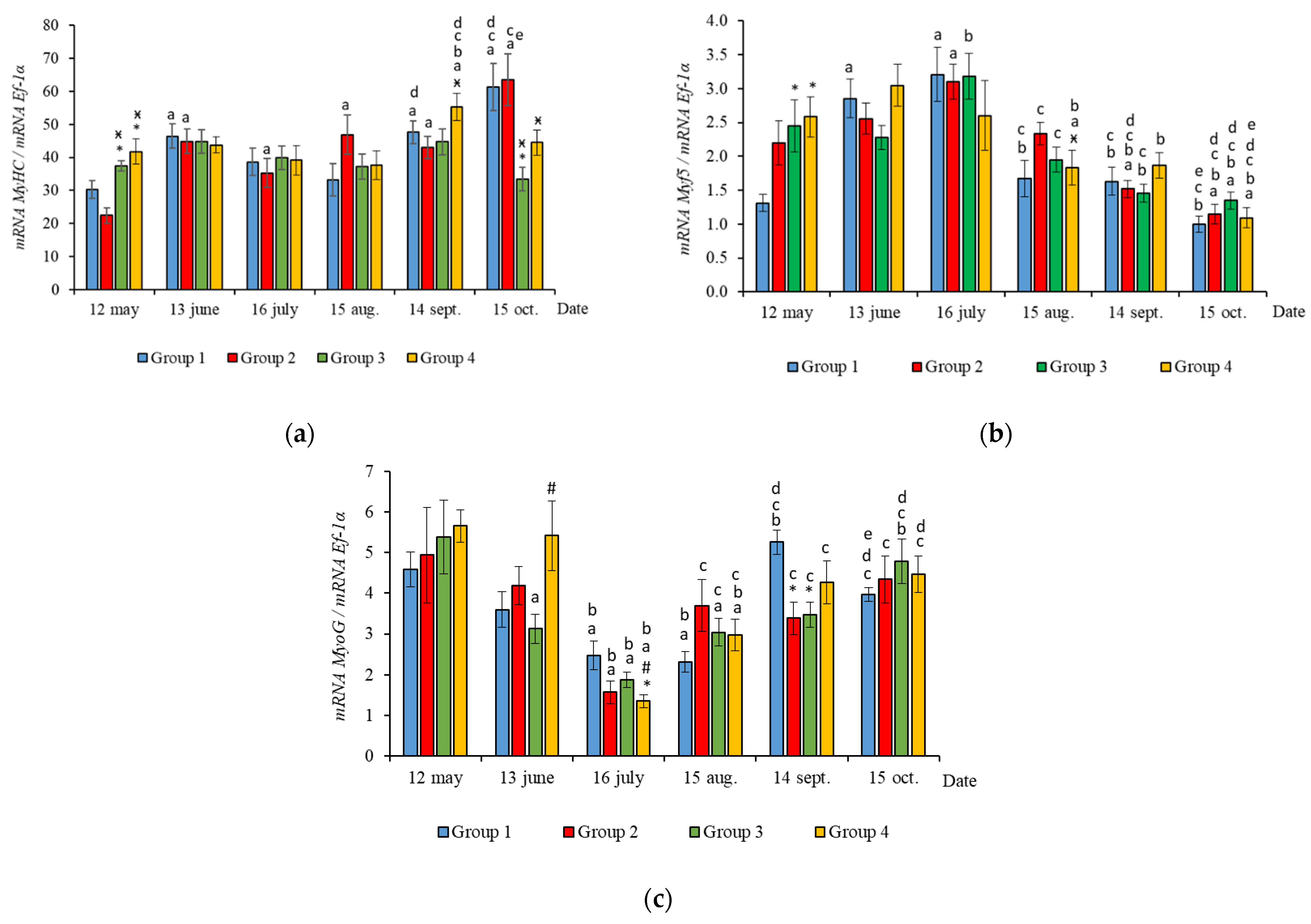
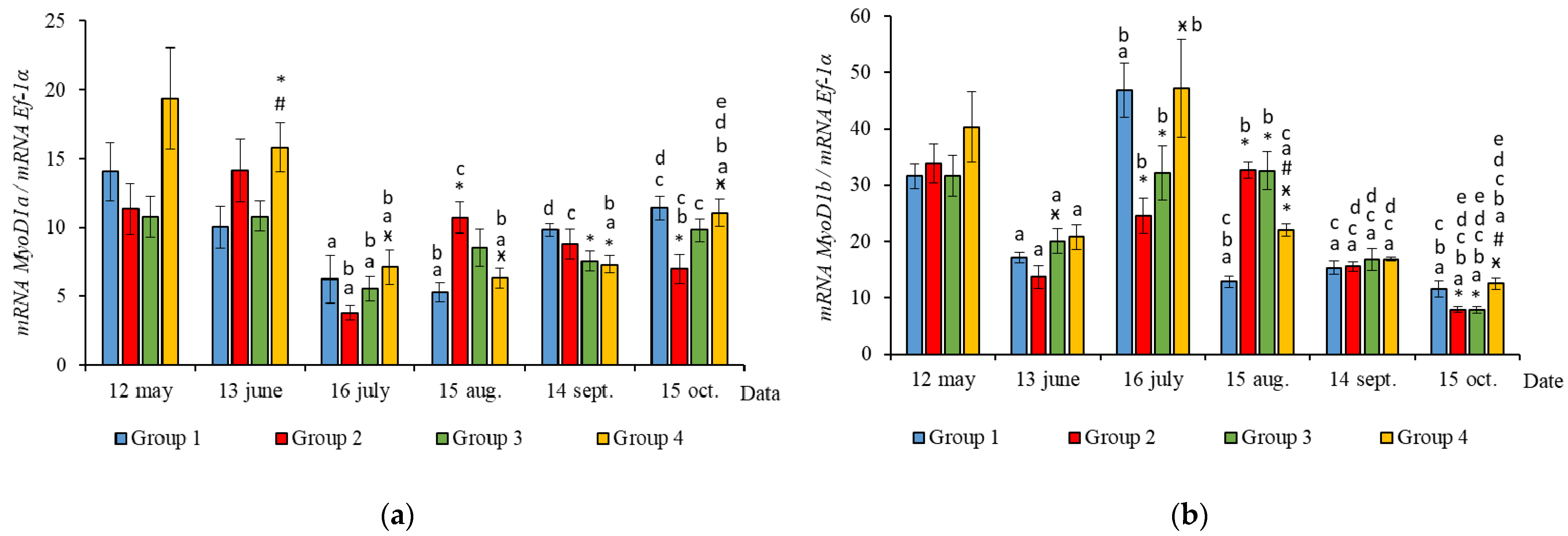
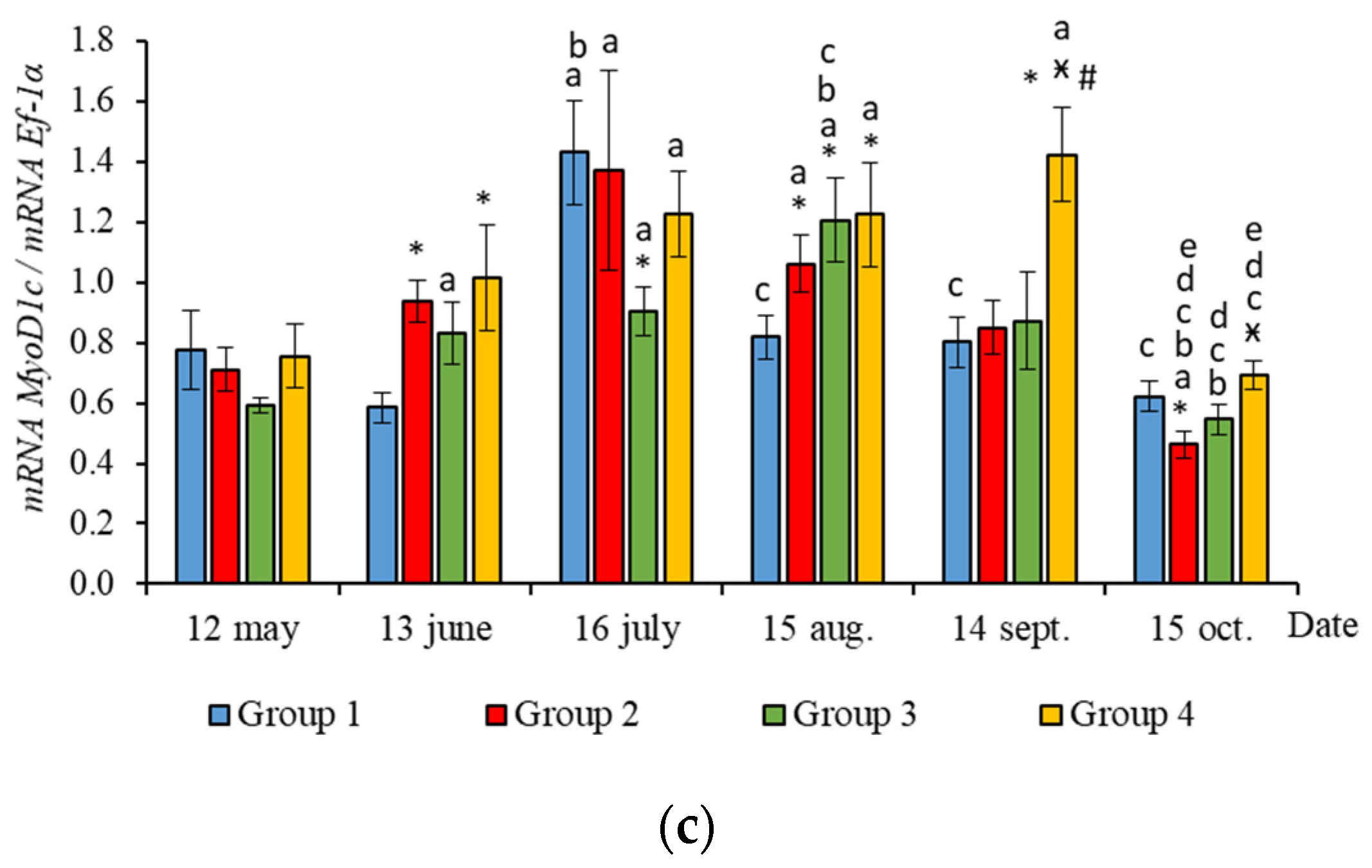
2.2. Muscle-Specific Gene Expression
2.3. The Relationship with the Growth Rate (SGR)
2.4. Correlation Analysis
3. Discussion
3.1. Weight Gain and Growth Rate
3.2. Muscle-Specific Gene mRNA Expression
4. Materials and Methods
4.1. Experimental Design
4.2. Muscle-Specific Gene mRNA Expression
4.3. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Johnston, I.A. Environment and plasticity of myogenesis in teleost fish. J. Exp. Biol. 2006, 209, 2249–2264. [Google Scholar] [CrossRef]
- Watabe, S. Myogenic regulatory factors. In Fish Physiology-Muscle Development and Growth; Johnston, I.A., Ed.; Academic Press: San Diego, CA, USA, 2001; Volume 18, pp. 19–41. [Google Scholar]
- Johansen, K.A.; Overturf, K. Quantitative expression analysis of genes affecting muscle growth during development of rainbow trout (Oncorhynchus mykiss). Mar. Biotech. 2005, 7, 576–587. [Google Scholar] [CrossRef] [PubMed]
- Watabe, S.; Ikeda, D. Diversity of the pufferfish Takifugu rubripes fast skeletal myosin heavy chain genes. Comp. Biochem. Physiol. Part. D Gen. Prot. 2006, 1, 28–34. [Google Scholar] [CrossRef] [PubMed]
- Hevrøy, E.M.; Jordal, A.O.; Hordvik, I.; Espe, M.; Hemrea, G.-I.; Olsvik, P.A. Myosin heavy chain mRNA expression correlates higher with muscle protein accretion than growth in Atlantic salmon Salmo salar. Aquaculture 2006, 252, 453–461. [Google Scholar] [CrossRef]
- Dhillon, R.S.; Esbaugh, Y.S.; Wang, B.L. Characterization and expression of a myosin heavy–chain isoform in juvenile walleye Sander vitreus. J. Fish. Biol. 2009, 75, 1048–1062. [Google Scholar] [CrossRef] [PubMed]
- Gabillard, J.C.; Biga, P.R.; Rescan, P.Y.; Seiliez, I. Revisiting the paradigm of myostatin in vertebrates: Insights from fishes. Gen. Comp. Endocrinol. 2013, 194, 45–54. [Google Scholar] [CrossRef] [PubMed]
- McCroskery, S.; Thomas, M.; Maxwell, L.; Sharma, M.; Kambadur, R. Myostatin negatively regulates satellite cell activation and self-renewal. J. Cell Biol. 2003, 162, 1135–1147. [Google Scholar] [CrossRef] [PubMed]
- Ostaszewska, T.; Dabrowski, K.; Wegner, A.; Krawiec, M. The effects of feeding on muscle growth dynamics and the proliferation of myogenic progenitor cells during pike perch development (Sander lucioperca). J. World Aquac. Soc. 2008, 39, 184–195. [Google Scholar] [CrossRef]
- Chapalamadugu, K.C.; Robison, B.D.; Drew, R.E.; Powell, M.S.; Hill, R.A.; Amberg, J.J.; Rodnick, K.J.; Hardy, R.W.; Hill, M.L.; Murdoch, G.K. Dietary carbohydrate level affects transcription factor expression that regulates skeletal muscle myogenesis in rainbow trout. Comp. Biochem. Physiol. B 2009, 153, 66–72. [Google Scholar] [CrossRef]
- Hagen, Ø.; Fernandes, J.M.O.; Solberg, C.; Johnston, I.A. Expression of growth-related genes in muscle during fasting and refeeding of juvenile Atlantic halibut, Hippoglossus hippoglossus L. Comp. Biochem. Physiol. B 2009, 152, 47–53. [Google Scholar] [CrossRef]
