Classification of the Biogenicity of Complex Organic Mixtures for the Detection of Extraterrestrial Life
Abstract
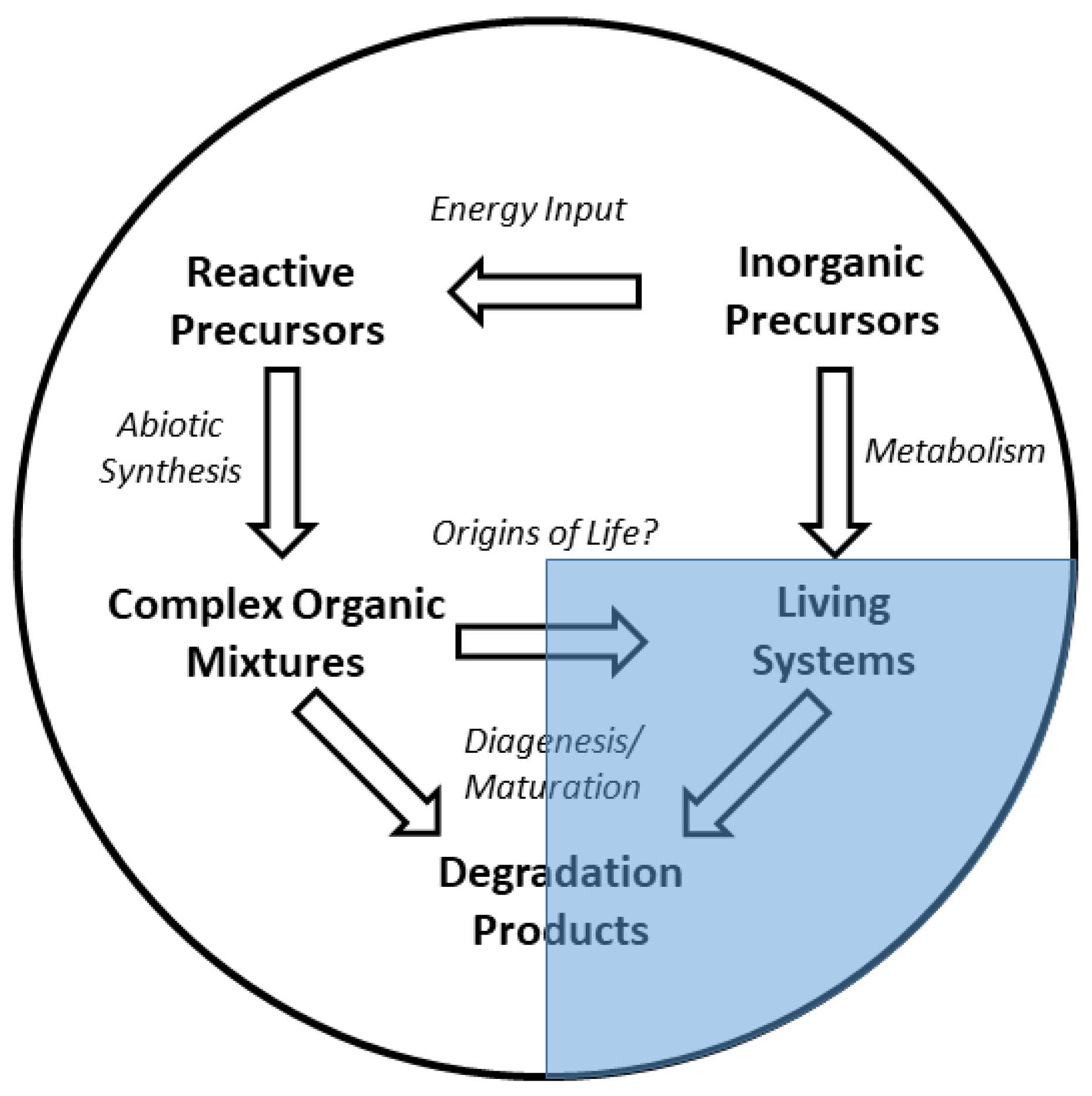
1. Introduction
2. Materials and Methods
2.1. General
2.2. Laboratory Abiological Simulants
- The reaction of aqueous NH3/HCHO/CH3CHO with or without glycolaldehyde (HOCH2CHO) as described in [49], which has been shown to produce a variety of amino acids observed in carbonaceous meteorites.
- Maillard reactions of glucose and glycine prepared by the methods of [52], heated either in aqueous solution or the dry state. As modern terrestrial organisms are predominantly composed of protein by dry weight and also contain significant amounts of sugar-containing materials [53], the reactions of these two compounds are perhaps good, though somewhat simplistic, general analogues for thermal alteration of terrestrial biological material.
- Maillard reactions of glucose and NH4OH heated in aqueous solution at 85 °C. As modern terrestrial organisms are predominantly composed of protein by dry weight and also contain significant amounts of sugar-containing materials [53], this model reaction examines the contribution of ammonia species to complex formose reaction products.
- Caramelized glucose, which may simulate the thermal alteration of formose-derived sugars, as well as glucose heated in water at 85 °C or 150 °C.
- Formose reactions prepared from the reaction for aqueous glycolaldehyde and formaldehyde, which have been suggested to produce good analogues for the macromolecular organic matter observed in carbonaceous meteorites [48].
- A commercial 70% (~13.2 M) aqueous glycolonitrile solution which had been left sealed from the manufacturer on the laboratory shelf at room temperature (~25 °C) for approximately 20 years. This reaction explores the long term outcome of combined HCN/HCHO chemistry.
- Aqueous or neat pyruvic acid (5.7 M) subjected to heating at 150 °C. Pyruvic acid has been implicated as an intermediate in the abiotic synthesis of tricarboxylic acid cycle components [54].
