Measuring Proviral HIV-1 DNA: Hurdles and Improvements to an Assay Monitoring Integration Events Utilising Human Alu Repeat Sequences
Abstract
1. Introduction
2. Materials and Methods
2.1. Integrated HIV-1 DNA Quantification by qPCR
2.2. Patients Tested with the Alu-gag and Alu-LTR Assay
2.3. Standard for Integrated HIV-1 DNA Assay
2.4. Statistics
3. Results
3.1. Design an Improved HIV-1 Integration Assay (Alu-5LTR)
3.2. Testing of Alu-5LTR in Patients with Undetectable Viral Load Undergoing Suppressive Antiretroviral Therapy
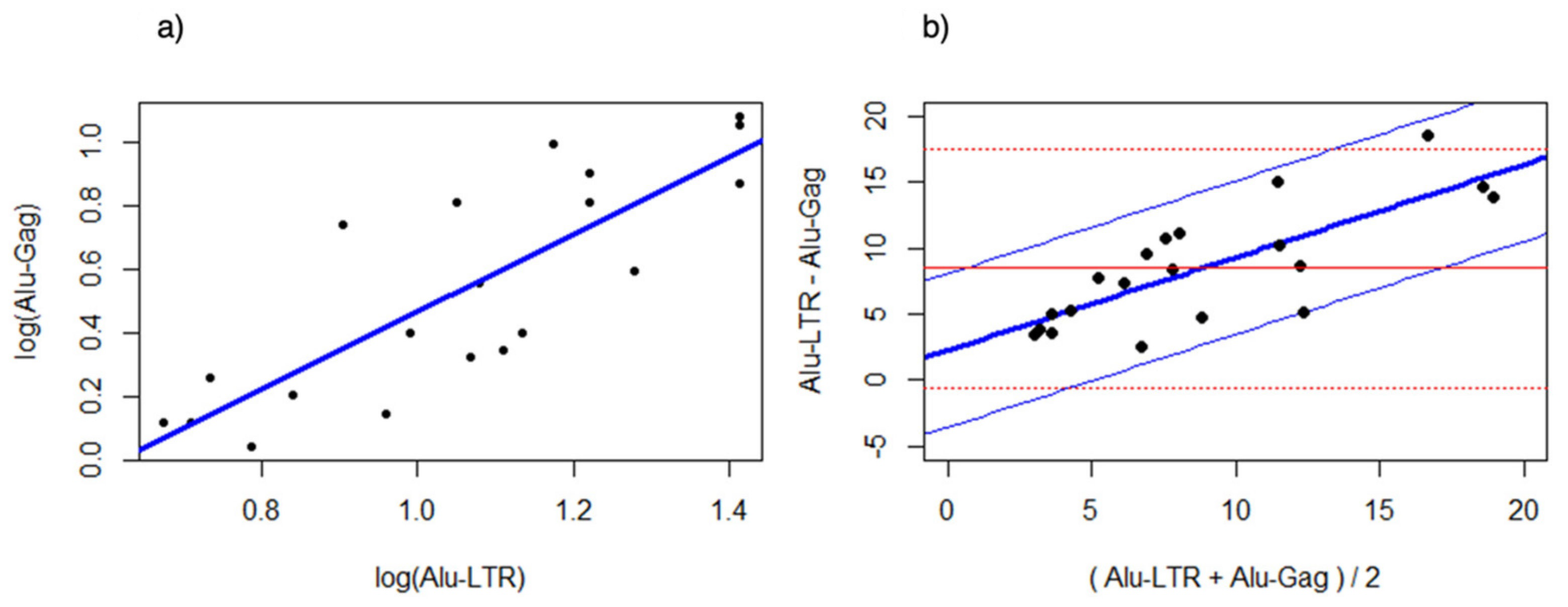
3.3. Bland Altman Analysis of the Alu-5LTR vs. Alu-gag Assays
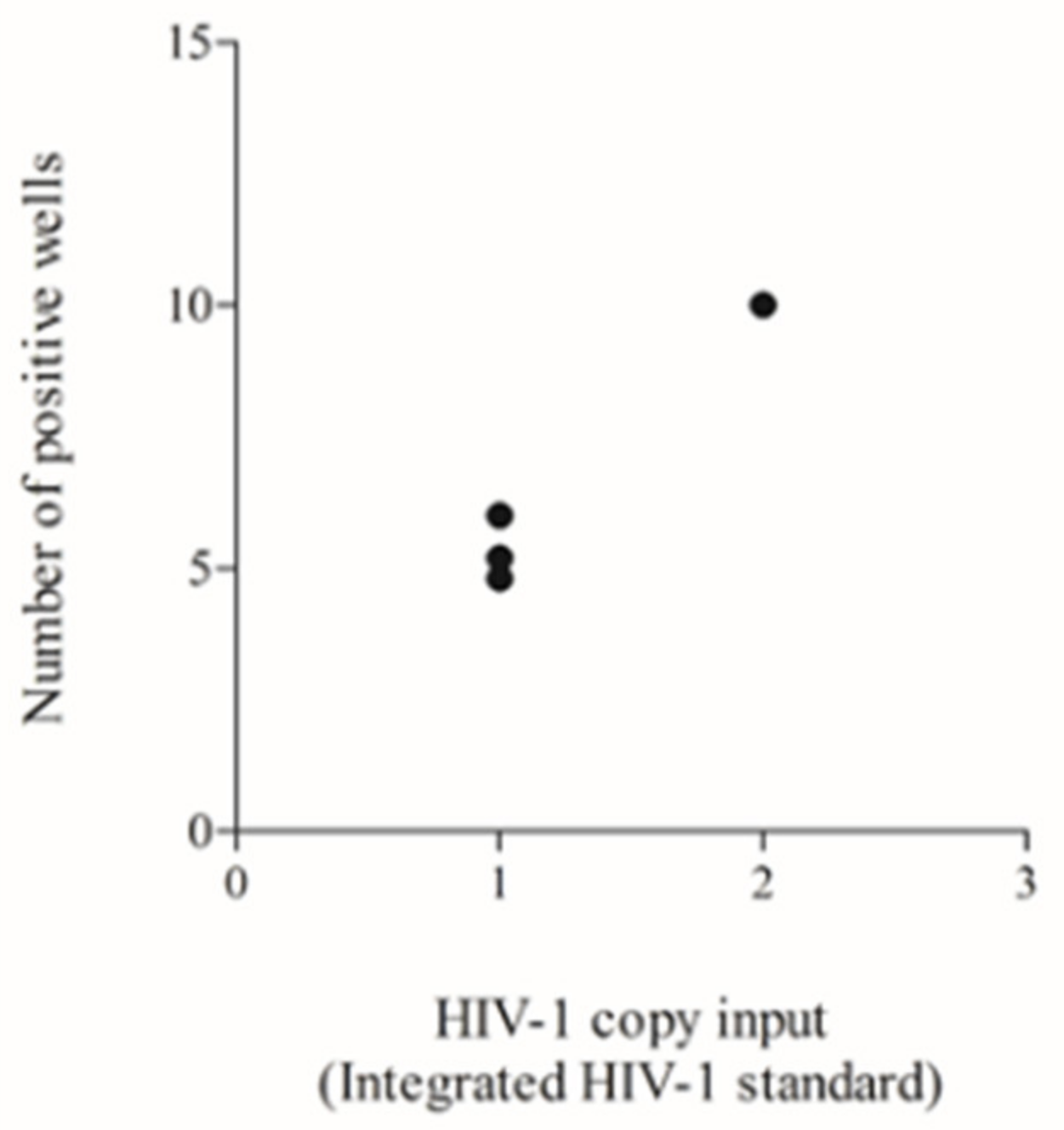
3.4. Testing of a Previously Used HIV-1 DNA Standard to Define the Lower Input Needed to Allow Reliable Poisson Quantification
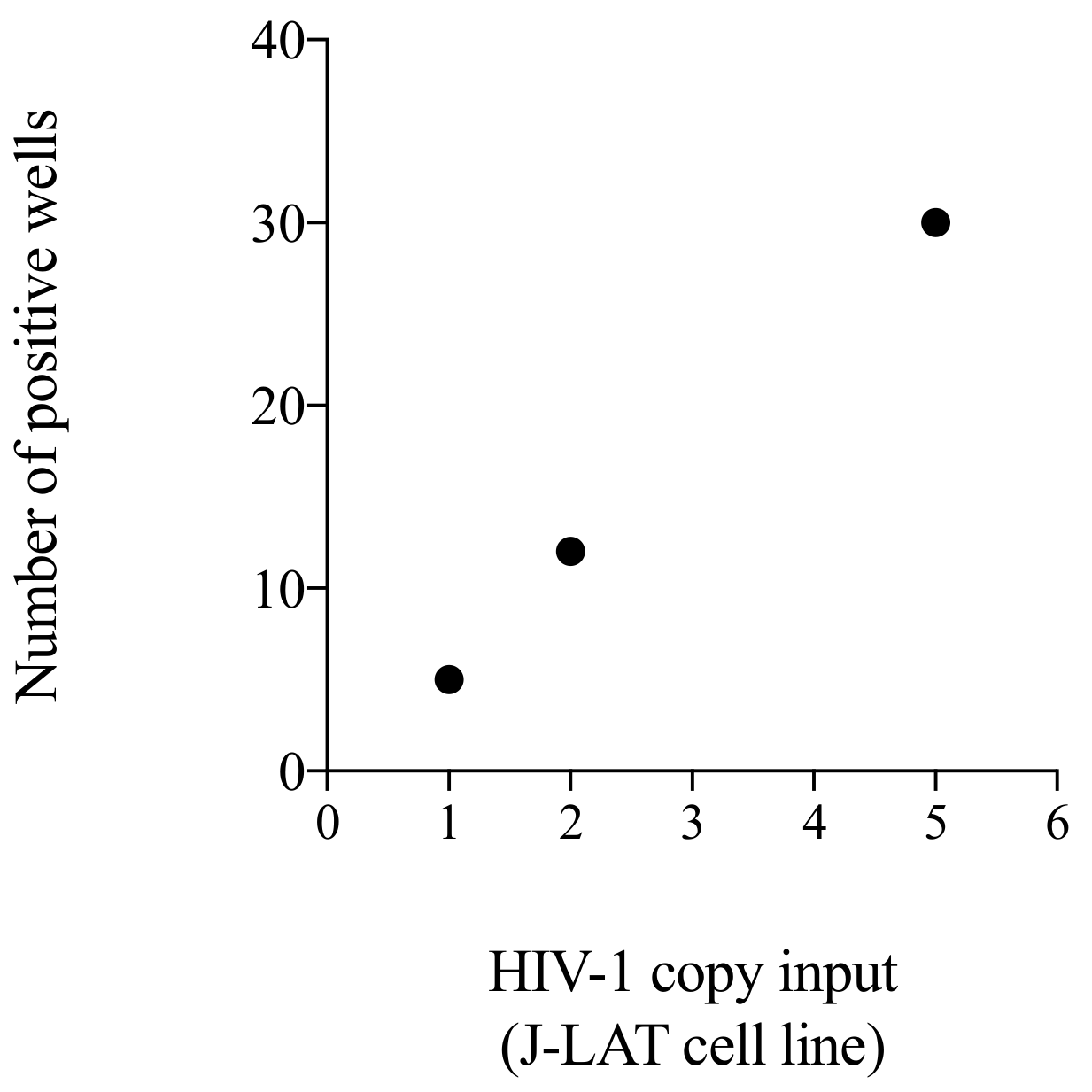
3.5. Testing of a Cellular Standard to Be Used for the Initial Setting of the Alu-5LTR Assay: Towards Cross-Laboratory Harmonisation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Palmer, S.; Maldarelli, F.; Wiegand, A.; Bernstein, B.; Hanna, G.J.; Brun, S.C.; Kempf, D.J.; Mellors, J.W.; Coffin, J.M.; King, M.S. Low-level viremia persists for at least 7 years in patients on suppressive antiretroviral therapy. Proc. Natl. Acad. Sci. USA 2008, 105, 3879–3884. [Google Scholar] [CrossRef] [PubMed]
- Yukl, S.A.; Shergill, A.K.; Ho, T.; Killian, M.; Girling, V.; Epling, L.; Li, P.; Wong, L.K.; Crouch, P.; Deeks, S.G.; et al. The distribution of HIV DNA and RNA in cell subsets differs in gut and blood of HIV-positive patients on ART: Implications for viral persistence. J. Infect. Dis. 2013, 208, 1212–1220. [Google Scholar] [CrossRef] [PubMed]
- Sturdevant, C.B.; Joseph, S.B.; Schnell, G.; Price, R.W.; Swanstrom, R.; Spudich, S. Compartmentalized replication of R5 T cell-tropic HIV-1 in the central nervous system early in the course of infection. PLoS Pathog. 2015, 11, e1004720. [Google Scholar] [CrossRef] [PubMed]
- Alexaki, A.; Liu, Y.; Wigdahl, B. Cellular reservoirs of HIV-1 and their role in viral persistence. Curr. HIV Res. 2008, 6, 388–400. [Google Scholar] [CrossRef]