- Johnston, I.A.; McLay, H.A.; Abercromby, M.; Robins, D. Early thermal experience has different effects on growth and muscle fibre recruitment in spring- and autumn-running Atlantic salmon populations. J. Exp. Biol. 2000, 203, 2553–2564. [Google Scholar]
- Wilkes, D.; Xie, S.Q.; Stickland, N.C.; Alami-Durante, H.; Goldspink, G. Temperature and myogenic factor transcript levels during early development determines muscle growth potential in rainbow trout (Oncorhynchus mykiss) and sea bass (Dicentrarchus labrax). J. Exp. Biol. 2001, 204, 2763–2771. [Google Scholar]
- López-Albors, O.; Abdel, I.; Periago, M.J.; Ayala, M.D.; Alcazar, A.G.; Graciá, C.M.; Nathanailides, C.; Vázquez, J.M. Temperature influence on the white muscle growth dynamics of the sea bass Dicentrarchus labrax, L. Flesh quality implications at commercial size. Aquaculture 2008, 277, 39–51. [Google Scholar] [CrossRef]
- Johnston, I.A.; Manthri, S.; Smart, A.; Campbell, P.; Nickell, D.; Alderson, R. Plasticity of muscle fibre number in seawater stages of Atlantic salmon in response to photoperiod manipulation. J. Exp. Biol. 2003, 206, 3425–3435. [Google Scholar] [CrossRef] [PubMed]
- Nagasawa, K.; Giannetto, A.; Fernandes, J.M.O. Photoperiod influences growth and mll (mixed-lineage leukaemia) expression in Atlantic cod. PLoS ONE 2012, 7, e36908. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ayala, M.D.; Abellan, E.; Arizcun, M.; Garcia-Alcazar, A.; Navarro, F.; Blanco, A.; Lopez-Albors, O.M. Muscle development and body growth in larvae and early post-larvae of shi drum, Umbrina cirrosa L., reared under different larval photoperiod: Muscle structural and ultrastructural study. Fish. Physiol. Biochem. 2013, 39, 807–827. [Google Scholar] [CrossRef]
- Boeuf, G.; Falcon, J. Photoperiod and growth in fish. Vie Milieu 2002, 51, 237–246. [Google Scholar]
- Taylor, J.F.; North, B.P.; Porter, M.J.R.; Bromage, N.R.; Migaud, H. Photoperiod can be used to enhance growth and improve feeding efficiency in farmed rainbow trout, Oncorhynchus mykiss. Aquaculture 2006, 256, 216–234. [Google Scholar] [CrossRef]
- Taylor, J.; Migaud, H. Timing and duration of constant light affects rainbow trout (Oncorhynchus mykiss) growth during autumn–spring grow-out in freshwater. Aquac. Res. 2009, 40, 1551–1558. [Google Scholar] [CrossRef]
- Noori, A.; Mojazi Amiri, B.; Mirvaghefi, A.; Rafiee, G.; KalvaniNeitali, B. Enhanced growth and retarded gonadal development of farmed rainbow trout, Oncorhynchus mykiss (Walbaum) following a long-day photoperiod. Aquac. Res. 2015, 46, 2398–2406. [Google Scholar] [CrossRef]
- Imsland, A.K.D.; Roth, B.; Fjelldal, P.G.; Stefansson, S.O.; Handeland, S.; Mikalsen, B. The effect of continuous light at low temperatures on growth in Atlantic salmon reared in commercial size sea pens. Aquaculture 2017, 479, 645–651. [Google Scholar] [CrossRef]
- Saunders, R.L.; Henderson, E.B.; Harmon, P.R. Effects of photoperiod on juvenile growth and smolting of Atlantic salmon and subsequent survival and growth in sea cages. Aquaculture 1985, 45, 55–66. [Google Scholar] [CrossRef]