2.3. Petroleum Samples
2.4. Meteorite Samples
2.5. Biological Samples
2.6. FT-ICR MS
2.7. Ionization
2.8. Mass Calibration and Data Analysis
2.9. Computational Methods
3. Results and Discussion
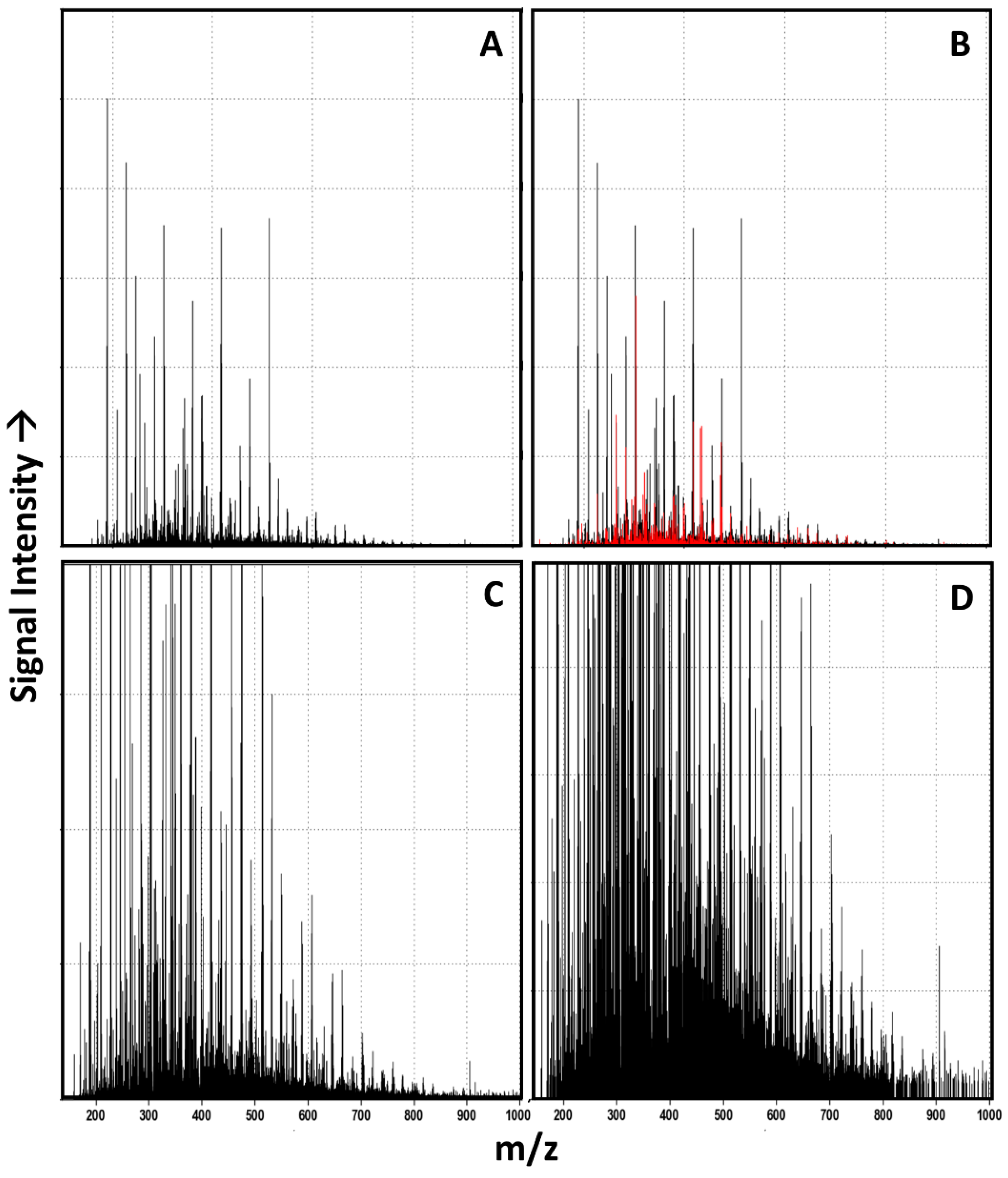
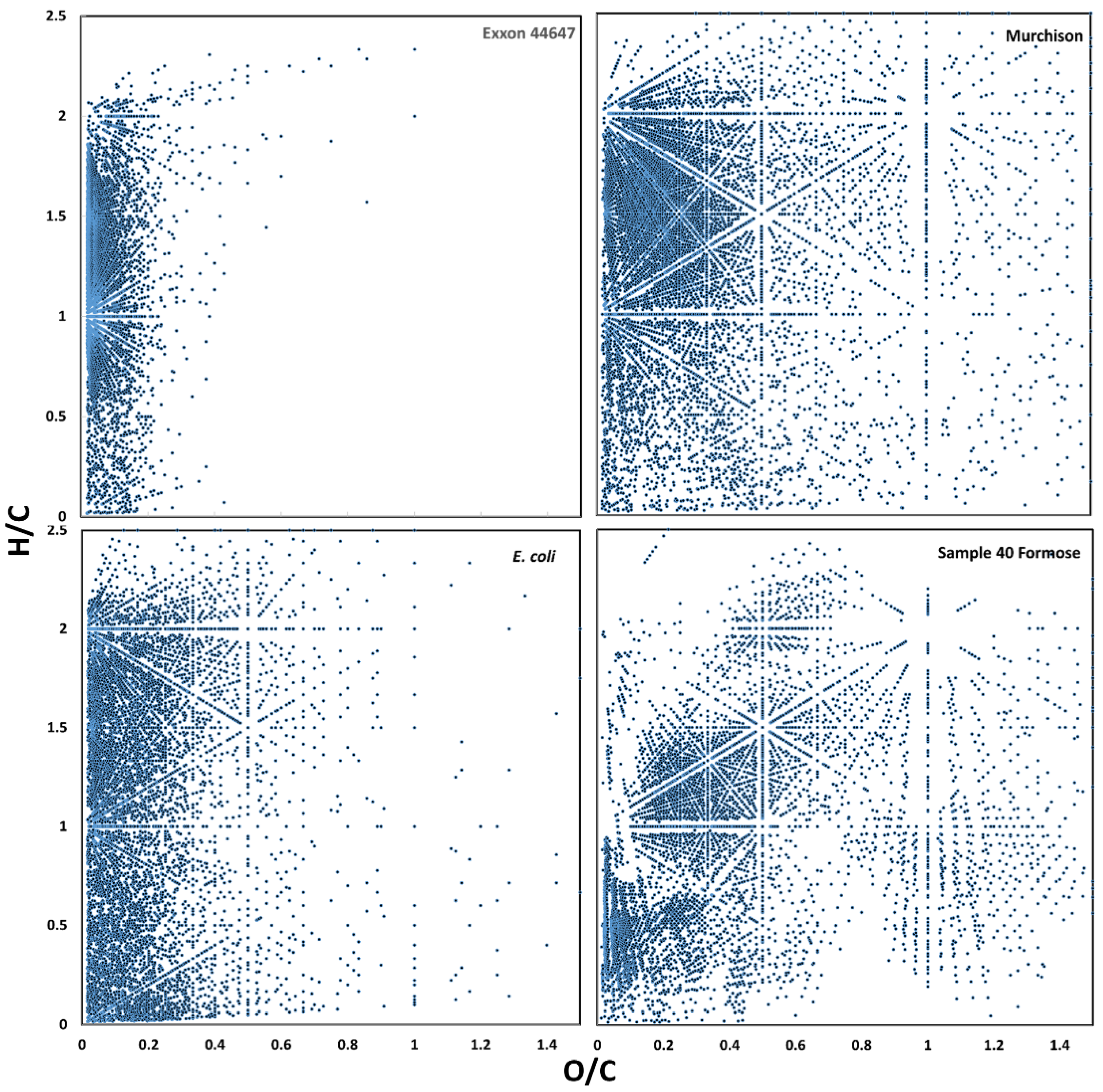
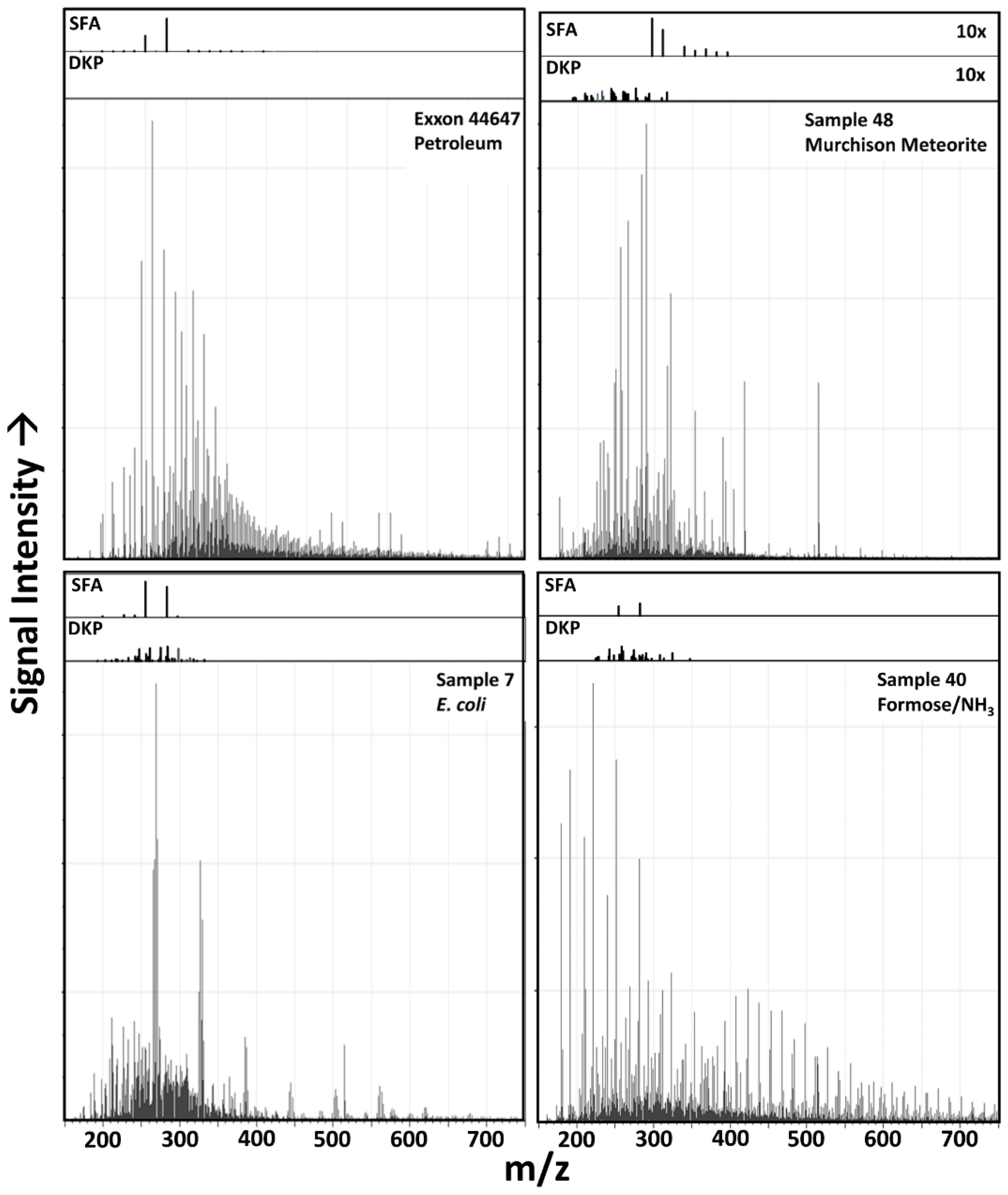
3.1. Raw Data Analysis
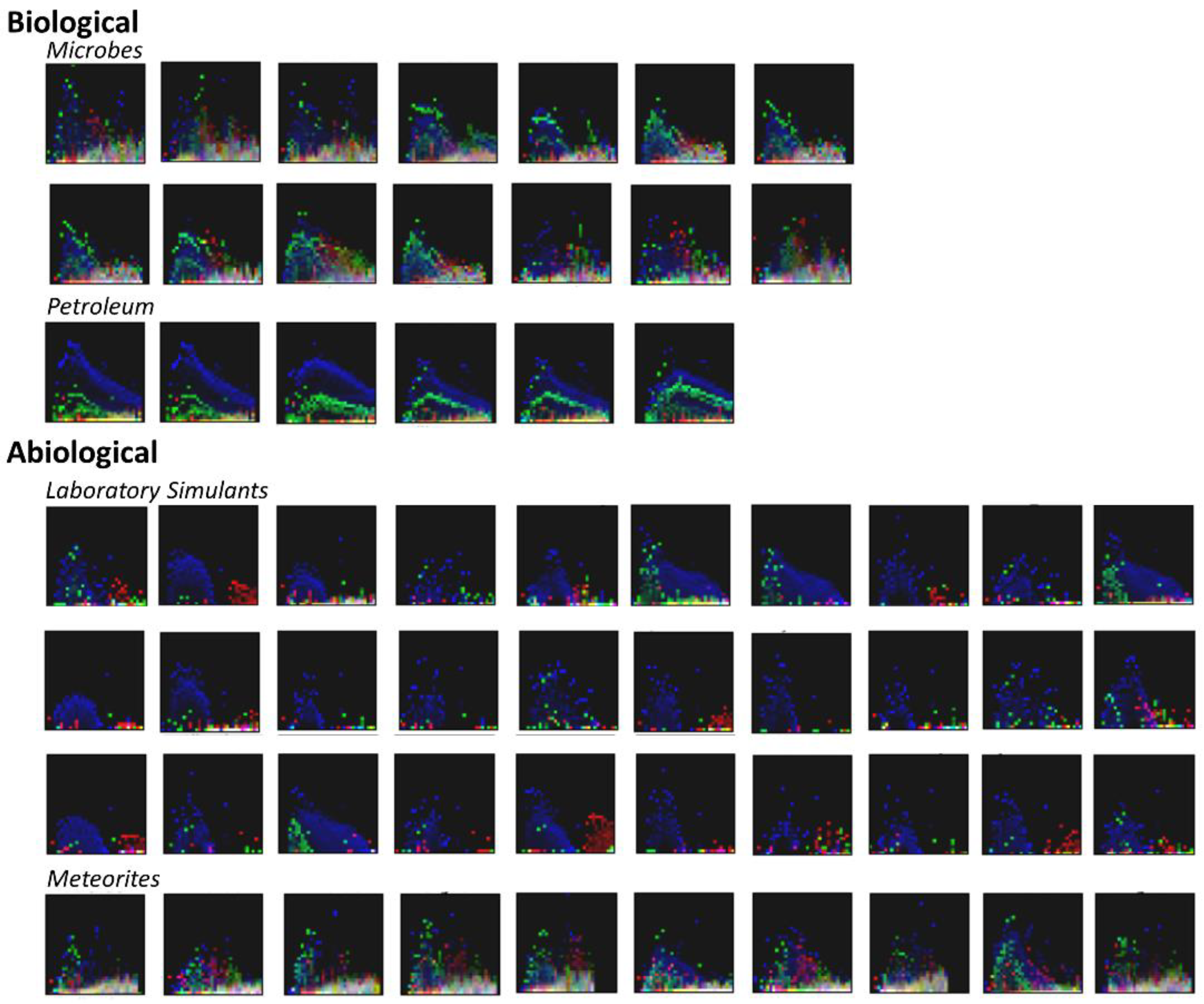
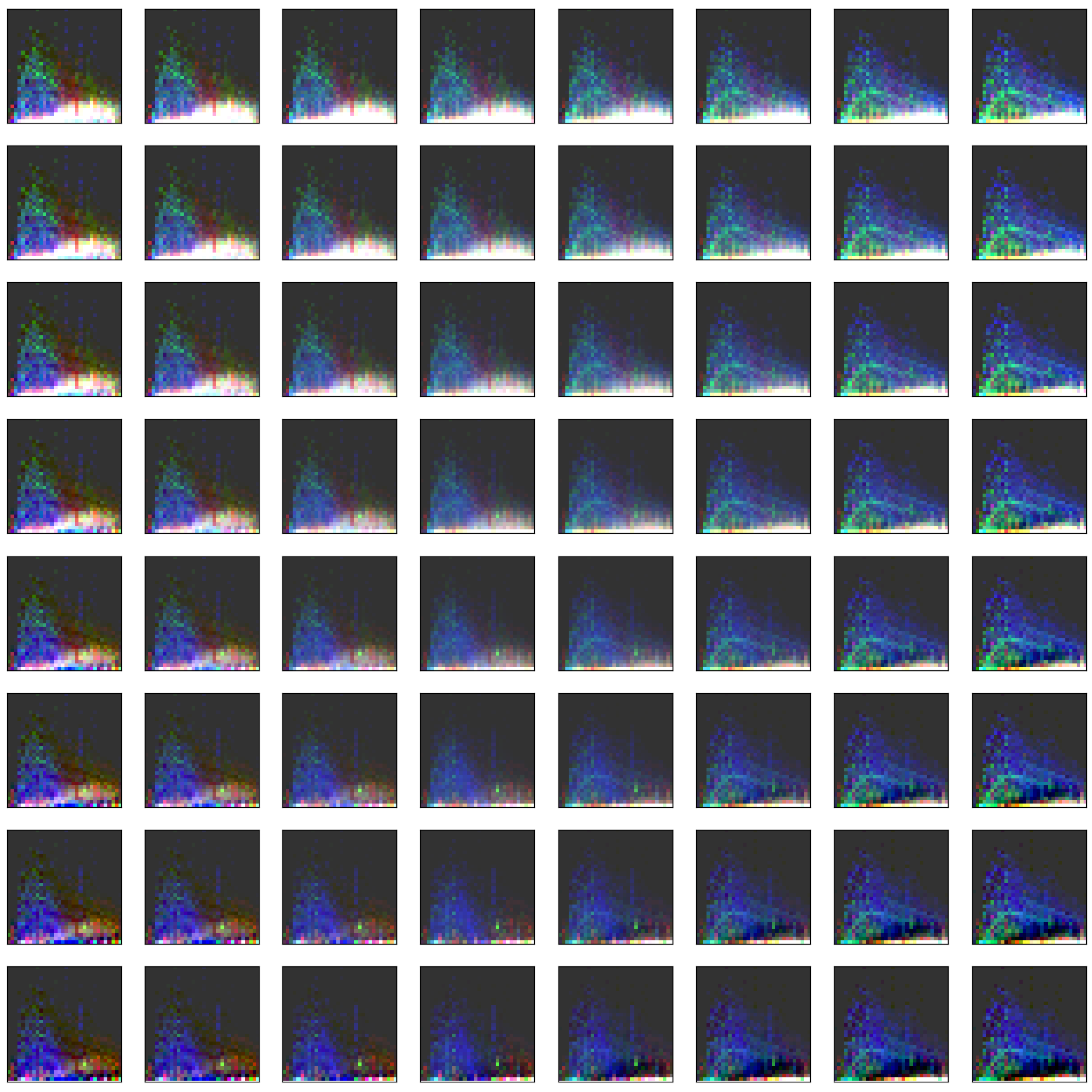
3.2. Visualization of Binned Data
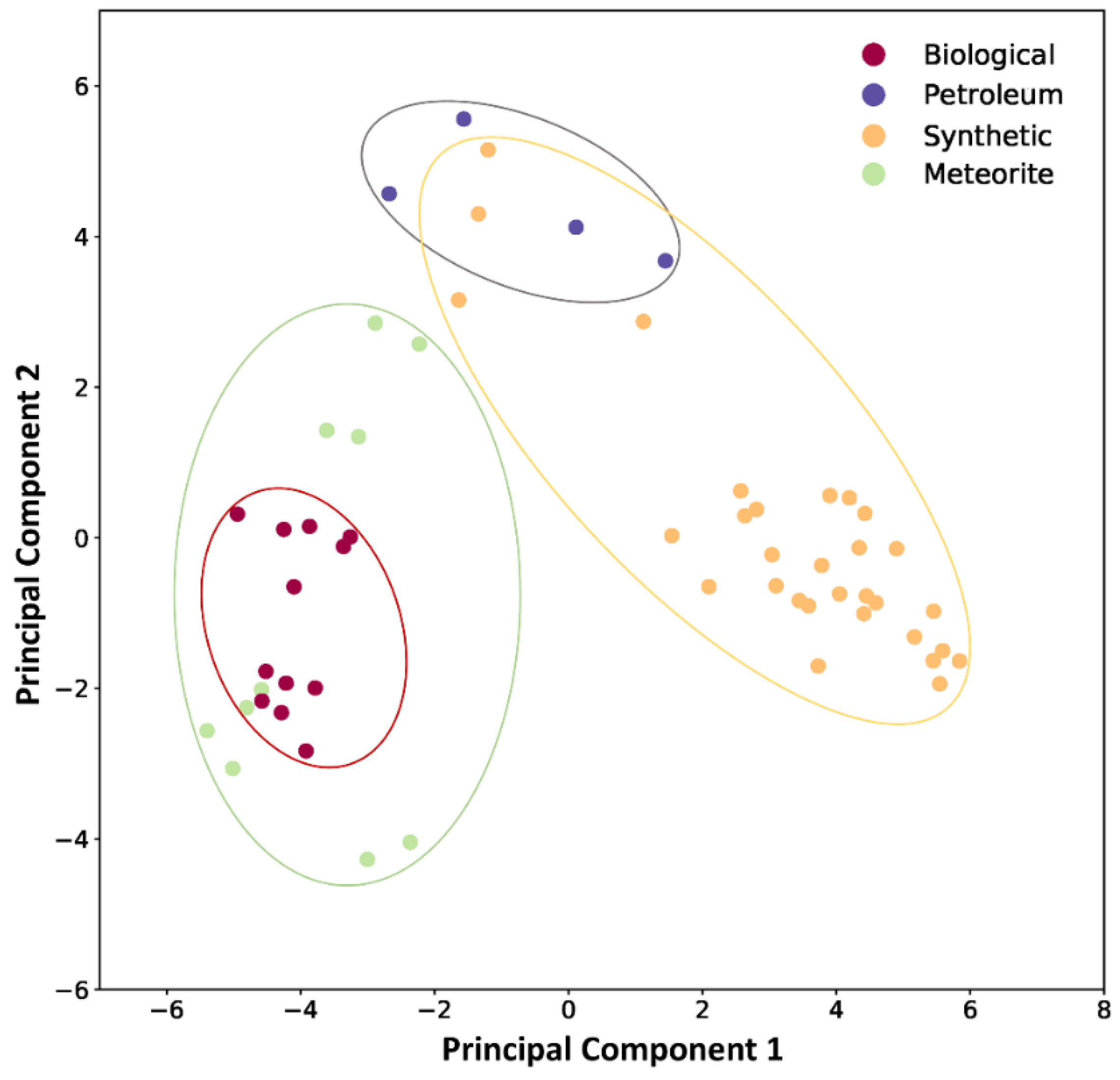
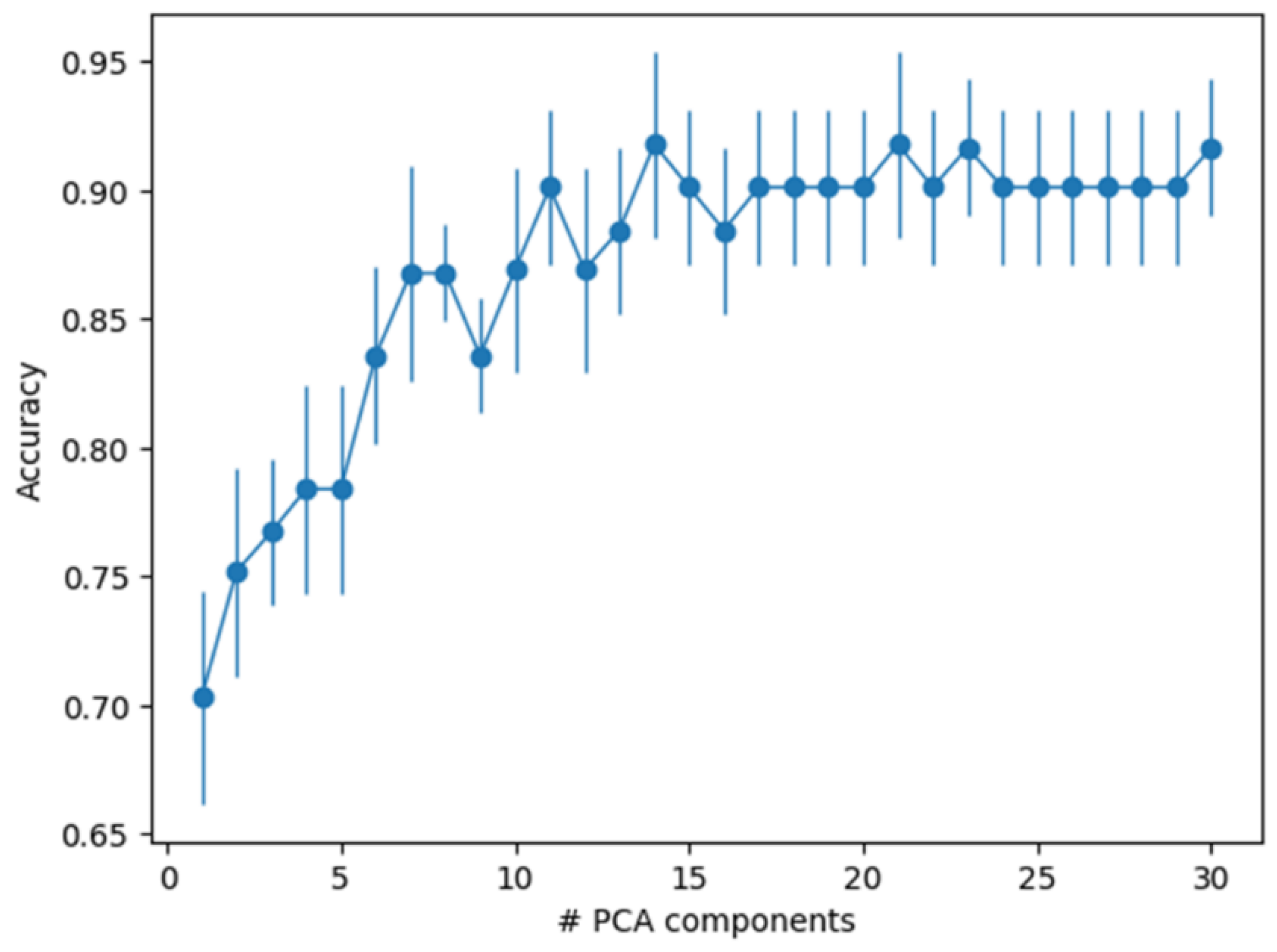
3.3. Principal Component Analysis
3.4. Machine Learning Sample Classification
3.5. Usefulness of Relational Binning Data to Detect Underlying Chemical Patterns
3.6. Possible Molecular Sources of Classification
3.7. Potential Application to Detection of Ancient Terrestrial Biomarkers
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Des Marais, D.J.; Nuth, J.A.; Allamandola, L.J.; Boss, A.P.; Farmer, J.D.; Hoehler, T.M.; Jakosky, B.M.; Meadows, V.S.; Pohorille, A.; Runnegar, B.; et al. The NASA astrobiology roadmap. Astrobiology 2008, 8, 715–730. [Google Scholar] [CrossRef] [PubMed]
- Neveu, M.; Hays, L.E.; Voytek, M.A.; New, M.H.; Schulte, M.D. The ladder of life detection. Astrobiology 2018, 18, 1375–1402. [Google Scholar] [CrossRef] [PubMed]
- Chan, M.A.; Hinman, N.W.; Potter-McIntyre, S.L.; Schubert, K.E.; Gillams, R.J.; Awramik, S.M.; Boston, P.J.; Bower, D.M.; Des Marais, D.J.; Farmer, J.D.; et al. Deciphering biosignatures in planetary contexts. Astrobiology 2019, 19, 1075–1102. [Google Scholar] [CrossRef] [PubMed]
- Walker, S.I.; Cronin, L.; Drew, A.; Domagal-Goldman, S.; Fisher, T.; Line, M.; Millsaps, C. Probabilistic frameworks for life detection. In Planetary Astrobiology (Space Science Series); Meadows, V., Des Marais, D.J., Arney, G., Schmidt, B., Eds.; University of Arizona Press: Phoenix, AZ, USA, 2020; pp. 447–505. [Google Scholar] [CrossRef]
- Mathis, C.; Carrick, E.; Keenan, G.; Cooper, G.; Graham, H.; Bame, J.; Craven, M.; Bell, N.; Gromski, P.S.; Swart, M.; et al. Identifying molecules as biosignatures with assembly theory and mass spectrometry. ChemArxiv 2020. preprint. [Google Scholar] [CrossRef]
- Killops, S.D.; Killops, V.J. Introduction to Organic Geochemistry; Wiley: Hoboken, NJ, USA, 2013. [Google Scholar]
- Cleaves, H.J. Prebiotic chemistry: What we know, what we don’t. Evol. Educ. Outreach 2012, 5, 342–360. [Google Scholar] [CrossRef]
- Lovelock, J.E. A physical basis for life detection experiments. Nature 1965, 207, 568–570. [Google Scholar] [CrossRef]
- Klein, H.P. The Viking mission and the search for life on Mars. Rev. Geophys. 1979, 17, 1655–1662. [Google Scholar] [CrossRef]