- Lee, E.; von Stockenstrom, S.; Morcilla, V.; Odevall, L.; Hiener, B.; Shao, W.; Hartogensis, W.; Bacchetti, P.; Milush, J.; Liegler, T.; et al. Impact of Antiretroviral Therapy Duration on HIV-1 Infection of T Cells within Anatomic Sites. J. Virol. 2020, 94, e01270-19. [Google Scholar] [CrossRef]
- Thomas, J.; Ruggiero, A.; Paxton, W.A.; Pollakis, G. Measuring the Success of HIV-1 Cure Strategies. Front. Cell. Infect. Microbiol. 2020, 10, 134. [Google Scholar] [CrossRef]
- Banga, R.; Procopio, F.A.; Noto, A.; Pollakis, G.; Cavassini, M.; Ohmiti, K.; Corpataux, J.M.; de Leval, L.; Pantaleo, G.; Perreau, M. PD-1(+) and follicular helper T cells are responsible for persistent HIV-1 transcription in treated aviremic individuals. Nat. Med. 2016, 22, 754–761. [Google Scholar] [CrossRef]
- Banga, R.; Procopio, F.A.; Ruggiero, A.; Noto, A.; Ohmiti, K.; Cavassini, M.; Corpataux, J.M.; Paxton, W.A.; Pollakis, G.; Perreau, M. Blood CXCR3(+) CD4 T Cells Are Enriched in Inducible Replication Competent HIV in Aviremic Antiretroviral Therapy-Treated Individuals. Front. Immunol. 2018, 9, 144. [Google Scholar] [CrossRef]
- Banga, R.; Rebecchini, C.; Procopio, F.A.; Noto, A.; Munoz, O.; Ioannidou, K.; Fenwick, C.; Ohmiti, K.; Cavassini, M.; Corpataux, J.M.; et al. Lymph node migratory dendritic cells modulate HIV-1 transcription through PD-1 engagement. PLoS Pathog. 2019, 15, e1007918. [Google Scholar] [CrossRef]
- Buzon, M.J.; Martin-Gayo, E.; Pereyra, F.; Ouyang, Z.; Sun, H.; Li, J.Z.; Piovoso, M.; Shaw, A.; Dalmau, J.; Zangger, N.; et al. Long-term antiretroviral treatment initiated at primary HIV-1 infection affects the size, composition, and decay kinetics of the reservoir of HIV-1-infected CD4 T cells. J. Virol. 2014, 88, 10056–10065. [Google Scholar] [CrossRef]
- Buzon, M.J.; Sun, H.; Li, C.; Shaw, A.; Seiss, K.; Ouyang, Z.; Martin-Gayo, E.; Leng, J.; Henrich, T.J.; Li, J.Z.; et al. HIV-1 persistence in CD4+ T cells with stem cell-like properties. Nat. Med. 2014, 20, 139–142. [Google Scholar] [CrossRef]