- Saunders, R.L.; Henderson, E.B. Effects of constant day length on sexual maturation and growth of Atlantic salmon (Salmo salar) parr. Can. J. Fish. Aquatic Sci. 1988, 45, 60–64. [Google Scholar] [CrossRef]
- Handeland, S.O.; Björnsson, B.T.; Arnesen, A.M.; Stefansson, S.O. Seawater adaptation and growth of post-smolt Atlantic salmon (Salmo salar L.) of wild and farmed strain. Aquaculture 2003, 220, 367–384. [Google Scholar] [CrossRef]
- Imsland, A.K.; Folkvord, A.; Stefansson, S.O. Growth, oxygen consumption and activity of juvenile turbot (Scopthalmus maximus L.) reared under different temperatures and photoperiods. Neth. J. Sea Res. 1995, 34, 149–159. [Google Scholar] [CrossRef]
- Petit, G.; Beauchaud, M.; Attia, J.; Buisson, B. Food intake and growth of largemouth bass (Micropterus salmoides) held under alternated light/dark cycle (12L:12D) or exposed to continuous light. Aquaculture 2003, 228, 397–401. [Google Scholar] [CrossRef]
- Jonassen, T.M.; Imsland, A.K.; Kadowaki, S.; Stefabsson, S.O. Interaction of temperature and photoperiod on growth of Atlantic halibut Hippoglossus hippoglossus L. Aquac. Res. 2000, 31, 219–227. [Google Scholar] [CrossRef]
- Kissil, G.W.; Lupatsch, I.; Elizur, A.; Zohar, Y. Long photoperiod delayed spawning and increased somatic growth in gilthead sea bream (Sparus aurata). Aquaculture 2001, 200, 363–379. [Google Scholar] [CrossRef]
- Nordgarden, U.; Oppedal, F.; Taranger, G.L.; Hemre, G.I.; Hansen, T. Seasonally changing metabolism in Atlantic salmon (Salmo salar L.) I–Growth and feed conversion ratio. Aquac. Nutr. 2003, 9, 287–293. [Google Scholar] [CrossRef]
- Björnsson, B.T.; Hemre, G.-I.; Bjørnevik, M.; Hansen, T. Photoperiod regulation of plasma growth hormone levels during induced smoltification of underyearling Atlantic salmon. Gen. Comp. Endocrinol. 2000, 119, 17–25. [Google Scholar] [CrossRef] [PubMed]
- McCormick, S.D.; Moriyama, S.; Björnsson, B.T. Low temperature limits photoperiod control of smolting in Atlantic salmon through endocrine mechanisms. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2000, 278, R1352–R1361. [Google Scholar] [CrossRef] [PubMed]
- Reinecke, M.; Björnsson, B.T.; Dickhoff, W.W.; McCormick, S.D.; Navarro, I.; Power, D.M.; Gutiérrez, J. Growth hormone and insulin-like growth factors in fish: Where we are and where to go. Gen. Comp. Endocrinol. 2005, 142, 20–24. [Google Scholar] [CrossRef] [PubMed]
- Kwasek, K.; Wick, M.; Dabrowski, K. Muscle protein characteristic and its association with faster growth in percids and other teleosts. In Biology and Culture of Percid Fishes, eBook; Kestemont, P., Dabrowski, K., Summerfelt, R.C., Eds.; Springer: New York, NY, USA, 2015; pp. 339–352. [Google Scholar]
- Hansen, T.; Stefansson, S.; Taranger, G.L. Growth and sexual maturation in Atlantic salmon, Salmon salar L., reared in sea cages at two different light regimes. Aquac. Res. 1992, 23, 275–280. [Google Scholar] [CrossRef]