- Sephton, M.A.; Hunter Waite, J.; Brockwell, T.G. Searching for life with mass spectrometry. Astron. Geophys. 2018, 59, 3–23. [Google Scholar] [CrossRef]
- Niemann, H.B.; Atreya, S.K.; Demick, J.E.; Gautier, D.; Haberman, J.A.; Harpold, D.N.; Kasprzak, W.T.; Lunine, J.I.; Owen, T.C.; Raulin, F. Composition of Titan’s lower atmosphere and simple surface volatiles as measured by the Cassini-Huygens probe gas chromatograph mass spectrometer experiment. J. Geophys. Res. Planets 2010, 115, E12006. [Google Scholar] [CrossRef]
- Goesmann, F.; Rosenbauer, H.; Bredehöft, J.H.; Cabane, M.; Ehrenfreund, P.; Gautier, T.; Giri, C.; Krüger, H.; Le Roy, L.; MacDermott, A.J.; et al. Organic compounds on comet 67P/Churyumov-Gerasimenko revealed by COSAC mass spectrometry. Science 2015, 349, aab0689. [Google Scholar] [CrossRef]
- Eigenbrode, J.L.; Summons, R.E.; Steele, A.; Freissinet, C.; Millan, M.; Navarro-González, R.; Sutter, B.; McAdam, A.C.; Franz, H.B.; Glavin, D.P.; et al. Organic matter preserved in 3-billion-year-old mudstones at Gale crater, Mars. Science 2018, 360, 1096–1101. [Google Scholar] [CrossRef] [PubMed]
- Postberg, F.; Khawaja, N.; Abel, B.; Choblet, G.; Glein, C.R.; Gudipati, M.S.; Henderson, B.L.; Hsu, H.W.; Kempf, S.; Klenner, F.; et al. Macromolecular organic compounds from the depths of Enceladus. Nature 2018, 558, 564. [Google Scholar] [CrossRef] [PubMed]
- O’Neil, W.J.; Cazaux, C. The Mars sample return project. Acta Astronaut. 2000, 47, 453–465. [Google Scholar] [CrossRef]
- Phillips, C.B.; Pappalardo, R.T. Europa clipper mission concept: Exploring Jupiter’s ocean moon. Eos 2014, 95, 165–167. [Google Scholar] [CrossRef]
- Lauretta, D.S.; Balram-Knutson, S.S.; Beshore, E.; Boynton, W.V.; Drouet d’Aubigny, C.; DellaGiustina, D.N.; Enos, H.L.; Gholish, D.R.; Hergenrother, C.W.; Howell, E.S.; et al. OSIRIS-REx: Sample return from asteroid (101955) Bennu. Space Sci. Rev. 2017, 212, 925–984. [Google Scholar] [CrossRef]
- Baum, D.A. The origin and early evolution of life in chemical composition space. J. Theor. Biol. 2018, 456, 295–304. [Google Scholar] [CrossRef]
- Surman, A.J.; Rodriguez-Garcia, M.; Abul-Haija, Y.M.; Cooper, G.J.T.; Gromski, P.S.; Turk-MacLeod, R.; Mullin, M.; Mathis, C.; Walker, S.I.; Cronin, L. Environmental control programs the emergence of distinct functional ensembles from unconstrained chemical reactions. Proc. Natl. Acad. Sci. USA 2019, 116, 5387–5392. [Google Scholar] [CrossRef]
- Pace, N.R. The universal nature of biochemistry. Proc. Natl. Acad. Sci. USA 2001, 98, 805–808. [Google Scholar] [CrossRef]