- Tran, T.A.; de Goer de Herve, M.G.; Hendel-Chavez, H.; Dembele, B.; Le Nevot, E.; Abbed, K.; Pallier, C.; Goujard, C.; Gasnault, J.; Delfraissy, J.F.; et al. Resting regulatory CD4 T cells: A site of HIV persistence in patients on long-term effective antiretroviral therapy. PloS ONE 2008, 3, e3305. [Google Scholar] [CrossRef]
- Alvarez, Y.; Tuen, M.; Shen, G.; Nawaz, F.; Arthos, J.; Wolff, M.J.; Poles, M.A.; Hioe, C.E. Preferential HIV infection of CCR6+ Th17 cells is associated with higher levels of virus receptor expression and lack of CCR5 ligands. J. Virol. 2013, 87, 10843–10854. [Google Scholar] [CrossRef]
- Christensen-Quick, A.; Lafferty, M.; Sun, L.; Marchionni, L.; DeVico, A.; Garzino-Demo, A. Human Th17 Cells Lack HIV-Inhibitory RNases and Are Highly Permissive to Productive HIV Infection. J. Virol. 2016, 90, 7833–7847. [Google Scholar] [CrossRef]
- Sun, H.; Kim, D.; Li, X.; Kiselinova, M.; Ouyang, Z.; Vandekerckhove, L.; Shang, H.; Rosenberg, E.S.; Yu, X.G.; Lichterfeld, M. Th1/17 Polarization of CD4 T Cells Supports HIV-1 Persistence during Antiretroviral Therapy. J. Virol. 2015, 89, 11284–11293. [Google Scholar] [CrossRef]
- Caruso, M.P.; Falivene, J.; Holgado, M.P.; Zurita, D.H.; Laufer, N.; Castro, C.; Nico, A.; Maeto, C.; Salido, J.; Perez, H.; et al. Impact of HIV-ART on the restoration of Th17 and Treg cells in blood and female genital mucosa. Sci. Rep. 2019, 9, 1978. [Google Scholar] [CrossRef]
- Darcis, G.; Kootstra, N.A.; Hooibrink, B.; van Montfort, T.; Maurer, I.; Groen, K.; Jurriaans, S.; Bakker, M.; van Lint, C.; Berkhout, B.; et al. CD32(+)CD4(+) T Cells Are Highly Enriched for HIV DNA and Can Support Transcriptional Latency. Cell Rep. 2020, 30, 2284–2296.E3. [Google Scholar] [CrossRef]
- Descours, B.; Petitjean, G.; Lopez-Zaragoza, J.L.; Bruel, T.; Raffel, R.; Psomas, C.; Reynes, J.; Lacabaratz, C.; Levy, Y.; Schwartz, O.; et al. CD32a is a marker of a CD4 T-cell HIV reservoir harbouring replication-competent proviruses. Nature 2017, 543, 564–567. [Google Scholar] [CrossRef]