- Bromage, N.; Randall, C.; Davies, B.; Thrush, M.; Duston, J.; Carillo, M.; Zanuy, S. Photoperiodism and the control of reproduction and development in farmed fish. Aquac. Fundam. Appl. Res. 1993, 43, 81–102. [Google Scholar]
- Nemova, N.N.; Nefedova, Z.A.; Pekkoeva, S.N.; Voronin, V.P.; Shulgina, N.S.; Churova, M.V.; Murzina, S.A. The Effect of the Photoperiod on the Fatty Acid Profile and Weight in Hatchery-Reared Underyearlings and Yearlings of Atlantic Salmon Salmo salar L. Biomolecules 2020, 10, 845. [Google Scholar] [CrossRef]
- Churova, M.V.; Meshcheryakova, O.V.; Veselov, A.E.; Nemova, N.N. Activity of enzymes involved in the energy and carbohydrate metabolism and the level of some molecular-genetic characteristics in young salmons (Salmo salar L.) with different age and weight. Russ. J. Dev. Biol. 2015, 46, 254–262. [Google Scholar] [CrossRef]
- Johnston, I.A.; Manthri, S.; Bickerdike, R.; Dingwall, A.; Luijkx, R.; Campbell, P.; Nickell, D.; Alderson, R. Growth performance, muscle structure and flesh quality in out-of-season Atlantic salmon (Salmo salar) smolts reared under two different photoperiod regimes. Aquaculture 2004, 237, 281–300. [Google Scholar] [CrossRef]
- Macqueen, D.J.; Johnston, I.A. A novel salmonid myoD gene is distinctly regulated during development and probably arose by duplication after the genome tetraploidization. FEBS Lett. 2006, 580, 4996–5002. [Google Scholar] [CrossRef] [PubMed]
- Bower, N.I.; Johnston, I.A. Paralogs of Atlantic salmon myoblast determination factor genes are distinctly regulated in proliferating and differentiating myogenic cells. Am. J. Phys. Regul. Integr. Comp. Phys. 2010, 298, R1615–R1626. [Google Scholar] [CrossRef]
- de Almeida, F.L.A.; Carvalho, R.F.; Pinhal, D.; Padovani, C.R.; Martins, C.; Dal Pai-Silva, M. Differential expression of myogenic regulatory factor MyoD in pacu skeletal muscle (Piaractus mesopotamicus Holmberg 1887: Serrasalminae, Characidae, Teleostei) during juvenile and adult growth phases. Micron 2008, 39, 1306–1311. [Google Scholar] [CrossRef]
- Takata, R.; Nakayama, C.L.; e Silva, W.D.S.; Bazzoli, N.; Luz, R.K. The effect of water temperature on muscle cellularity and gill tissue of larval and juvenile Lophiosilurus alexandri, a Neotropical freshwater fish. J. Therm. Biol. 2018, 76, 80–88. [Google Scholar] [CrossRef] [PubMed]
- Valente, L.M.; Bower, N.I.; Johnston, I.A. Postprandial expression of growth-related genes in Atlantic salmon (Salmo salar L.) juveniles fasted for 1 week and fed a single meal to satiation. Br. J. Nutr. 2012, 108, 2148–2157. [Google Scholar] [CrossRef] [PubMed]
- Danzmann, R.G.; Kocmarek, A.L.; Norman, J.D.; Rexroad, C.E.; Palti, Y. Transcriptome profiling in fast versus slow-growing rainbow trout across seasonal gradients. BMC Genom. 2016, 17, 60. [Google Scholar] [CrossRef] [PubMed]
- Rowlerson, A.; Veggetti, A. Cellular mechanisms of post-embryonic muscle growth in aquaculture species. In Fish Physiol.: Muscle Development and Growth; Johnston, I.A., Ed.; Academic Press: San Diego, CA, USA, 2001; Volume 18, pp. 103–140. [Google Scholar]