- Bains, W. Many chemistries could be used to build living systems. Astrobiology 2004, 4, 137–167. [Google Scholar] [CrossRef]
- McKay, C.P. What is life-and how do we search for it in other worlds? PLoS Biol. 2004, 2, E302. [Google Scholar] [CrossRef]
- Bedau, M.A.; Cleland, C.E. The Nature of Life; Cambridge University Press: Cambridge, UK, 2018. [Google Scholar]
- Cleland, C.E. The Quest for a Universal Theory of Life: Searching for Life as We Don’t Know It; Volume 11 of Cambridge Astrobiology; Cambridge University Press: Cambridge, UK, 2019. [Google Scholar]
- Bar-Even, A.; Noor, E.; Savir, Y.; Liebermeister, W.; Davidi, D.; Tawfik, D.S.; Milo, R. The moderately efficient enzyme: Evolutionary and physicochemical trends shaping enzyme parameters. Biochemistry 2011, 50, 4402–4410. [Google Scholar] [CrossRef]
- Ilardo, M.; Meringer, M.; Freeland, S.; Rasulev, B.; Cleaves, H.J., II. Extraordinarily adaptive properties of the genetically encoded amino acids. Sci. Rep. 2015, 5, 9414. [Google Scholar] [CrossRef]
- Meringer, M.; Cleaves, H.J. Exploring astrobiology using in silico molecular structure generation. Philos. Trans. R. Soc. A Math. Phys. Eng. Sci. 2017, 375, 20160344. [Google Scholar] [CrossRef]
- Dunn, I.S. Searching for Molecular Solutions: Empirical Discovery and Its Future; John Wiley & Sons: Hoboken, NJ, USA, 2010. [Google Scholar]
- Marlière, P.; Patrouix, J.; Döring, V.; Herdewijn, P.; Tricot, S.; Cruveiller, S.; Bouzon, M.; Mutzel, R. Chemical evolution of a bacterium’s genome. Angew. Chem. Int. Ed. 2011, 50, 7109–7114. [Google Scholar] [CrossRef]
- Zhang, Y.; Jerod, L.; Ptacin, J.L.; Fischer, E.C.; Aerni, H.R.; Caffaro, C.E.; San Jose, K.; Feldman, A.W.; Turner, C.R.; Romesberg, F.E. A semi-synthetic organism that stores and retrieves increased genetic information. Nature 2017, 551, 644. [Google Scholar] [CrossRef]
- Hoshika, S.; Leal, N.A.; Kim, M.-J.; Kim, M.-S.; Karalkar, N.B.; Kim, H.-J.; Bates, A.M.; Watkins, N.E.; SantaLucia, H.A.; Meyer, A.J.; et al. Hachimoji DNA and RNA: A genetic system with eight building blocks. Science 2019, 363, 884–887. [Google Scholar] [CrossRef]
- Johnson, S.S.; Anslyn, E.V.; Graham, H.V.; Mahaffy, P.R.; Ellington, A.D. Fingerprinting non-terran biosignatures. Astrobiology 2018, 18, 915–922. [Google Scholar] [CrossRef]
- Guttenberg, N.; Virgo, N.; Chandru, K.; Scharf, C.; Mamajanov, I. Bulk measurements of messy chemistries are needed for a theory of the origins of life. Philos. Trans. R. Soc. A Math. Phys. Eng. Sci. 2017, 375, 20160347. [Google Scholar] [CrossRef]