- Abdel-Mohsen, M.; Kuri-Cervantes, L.; Grau-Exposito, J.; Spivak, A.M.; Nell, R.A.; Tomescu, C.; Vadrevu, S.K.; Giron, L.B.; Serra-Peinado, C.; Genesca, M.; et al. CD32 is expressed on cells with transcriptionally active HIV but does not enrich for HIV DNA in resting T cells. Sci. Transl. Med. 2018, 10. [Google Scholar] [CrossRef]
- Badia, R.; Ballana, E.; Castellvi, M.; Garcia-Vidal, E.; Pujantell, M.; Clotet, B.; Prado, J.G.; Puig, J.; Martinez, M.A.; Riveira-Munoz, E.; et al. CD32 expression is associated to T-cell activation and is not a marker of the HIV-1 reservoir. Nat. Commun. 2018, 9, 2739. [Google Scholar] [CrossRef]
- Bertagnolli, L.N.; White, J.A.; Simonetti, F.R.; Beg, S.A.; Lai, J.; Tomescu, C.; Murray, A.J.; Antar, A.A.R.; Zhang, H.; Margolick, J.B.; et al. The role of CD32 during HIV-1 infection. Nature 2018, 561, E17–E19. [Google Scholar] [CrossRef] [PubMed]
- Martin, G.E.; Pace, M.; Thornhill, J.P.; Phetsouphanh, C.; Meyerowitz, J.; Gossez, M.; Brown, H.; Olejniczak, N.; Lwanga, J.; Ramjee, G.; et al. CD32-Expressing CD4 T Cells Are Phenotypically Diverse and Can Contain Proviral HIV DNA. Front. Immunol. 2018, 9, 928. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, G.; Xu, X.; Chermann, J.C.; Hirsch, I. Accumulation of defective viral genomes in peripheral blood mononuclear cells of human immunodeficiency virus type 1-infected individuals. J. Virol. 1997, 71, 2233–2240. [Google Scholar] [CrossRef] [PubMed]
- Ho, Y.C.; Shan, L.; Hosmane, N.N.; Wang, J.; Laskey, S.B.; Rosenbloom, D.I.; Lai, J.; Blankson, J.N.; Siliciano, J.D.; Siliciano, R.F. Replication-competent noninduced proviruses in the latent reservoir increase barrier to HIV-1 cure. Cell 2013, 155, 540–551. [Google Scholar] [CrossRef]
- Bruner, K.M.; Murray, A.J.; Pollack, R.A.; Soliman, M.G.; Laskey, S.B.; Capoferri, A.A.; Lai, J.; Strain, M.C.; Lada, S.M.; Hoh, R.; et al. Defective proviruses rapidly accumulate during acute HIV-1 infection. Nat. Med. 2016, 22, 1043–1049. [Google Scholar] [CrossRef]