- Ayala, M.D.; Lopez-Albors, O.; Gil, F.; Garcıa-Alcazar, A.; Abellán, E.; Alarcon, J.A.; Álvarez, M.C.; Ramírez-Zarzosa, G.; Moreno, F. Temperature effects on muscle growth in two populations (Atlantic and Mediterranean) of sea bass, Dicentrarchus labrax L. Aquaculture 2001, 202, 359–370. [Google Scholar] [CrossRef]
- Berkes, C.A.; Tapscott, S.J. MyoD and the transcriptional control of myogenesis. Semin. Cell Dev. Biol. 2005, 16, 585–595. [Google Scholar] [CrossRef]
- Churova, M.V.; Meshcheryakova, O.V.; Veselov, A.E.; Efremov, D.A.; Nemova, N.N. Activity of metabolic enzymes and muscle-specific gene expression in parr and smolts Atlantic salmon Salmo salar L. of different age groups. Fish Physiol. Biochem. 2017, 43, 1117–1130. [Google Scholar] [CrossRef]
- De Almeida, F.L.A.; Pessotti, N.S.; Pinhal, D.; Padovani, C.R.; de Jesus Leitão, N.; Carvalho, R.F.; Martins, C.; Portella, M.C.; Pai-Silva, M.D. Quantitative expression of myogenic regulatory factors MyoD and myogenin in pacu (Piaractus mesopotamicus) skeletal muscle during growth. Micron 2010, 41, 997–1004. [Google Scholar] [CrossRef] [PubMed]
- Churova, M.V.; Shulgina, N.S.; Kuritsyn, A.; Krupnova, M.Y.; Nemova, N.N. Muscle-specific gene expression and metabolic enzyme activities in Atlantic salmon Salmo salar L. fry reared under different photoperiod regimes. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2020, 239, 110330. [Google Scholar] [CrossRef]
- Johnston, I.A. Genetic and environmental determinants of muscle growth patterns. In Fish Physiology; Johnston, I.A., Ed.; Academic Press: Cambridge, MA, USA, 2001; Volume 18, pp. 141–186. [Google Scholar]
- Weber, T.E.; Bosworth, B.G. Effects of 28 day exposure to cold temperature or feed restriction on growth, body composition, and expression of genes related to muscle growth and metabolism in channel catfish. Aquaculture 2005, 246, 483–492. [Google Scholar] [CrossRef]
- Garikipati, D.K.; Rodgers, B.D. Myostatin inhibits myosatellite cell proliferation and consequently activates differentiation: Evidence for endocrine-regulated transcript processing. J. Endocrinol. 2012, 215, 177–187. [Google Scholar] [CrossRef] [PubMed]
- Seiliez, I.; Sabin, N.; Gabillard, J.C. Myostatin inhibits proliferation but not differentiation of trout myoblasts. Mol. Cell. Endocrinol. 2012, 351, 220–226. [Google Scholar] [CrossRef]
- Joulia, D.; Bernardi, H.; Garandel, V.; Rabenoelina, F.; Vernus, B.; Cabello, G. Mechanisms involved in the inhibition of myoblast proliferation and differentiation by myostatin. Exp. Cell Res. 2003, 286, 263–275. [Google Scholar] [CrossRef]
- Johnston, I.A.; Macqueen, D.J.; Watabe, S. Molecular biotechnology of development and growth in fish muscle. In Fisheries for Global Welfare and Environment: Memorial Book of the 5th World Fisheries Congress; Tsukamoto, K., Kawamura, T., Takeuchi, T., Beard, T.D., Kaiser, M.J., Jr., Eds.; Terrapub: Tokyo, Japan, 2008; pp. 241–262. [Google Scholar]