- Marshall, S.M.; Murray, A.R.; Cronin, L. A probabilistic framework for identifying biosignatures using pathway complexity. Philos. Trans. R. Soc. A Math. Phys. Eng. Sci. 2017, 375, 20160342. [Google Scholar] [CrossRef]
- Bonchev, D.D.; Rouvray, D. Complexity in Chemistry, Biology, and Ecology; Springer Science & Business Media: Berlin, Germany, 2007. [Google Scholar]
- Cleaves, H.J. The origin of the biologically coded amino acids. J. Theor. Biol. 2010, 263, 490–498. [Google Scholar] [CrossRef]
- Miller, S.L. A production of amino acids under possible primitive earth conditions. Science 1953, 117, 528–529. [Google Scholar] [CrossRef]
- Pizzarello, S.; Shock, E. The organic composition of carbonaceous meteorites: The evolutionary story ahead of biochemistry. Cold Spring Harb. Perspect. Biol. 2010, 2, a002105. [Google Scholar] [CrossRef]
- McCollom, T.M. Abiotic methane formation during experimental serpentinization of olivine. Proc. Natl. Acad. Sci. USA 2016, 113, 13965–13970. [Google Scholar] [CrossRef]
- Callahan, M.P.; Smith, K.E.; Cleaves, H.J.; Ruzicka, J.; Stern, J.C.; Glavin, D.P.; House, C.H.; Dworkin, J.P. Carbonaceous meteorites contain a wide range of extraterrestrial nucleobases. Proc. Natl. Acad. Sci. USA 2011, 108, 13995–13998. [Google Scholar] [CrossRef] [PubMed]
- Schmitt-Kopplin, P.; Gabelica, Z.; Gougeon, R.D.; Fekete, A.; Kanawati, B.; Harir, M.; Gebefuegi, I.; Eckel, G.; Norbert Hertkorn, N. High molecular diversity of extraterrestrial organic matter in Murchison meteorite revealed 40 years after its fall. Proc. Natl. Acad. Sci. USA 2010, 107, 2763–2768. [Google Scholar] [CrossRef]
- Parker, E.T.; Cleaves, H.J.; Bada, J.L.; Fernández, F.M. Quantitation of α-hydroxy acids in complex prebiotic mixtures via liquid chromatography/tandem mass spectrometry. Rapid Commun. Mass Spectrom. 2016, 30, 2043–2051. [Google Scholar] [CrossRef]
- Louie, A.H. Relational biology and Church’s thesis. Biosystems 2020, 197, 104179. [Google Scholar] [CrossRef]
- Wittgenstein, L. Philosophical Investigations; Blackwell Publishing: Hongkong, China, 1953/2001; ISBN 0-631-23127-7. [Google Scholar]
- Wolman, Y.; Haverland, W.J.; Miller, S.L. Nonprotein amino acids from spark discharges and their comparison with the Murchison meteorite amino acids. Proc. Natl. Acad. Sci. USA 1972, 69, 809–811. [Google Scholar] [CrossRef] [PubMed]
- Ferris, J.P.; Hagan, W.J., Jr. HCN and chemical evolution: The possible role of cyano compounds in prebiotic synthesis. Tetrahedron 1984, 40, 1093–1120. [Google Scholar] [CrossRef]
- Matthews, C.N.; Minard, R.D. Hydrogen cyanide polymers, comets and the origin of life. Faraday Disc. 2006, 133, 393–401. [Google Scholar] [CrossRef] [PubMed]