- Azzoni, L.; Foulkes, A.S.; Papasavvas, E.; Mexas, A.M.; Lynn, K.M.; Mounzer, K.; Tebas, P.; Jacobson, J.M.; Frank, I.; Busch, M.P.; et al. Pegylated Interferon alfa-2a monotherapy results in suppression of HIV type 1 replication and decreased cell-associated HIV DNA integration. J. Infect. Dis. 2013, 207, 213–222. [Google Scholar] [CrossRef]
- Chun, T.W.; Davey, R.T., Jr.; Engel, D.; Lane, H.C.; Fauci, A.S. Re-emergence of HIV after stopping therapy. Nature 1999, 401, 874–875. [Google Scholar] [CrossRef]
- Davey, R.T., Jr.; Bhat, N.; Yoder, C.; Chun, T.W.; Metcalf, J.A.; Dewar, R.; Natarajan, V.; Lempicki, R.A.; Adelsberger, J.W.; Miller, K.D.; et al. HIV-1 and T cell dynamics after interruption of highly active antiretroviral therapy (HAART) in patients with a history of sustained viral suppression. Proc. Natl. Acad. Sci. USA 1999, 96, 15109–15114. [Google Scholar] [CrossRef]
- Li, J.Z.; Etemad, B.; Ahmed, H.; Aga, E.; Bosch, R.J.; Mellors, J.W.; Kuritzkes, D.R.; Lederman, M.M.; Para, M.; Gandhi, R.T. The size of the expressed HIV reservoir predicts timing of viral rebound after treatment interruption. Aids 2016, 30, 343–353. [Google Scholar] [CrossRef]
- Siliciano, J.D.; Siliciano, R.F. Enhanced culture assay for detection and quantitation of latently infected, resting CD4+ T-cells carrying replication-competent virus in HIV-1-infected individuals. Methods Mol. Biol. 2005, 304, 3–15. [Google Scholar] [CrossRef]
- Laird, G.M.; Eisele, E.E.; Rabi, S.A.; Lai, J.; Chioma, S.; Blankson, J.N.; Siliciano, J.D.; Siliciano, R.F. Rapid quantification of the latent reservoir for HIV-1 using a viral outgrowth assay. PLoS Pathog. 2013, 9, e1003398. [Google Scholar] [CrossRef]
- Rosenbloom, D.I.; Elliott, O.; Hill, A.L.; Henrich, T.J.; Siliciano, J.M.; Siliciano, R.F. Designing and Interpreting Limiting Dilution Assays: General Principles and Applications to the Latent Reservoir for Human Immunodeficiency Virus-1. Open Forum Infect. Dis. 2015, 2, ofv123. [Google Scholar] [CrossRef]