- Johansen, K.A.; Overturf, K. Alterations in expression of genes associated with muscle metabolism and growth during nutritional restriction and refeeding in rainbow trout. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2006, 144, 119–127. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]



| Variable (y) | Group | SGR (x) | Coefficient of Correlation (SGR) |
|---|---|---|---|
| MyHC | 1 | y = 51.94 − 4.76x R2 = 0.057 | −0.24 |
| 2 | y = 60.53 − 10.66x R2 = 0.378 * | −0.61 * | |
| 3 | y = 38.36 + 1.42x R2 = 0.014 | 0.12 | |
| 4 | y = 46.12 − 1.53x R2 = 0.017 | −0.13 | |
| MyoG | 1 | y = 3.98 − 0.35x R2 = 0.036 | −0.19 |
| 2 | y = 4.26 − 0.67x R2 = 0.138 * | −0.37 * | |
| 3 | y = 4.09 − 0.71x R2 = 0.246 * | −0.50 * | |
| 4 | y = 4.29 − 0.47x R2 = 0.056 | −0.24 | |
| Myf5 | 1 | y = 1.60 + 0.36x R2 = 0.061 | 0.25 |
| 2 | y = 1.37 + 0.65x R2 = 0.470 * | 0.69 * | |
| 3 | y = 1.52 + 0.42x R2 = 0.207 * | 0.45 * | |
| 4 | y = 1.46 + 0.45x R2 = 0.184 * | 0.43 * | |
| MyoD1a | 1 | y = 10.23 − 1.35x R2 = 0.086 | −0.29 |
| 2 | y = 9.07 − 0.30x R2 = 0.004 | −0.06 | |
| 3 | y = 8.90 − 0.34x R2 = 0.010 | −0.10 | |
| 4 | y = 9.23 + 0.05x R2 = 0.00 | 0.01 | |
| MyoD1b | 1 | y = 9.78 + 7.78x R2 = 0.178 * | 0.42 * |
| 2 | y = 14.75 + 3.44x R2 = 0.104 | 0.32 | |
| 3 | y = 17.89 + 3.39x R2 = 0.069 | 0.26 | |
| 4 | y = 14.27 + 7.50x R2 = 0.227 * | 0.48 * | |
| MyoD1c | 1 | y = 0.63 + 0.17x R2 = 0.109 * | 0.33 * |
| 2 | y = 0.72 + 0.17x R2 = 0.089 | 0.30 | |
| 3 | y = 0.82 + 0.04x R2 = 0.011 | 0.10 | |
| 4 | y = 1.10 + 0.01x R2 = 0.001 | 0.03 | |
| MSTN1a | 1 | y = 0.66 − 0.05x R2 = 0.027 | −0.16 |
| 2 | y = 0.63 − 0.06x R2 = 0.041 | −0.20 | |
| 3 | y = 0.47 − 0.05x R2 = 0.092 | −0.30 | |
| 4 | y = 0.53 + 0.00x R2 = 0.001 | 0.02 | |
| MSTN1b | 1 | y = 22.37 − 5.04x R2 = 0.300 * | −0.55 * |
| 2 | y = 15.78 − 1.54x R2 = 0.064 | −0.25 | |
| 3 | y = 12.71 − 1.09x R2 = 0.048 | −0.22 | |
| 4 | y = 12.55 + 2.12x R2 = 0.047 | 0.22 |
| Parameter | MyoG | Myf5 | MyoD1a | MyoD1b | MyoD1c | MSTN1a | MSTN1b |
|---|---|---|---|---|---|---|---|
| MyHC | 0.23 | −0.12 | 0.36 * | −0.23 | 0.01 | 0.27 | 0.51 * |
| (p = 0.13) | (p = 0.43) | (p = 0.04) | (p = 0.18) | (p = 0.97) | (p = 0.10) | (p = 0.00) | |
| MyoG | −0.17 | 0.64 * | −0.20 | −0.24 | −0.35 * | 0.27 | |
| (p = 0.27) | (p = 0.00) | (p = 0.22) | (p = 0.11) | (p = 0.03) | (p = 0.10) | ||
| Myf5 | −0.19 | 0.47 * | 0.51 * | 0.26 | −0.13 | ||
| (p = 0.27) | (p = 0.00) | (p = 0.00) | (p = 0.10) | (p = 0.43) | |||
| MyoD1a | 0.20 | −0.17 | −0.39 * | 0.25 | |||
| (p = 0.28) | (p = 0.34) | (p = 0.03) | (p = 0.18) | ||||
| MyoD1b | 0.66 * | 0.01 | −0.19 | ||||
| (p = 0.00) | (p = 0.95) | (p = 0.28) | |||||
| MyoD1c | 0.41 * | −0.04 | |||||
| (p = 0.01) | (p = 0.80) | ||||||
| MSTN1a | 0.46 * | ||||||
| (p = 0.01) |
| Parameter | MyoG | Myf5 | MyoD1a | MyoD1b | MyoD1c | MSTN1a | MSTN1b |
|---|---|---|---|---|---|---|---|
| MyHC | 0.33 * | −0.39 * | 0.20 | −0.37 * | −0.02 | 0.35 * | 0.35 * |
| (p = 0.04) | (p = 0.01) | (p = 0.25) | (p = 0.03) | (p = 0.90) | (p = 0.03) | (p = 0.04) | |
| MyoG | −0.12 | 0.69 * | 0.18 | −0.21 | −0.07 | 0.29 | |