- Kebukawa, Y.; Kilcoyne, A.L.D.; Cody, G.D. Exploring the potential formation of organic solids in chondrites and comets through polymerization of interstellar formaldehyde. Astrophys. J. 2013, 771, 19. [Google Scholar] [CrossRef]
- Koga, T.; Naraoka, H. A new family of extraterrestrial amino acids in the Murchison meteorite. Sci. Rep. 2017, 7, 1–8. [Google Scholar] [CrossRef]
- Sasselov, D.D.; Grotzinger, J.P.; Sutherland, J.D. The origin of life as a planetary phenomenon. Sci. Adv. 2020, 6, eaax3419. [Google Scholar] [CrossRef]
- Levy, M.; Miller, S.L.; Brinton, K.; Bada, J.L. Prebiotic synthesis of adenine and amino acids under Europa-like conditions. Icarus 2000, 145, 609–613. [Google Scholar] [CrossRef]
- Golon, A.; Kropf, C.; Vockenroth, I.; Kuhnert, N. An investigation of the complexity of Maillard reaction product profiles from the thermal reaction of amino acids with sucrose using high resolution mass spectrometry. Foods 2014, 3, 461–475. [Google Scholar] [CrossRef]
- Milo, R.; Phillips, R. Cell Biology by the Numbers; Garland Science: New York, NY, USA, 2015; ISBN 1317230698, 9781317230694. [Google Scholar]
- Muchowska, K.B.; Varma, S.J.; Moran, J. Nonenzymatic metabolic reactions and life’s origins. Chem. Rev. 2020, 120, 7708–7744. [Google Scholar] [CrossRef] [PubMed]
- Lobodin, V.V.; Juyal, P.; McKenna, A.M.; Rodgers, R.P.; Marshall, A.G. Tetramethylammonium hydroxide as a reagent for complex mixture analysis by negative ion electrospray ionization mass spectrometry. Anal. Chem. 2013, 85, 7803–7808. [Google Scholar] [CrossRef]
- Kebukawa, Y.; Nakashima, S.; Zolensky, M.E. Kinetics of organic matter degradation in the Murchison meteorite for the evaluation of parent-body temperature history. Meteorit. Planet. Sci. 2010, 45, 99–113. [Google Scholar] [CrossRef]
- Gordadze, G.N.; Poshibaeva, A.R.; Giruts, M.V.; Perevalova, A.A.; Koshelev, V.N. Formation of petroleum hydrocarbons from prokaryote biomass: 1. Formation of petroleum biomarker hydrocarbons from Thermoplasma sp. archaea biomass. Pet. Chem. 2018, 58, 186–189. [Google Scholar] [CrossRef]
- Kaiser, N.K.; Quinn, J.P.; Blakney, G.T.; Hendrickson, C.L.; Marshall, A.G. A novel 9.4 Tesla FTICR mass spectrometer with improved sensitivity, mass resolution, and mass range. J. Am. Soc. Mass Spectrom. 2011, 22, 1343–1351. [Google Scholar] [CrossRef] [PubMed]
- Blakney, G.T.; Hendrickson, C.L.; Marshall, A.G. Predator data station: A fast data acquisition system for advanced FT-ICR MS experiments. Int. J. Mass Spectrom. 2011, 306, 246–252. [Google Scholar] [CrossRef]
- Kaiser, N.K.; Savory, J.J.; Hendrickson, C.L. Controlled ion ejection from an external trap for extended m/z range in FT-ICR mass spectrometry. J. Am. Soc. Mass Spectrom. 2014, 25, 943–949. [Google Scholar] [CrossRef]