- Fun, A.; Mok, H.P.; Wills, M.R.; Lever, A.M. A highly reproducible quantitative viral outgrowth assay for the measurement of the replication-competent latent HIV-1 reservoir. Sci. Rep. 2017, 7, 43231. [Google Scholar] [CrossRef]
- Passaes, C.P.B.; Bruel, T.; Decalf, J.; David, A.; Angin, M.; Monceaux, V.; Muller-Trutwin, M.; Noel, N.; Bourdic, K.; Lambotte, O.; et al. Ultrasensitive HIV-1 p24 Assay Detects Single Infected Cells and Differences in Reservoir Induction by Latency Reversal Agents. J. Virol. 2017, 91, e02296-16. [Google Scholar] [CrossRef]
- Massanella, M.; Yek, C.; Lada, S.M.; Nakazawa, M.; Shefa, N.; Huang, K.; Richman, D.D. Improved assays to measure and characterize the inducible HIV reservoir. Ebiomedicine 2018, 36, 113–121. [Google Scholar] [CrossRef]
- Sanyal, A.; Mailliard, R.B.; Rinaldo, C.R.; Ratner, D.; Ding, M.; Chen, Y.; Zerbato, J.M.; Giacobbi, N.S.; Venkatachari, N.J.; Patterson, B.K.; et al. Novel assay reveals a large, inducible, replication-competent HIV-1 reservoir in resting CD4(+) T cells. Nat. Med. 2017, 23, 885–889. [Google Scholar] [CrossRef]
- Stone, M.; Rosenbloom, D.; Bacchetti, P.; Deng, X.; Dimapasoc, M.; Keating, S.; Bakkour, S.; Richman, D.; Mellors, J.; Deeks, S.; et al. Assessing suitability of next-generation viral outgrowth assays as proxies for classic QVOA to measure HIV-1 latent reservoir size. J. Infect. Dis. 2020, 224, 1209–1218. [Google Scholar] [CrossRef]
- Rosenbloom, D.I.S.; Bacchetti, P.; Stone, M.; Deng, X.; Bosch, R.J.; Richman, D.D.; Siliciano, J.D.; Mellors, J.W.; Deeks, S.G.; Ptak, R.G.; et al. Assessing intra-lab precision and inter-lab repeatability of outgrowth assays of HIV-1 latent reservoir size. PLoS Comput. Biol. 2019, 15, e1006849. [Google Scholar] [CrossRef]
- Procopio, F.A.; Fromentin, R.; Kulpa, D.A.; Brehm, J.H.; Bebin, A.G.; Strain, M.C.; Richman, D.D.; O’Doherty, U.; Palmer, S.; Hecht, F.M.; et al. A Novel Assay to Measure the Magnitude of the Inducible Viral Reservoir in HIV-infected Individuals. Ebiomedicine 2015, 2, 874–883. [Google Scholar] [CrossRef]