| (p = 0.45) | (p = 0.00) | (p = 0.29) | (p = 0.17) | (p = 0.68) | (p = 0.08) | ||
| Myf5 | 0.26 | 0.45 * | 0.30 * | −0.03 | −0.16 | ||
| (p = 0.12) | (p = 0.00) | (p = 0.04) | (p = 0.84) | (p = 0.35) | |||
| MyoD1a | 0.14 | 0.22 | −0.15 | 0.10 | |||
| (p = 0.44) | (p = 0.19) | (p = 0.39) | (p = 0.56) | ||||
| MyoD1b | 0.53 * | 0.23 | 0.40 * | ||||
| (p = 0.00) | (p = 0.19) | (p = 0.02) | |||||
| MyoD1c | 0.30 * | 0.38 * | |||||
| (p = 0.047) | (p = 0.02) | ||||||
| MSTN1a | 0.56 * | ||||||
| (p = 0.00) |
| Parameter | MyoG | Myf5 | MyoD1a | MyoD1b | MyoD1c | MSTN1a | MSTN1b |
|---|---|---|---|---|---|---|---|
| MyHC | 0.03 | 0.14 | 0.35 * | 0.28 | 0.25 | 0.40 * | −0.13 |
| (p = 0.87) | (p = 0.41) | (p = 0.04) | (p = 0.14) | (p = 0.12) | (p = 0.02) | (p = 0.46) | |
| MyoG | −0.19 | 0.65 * | 0.10 | −0.29 | −0.23 | 0.37 * | |
| (p = 0.25) | (p = 0.00) | (p = 0.59) | (p = 0.07) | (p = 0.18) | (p = 0.03) | ||
| Myf5 | −0.08 | 0.71 * | 0.23 | −0.12 | −0.02 | ||
| (p = 0.63) | (p = 0.00) | (p = 0.12) | (p = 0.48) | (p = 0.92) | |||
| MyoD1a | 0.06 | −0.08 | 0.03 | 0.11 | |||
| (p = 0.77) | (p = 0.61) | (p = 0.87) | (p = 0.54) | ||||
| MyoD1b | 0.46 * | 0.11 | −0.04 | ||||
| (p = 0.01) | (p = 0.54) | (p = 0.83) | |||||
| MyoD1c | 0.35 * | −0.10 | |||||
| (p = 0.02) | (p = 0.56) | ||||||
| MSTN1a | 0.15 | ||||||
| (p = 0.42) |
| Parameter | MyoG | Myf5 | MyoD1a | MyoD1b | MyoD1c | MSTN1a | MSTN1b |
|---|---|---|---|---|---|---|---|
| MyHC | 0.32 * | 0.19 | 0.32 | 0.17 | 0.27 | 0.45 * | 0.63 * |
| (p = 0.04) | (p = 0.26) | (p = 0.06) | (p = 0.33) | (p = 0.10) | (p = 0.00) | (p = 0.00) | |
| MyoG | 0.18 | 0.56 * | −0.15 | −0.13 | −0.08 | 0.53 * | |
| (p = 0.26) | (p = 0.00) | (p = 0.39) | (p = 0.40) | (p = 0.61) | (p = 0.00) | ||
| Myf5 | 0.41 * | 0.58 * | 0.17 | 0.06 | 0.40 * | ||
| (p = 0.02) | (p = 0.00) | (p = 0.30) | (p = 0.71) | (p = 0.02) | |||
| MyoD1a | 0.59 * | −0.36 * | −0.17 | 0.62 * | |||
| (p = 0.00) | (p = 0.04) | (p = 0.36) | (p = 0.00) | ||||
| MyoD1b | 0.04 | 0.17 | 0.58 * | ||||
| (p = 0.80) | (p = 0.32) | (p = 0.00) | |||||
| MyoD1c | 0.29 | 0.08 | |||||
| (p = 0.08) | (p = 0.67) | ||||||
| MSTN1a | 0.21 | ||||||
| (p = 0.23) |
| Date | Group | Regime | n | Average Weight, g | Average Length, cm |
|---|---|---|---|---|---|
| 12 May | 1 | HL | 10 | 4.20 ± 0.16 | 7.75 ± 0.08 |
| 2 | LD 24:0 (May–October 2018) | 10 | 4.22 ± 0.16 | 7.66 ± 0.11 | |
| 3 | LD 24:0 (August–November 2017 and May–October 2018) | 10 | 4.23 ± 0.19 | 7.76 ± 0.13 | |
| 4 | LD 24:0 (August 2017–October 2018) | 10 | 4.22 ± 0.17 | 7.64 ± 0.10 | |
| 13 June | 1 | HL | 10 | 7.55 ± 0.45 | 8.93 ± 0.13 |
| 2 | LD 24:0 (May–October 2018) | 10 | 8.15 ± 0.39 | 9.13 ± 0.15 | |
| 3 | LD 24:0 (August–November 2017 and May–October 2018) | 10 | 8.17 ± 0.40 | 9.17 ± 0.20 | |
| 4 | LD 24:0 (August 2017–October 2018) | 10 | 8.39 ± 0.33 | 9.19 ± 0.14 | |
| 16 July | 1 | HL | 10 | 14.15 ± 0.74 | 10.53 ± 0.17 |
| 2 | LD 24:0 (May–October 2018) | 10 | 17.06 ± 0.85 | 11.21± 0.18 | |