- Kaiser, N.K.; Savory, J.J.; McKenna, A.M.; Quinn, J.P.; Hendrickson, C.L.; Marshall, A.G. Electrically compensated Fourier transform ion cyclotron resonance cell for complex mixture mass analysis. Anal. Chem. 2011, 83, 6907–6910. [Google Scholar] [CrossRef] [PubMed]
- Tolmachev, A.V.; Robinson, E.W.; Wu, S.; Smith, R.D.; Pasa-Tolic, L. Trapping radial electric field optimization in compensated FTICR cells. J. Am. Soc. Mass. Spectrom. 2011, 22, 1334–1342. [Google Scholar] [CrossRef] [PubMed]
- Beu, S.C.; Blakney, G.T.; Quinn, J.P.; Hendrickson, C.L.; Marshall, A.G. Broadband phase correction of FT-ICR mass spectra via simultaneous excitation and detection. Anal. Chem. 2004, 76, 5756–5761. [Google Scholar] [CrossRef] [PubMed]
- Xian, F.; Hendrickson, C.L.; Blakney, G.T.; Beu, S.C.; Marshall, A.G. Automated broadband phase correction of Fourier transform ion cyclotron resonance mass spectra. Anal. Chem. 2010, 82, 8807–8812. [Google Scholar] [CrossRef] [PubMed]
- Emmett, M.R.; White, F.M.; Hendrickson, C.L.; Shi, S.D.H.; Marshall, A.G. Application of micro-electrospray liquid chromatography techniques to FT-ICR MS to enable high-sensitivity biological analysis. J. Am. Soc. Mass. Spectrom. 1998, 9, 333–340. [Google Scholar] [CrossRef]
- Hannis, J.C.; Muddiman, D.C. A dual electrospray ionization source combined with hexapole accumulation to achieve high mass accuracy of biopolymers in fourier transform ion cyclotron resonance mass spectrometry. J. Am. Soc. Mass. Spectrom. 2000, 11, 876–883. [Google Scholar] [CrossRef]
- Shi, S.D.H.; Drader, J.J.; Freitas, M.A.; Hendrickson, C.L.; Marshall, A.G. Comparison and interconversion of the two most common frequency-to-mass calibration functions for Fourier transform ion cyclotron resonance mass spectrometry. Int. J. Mass Spectrom. 2000, 195, 591–598. [Google Scholar] [CrossRef]
- Grosshans, P.B.; Shields, P.J.; Marshall, A.G. Comprehensive theory of the Fourier-transform ion-cyclotron resonance signal for all ion trap geometries. J. Chem. Phys. 1991, 94, 5341–5352. [Google Scholar] [CrossRef]
- Savory, J.J.; Kaiser, N.K.; McKenna, A.M.; Xian, F.; Blakney, G.T.; Rodgers, R.P.; Hendrickson, C.L.; Marshall, A.G. Parts-per-billion fourier transform ion cyclotron resonance mass measurement accuracy with a “walking” calibration equation. Anal. Chem. 2011, 83, 1732–1736. [Google Scholar] [CrossRef]
- Corilo, Y.E. Florida State University, 2017; All Rights Reserved. Available online: https://nationalmaglab.org/user-facilities/icr/icr-software (accessed on 7 March 2021).