- Frank, I.; Acharya, A.; Routhu, N.K.; Aravantinou, M.; Harper, J.L.; Maldonado, S.; Sole Cigoli, M.; Semova, S.; Mazel, S.; Paiardini, M.; et al. A Tat/Rev Induced Limiting Dilution Assay to Measure Viral Reservoirs in Non-Human Primate Models of HIV Infection. Sci. Rep. 2019, 9, 12078. [Google Scholar] [CrossRef]
- Bertoldi, A.; D’Urbano, V.; Bon, I.; Verbon, A.; Rokx, C.; Boucher, C.; van Kampen, J.J.A.; Gruters, R.A.; Gallinella, G.; Calza, L.; et al. Development of C-TILDA: A modified TILDA method for reservoir quantification in long term treated patients infected with subtype C HIV-1. J. Virol. Methods 2020, 276, 113778. [Google Scholar] [CrossRef] [PubMed]
- Hiener, B.; Horsburgh, B.A.; Eden, J.S.; Barton, K.; Schlub, T.E.; Lee, E.; von Stockenstrom, S.; Odevall, L.; Milush, J.M.; Liegler, T.; et al. Identification of Genetically Intact HIV-1 Proviruses in Specific CD4(+) T Cells from Effectively Treated Participants. Cell Rep. 2017, 21, 813–822. [Google Scholar] [CrossRef] [PubMed]
- Einkauf, K.B.; Lee, G.Q.; Gao, C.; Sharaf, R.; Sun, X.; Hua, S.; Chen, S.M.; Jiang, C.; Lian, X.; Chowdhury, F.Z.; et al. Intact HIV-1 proviruses accumulate at distinct chromosomal positions during prolonged antiretroviral therapy. J. Clin. Investig. 2019, 129, 988–998. [Google Scholar] [CrossRef] [PubMed]
- Kuo, H.H.; Banga, R.; Lee, G.Q.; Gao, C.; Cavassini, M.; Corpataux, J.M.; Blackmer, J.E.; Zur Wiesch, J.S.; Yu, X.G.; Pantaleo, G.; et al. Blood and lymph node dissemination of clonal genome-intact HIV-1 DNA sequences during suppressive antiretroviral therapy. J. Infect. Dis. 2020, 222, 655–660. [Google Scholar] [CrossRef]
- Pinzone, M.R.; Bertuccio, M.P.; VanBelzen, D.J.; Zurakowski, R.; O’Doherty, U. Next-Generation Sequencing in a Direct Model of HIV Infection Reveals Important Parallels to and Differences from In Vivo Reservoir Dynamics. J. Virol. 2020, 94, e01900-19. [Google Scholar] [CrossRef]
- Lambrechts, L.; Cole, B.; Rutsaert, S.; Trypsteen, W.; Vandekerckhove, L. Emerging PCR-Based Techniques to Study HIV-1 Reservoir Persistence. Viruses 2020, 12, 149. [Google Scholar] [CrossRef]