| 3 | LD 24:0 (August–November 2017 and May–October 2018) | 10 | 17.39 ± 1.03 | 11.16 ± 0.23 | |
| 4 | LD 24:0 (August 2017–October 2018) | 10 | 16.85 ± 0.77 | 11.09 ± 0.16 | |
| 15 August | 1 | HL | 10 | 20.10 ± 1.23 | 11.94 ± 0.27 |
| 2 | LD 24:0 (May–October 2018) | 10 | 22.94 ± 0.93 | 12.33 ± 0.14 | |
| 3 | LD 24:0 (August–November 2017 and May–October 2018) | 10 | 23.00 ± 1.20 | 12.24 ± 0.23 | |
| 4 | LD 24:0 (August 2017–October 2018) | 10 | 21.86 ± 2.44 | 12.13 ± 0.42 | |
| 14 September | 1 | HL | 10 | 30.31 ± 2.34 | 13.91 ± 0.37 |
| 2 | LD 24:0 (May–October 2018) | 10 | 31.18 ± 1.43 | 13.89 ± 0.25 | |
| 3 | LD 24:0 (August–November 2017 and May–October 2018) | 10 | 32.80 ± 2.28 | 14.11 ± 0.35 | |
| 4 | LD 24:0 (August 2017–October 2018) | 10 | 32.55 ± 2.44 | 14.08 ± 0.33 | |
| 15 October | 1 | HL | 10 | 33.58 ± 2.56 | 15.85 ± 0.29 |
| 2 | LD 24:0 (May–October 2018) | 10 | 31.90 ± 2.82 | 14.13 ± 0.41 | |
| 3 | LD 24:0 (August–November 2017 and May–October 2018) | 10 | 33.24 ± 2.58 | 14.21 ± 0.40 | |
| 4 | LD 24:0 (August 2017–October 2018) | 10 | 34.74 ± 2.19 | 14.36 ± 0.29 |
| Gene | Primer Sequence (5′–3′) | Size of Amplified Fragment, bp | GenBank Accession No. |
|---|---|---|---|
| Ef-1α | F: TTGCTGGTGGTGTTGGTGAG R: AAACGCTTCTGGCTGTAGGG | 154 | AF321836.1 |
| MyoG | F: GTGGAGATCCTGAGGAGTGC R: CTCACTCGACGACGAGACC | 147 | DQ452070 |
| MyoD1a | F: TGGACTGCCTATCAAACATCC R: TCTCACTCGCTATGGAACC | 123 | AJ557148 |
| MyoD1b | F: ATTTCGTTCCCTGTCACCTCTG R: ATGTGTTCGTCTTCGTTGTAATGG | 152 | AJ557150 |
| MyoD1c | F: ACGGCGAAAACTACTACCCTTC R: TAGCTGCTTCGTCTTGCGGA | 133 | DQ366709.1 |
| Myf5 | F: ACGCCATCCAGTACATCGAG R: AGTCAACCATGCTGTCGGAG | 132 | DQ452070 |
| MyHC | F: TCTCATCCATAGACGCCATC R: AGTTGACTGCCAAGAAGAGG | 159 | DN164736 |
| MSTN1a | F: GATTACACGCCATCAAGTCC R: CTCCATCCTTATTGTCATCTCC | 159 | AJ344158 |
| MSTN1b | F: TCTGAGTTTTATGGTTGCTTTCGG R: TTGTGACTTGATGGCGTGTAATC | 151 | NM_001123634.1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shulgina, N.S.; Churova, M.V.; Murzina, S.A.; Krupnova, M.Y.; Nemova, N.N. The Effect of Continuous Light on Growth and Muscle-Specific Gene Expression in Atlantic Salmon (Salmo salar L.) Yearlings. Life 2021, 11, 328. https://doi.org/10.3390/life11040328
Shulgina NS, Churova MV, Murzina SA, Krupnova MY, Nemova NN. The Effect of Continuous Light on Growth and Muscle-Specific Gene Expression in Atlantic Salmon (Salmo salar L.) Yearlings. Life. 2021; 11(4):328. https://doi.org/10.3390/life11040328
Chicago/Turabian StyleShulgina, Natalia S., Maria V. Churova, Svetlana A. Murzina, Marina Yu. Krupnova, and Nina N. Nemova. 2021. "The Effect of Continuous Light on Growth and Muscle-Specific Gene Expression in Atlantic Salmon (Salmo salar L.) Yearlings" Life 11, no. 4: 328. https://doi.org/10.3390/life11040328
APA StyleShulgina, N. S., Churova, M. V., Murzina, S. A., Krupnova, M. Y., & Nemova, N. N. (2021). The Effect of Continuous Light on Growth and Muscle-Specific Gene Expression in Atlantic Salmon (Salmo salar L.) Yearlings. Life, 11(4), 328. https://doi.org/10.3390/life11040328