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Harayama, T.; Riezman, H. Understanding the diversity of membrane lipid composition. Nat. Ecol. Evol. 2018, 19, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Ming, D.W.; Archer, P.D.; Glavin, D.P.; Eigenbrode, J.L.; Franz, H.B.; Sutter, B.; Brunner, A.E.; Stern, J.C.; Freissinet, C.; McAdam, A.C.; et al. Volatile and organic compositions of sedimentary rocks in Yellowknife Bay, Gale Crater, Mars. Science 2014, 343, 1245267. [Google Scholar] [CrossRef] [PubMed]
- Mumma, M.J.; Charnley, S.B. The chemical composition of comets-emerging taxonomies and natal heritage. Ann. Rev. Astron. Astrophys. 2011, 49, 471–524. [Google Scholar] [CrossRef]
- Alexander, C.O.D.; Fogel, M.; Yabuta, H.; Cody, G.D. The origin and evolution of chondrites recorded in the elemental and isotopic compositions of their macromolecular organic matter. Geochim. Et Cosmochim. Acta 2007, 71, 4380–4403. [Google Scholar] [CrossRef]
- Summons, R.E.; Albrecht, P.; McDonald, G.; Moldowan, J.M. Molecular biosignatures. Space Sci. Rev. 2008, 135, 133–159. [Google Scholar] [CrossRef]
- Dorn, E.D.; McDonald, G.D.; Storrie-Lombardi, M.C.; Nealson, K.H. Principal component analysis and neural networks for detection of amino acid biosignatures. Icarus 2003, 166, 403–409. [Google Scholar] [CrossRef]
- Glavin, D.P.; Callahan, M.P.; Dworkin, J.P.; Elsila, J.E. The effects of parent body processes on amino acids in carbonaceous chondrites. Meteorit. Planet. Sci. 2010, 45, 1948–1972. [Google Scholar] [CrossRef]
- Bada, J.L.; Wang, X.S.; Hamilton, H. Preservation of key biomolecules in the fossil record: Current knowledge and future challenges. Phil. Trans. R. Soc. Lond. B Biol. Sci. 1999, 354, 77–87. [Google Scholar] [CrossRef]
- Steinberg, S.; Bada, J.L. Diketopiperazine formation during investigations of amino acid racemization in dipeptides. Science 1981, 213, 544–545. [Google Scholar] [CrossRef]
- Ferris, J.P.; Wos, J.D.; Ryan, T.J.; Lobo, A.P.; Donner, D.B. Biomolecules from HCN. Orig. Life 1974, 5, 153–157. [Google Scholar] [CrossRef]
- Shimoyama, A.; Ogasawara, R. Dipeptides and diketopiperazines in the Yamato-791198 and Murchison carbonaceous chondrites. Orig. Life Evol. Biosph. 2002, 32, 165–179. [Google Scholar] [CrossRef] [PubMed]
- Fox, S.W.; Windsor, C.R. Synthesis of amino acids by the heating of formaldehyde and ammonia. Science 1970, 170, 984–986. [Google Scholar] [CrossRef] [PubMed]
- Kebukawa, Y.; Chan, Q.H.; Tachibana, S.; Kobayashi, K.; Zolensky, M.E. One-pot synthesis of amino acid precursors with insoluble organic matter in planetesimals with aqueous activity. Sci. Adv. 2017, 3, e1602093. [Google Scholar] [CrossRef]
- Meringer, M.; Cleaves, H.J.; Freeland, S.J. Beyond terrestrial biology: Charting the chemical universe of α-amino acid structures. J. Chem. Inf. Mod. 2013, 53, 2851–2862. [Google Scholar] [CrossRef]
- Goodwin, J.T.; Mehta, A.K.; Lynn, D.G. Digital and analog chemical evolution. Acc. Chem. Res. 2012, 45, 2189–2199. [Google Scholar] [CrossRef] [PubMed]
- Bada, J.L. Amino acid racemization dating of fossil bones. Ann. Rev. Earth Planet. Sci. 1985, 13, 241–268. [Google Scholar] [CrossRef]
- Schidlowski, M. Stable carbon isotopes: Possible clues to early life on Mars. Adv. Space Res. 1992, 12, 101–110. [Google Scholar] [CrossRef]
- Lewan, M.D. Laboratory simulation of petroleum formation. In Organic Geochemistry; Engel, M.H., Macko, S.A., Eds.; Springer: Boston, MA, USA, 1993; pp. 419–442. [Google Scholar]
- Vincent, L.; Berg, M.; Krismer, M.; Saghafi, S.T.; Cosby, J.; Sankari, T.; Vetsigian, K.; Cleaves, H.J.; Baum, D.A. Chemical ecosystem selection on mineral surfaces reveals long-term dynamics consistent with the spontaneous emergence of mutual catalysis. Life 2020, 9, 80. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guttenberg, N.; Chen, H.; Mochizuki, T.; Cleaves, H.J. Classification of the Biogenicity of Complex Organic Mixtures for the Detection of Extraterrestrial Life. Life 2021, 11, 234. https://doi.org/10.3390/life11030234
Guttenberg N, Chen H, Mochizuki T, Cleaves HJ. Classification of the Biogenicity of Complex Organic Mixtures for the Detection of Extraterrestrial Life. Life. 2021; 11(3):234. https://doi.org/10.3390/life11030234
Chicago/Turabian StyleGuttenberg, Nicholas, Huan Chen, Tomohiro Mochizuki, and H. James Cleaves. 2021. "Classification of the Biogenicity of Complex Organic Mixtures for the Detection of Extraterrestrial Life" Life 11, no. 3: 234. https://doi.org/10.3390/life11030234
APA StyleGuttenberg, N., Chen, H., Mochizuki, T., & Cleaves, H. J. (2021). Classification of the Biogenicity of Complex Organic Mixtures for the Detection of Extraterrestrial Life. Life, 11(3), 234. https://doi.org/10.3390/life11030234