- Gao, H.; Hawkins, T.; Jasti, A.; Chen, Y.H.; Mockaitis, K.; Dinauer, M.; Cornetta, K. Development and Evaluation of Quality Metrics for Bioinformatics Analysis of Viral Insertion Site Data Generated Using High Throughput Sequencing. Biomedicines 2014, 2, 195–210. [Google Scholar] [CrossRef]
- Wells, D.W.; Guo, S.; Shao, W.; Bale, M.J.; Coffin, J.M.; Hughes, S.H.; Wu, X. An analytical pipeline for identifying and mapping the integration sites of HIV and other retroviruses. BMC Genom. 2020, 21, 216. [Google Scholar] [CrossRef]
- Artesi, M.; Hahaut, V.; Cole, B.; Lambrechts, L.; Ashrafi, F.; Marcais, A.; Hermine, O.; Griebel, P.; Arsic, N.; van der Meer, F.; et al. PCIP-seq: Simultaneous sequencing of integrated viral genomes and their insertion sites with long reads. Genome Biol. 2021, 22, 97. [Google Scholar] [CrossRef]
- Schmidt, M.; Hoffmann, G.; Wissler, M.; Lemke, N.; Mussig, A.; Glimm, H.; Williams, D.A.; Ragg, S.; Hesemann, C.U.; von Kalle, C. Detection and direct genomic sequencing of multiple rare unknown flanking DNA in highly complex samples. Hum. Gene Ther. 2001, 12, 743–749. [Google Scholar] [CrossRef]
- Wu, C.; Jares, A.; Winkler, T.; Xie, J.; Metais, J.Y.; Dunbar, C.E. High efficiency restriction enzyme-free linear amplification-mediated polymerase chain reaction approach for tracking lentiviral integration sites does not abrogate retrieval bias. Hum. Gene Ther. 2013, 24, 38–47. [Google Scholar] [CrossRef]
- Vandergeeten, C.; Fromentin, R.; Merlini, E.; Lawani, M.B.; DaFonseca, S.; Bakeman, W.; McNulty, A.; Ramgopal, M.; Michael, N.; Kim, J.H.; et al. Cross-clade ultrasensitive PCR-based assays to measure HIV persistence in large-cohort studies. J. Virol. 2014, 88, 12385–12396. [Google Scholar] [CrossRef]
- Chun, T.W.; Carruth, L.; Finzi, D.; Shen, X.; DiGiuseppe, J.A.; Taylor, H.; Hermankova, M.; Chadwick, K.; Margolick, J.; Quinn, T.C.; et al. Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature 1997, 387, 183–188. [Google Scholar] [CrossRef]
- Gabriel, R.; Eckenberg, R.; Paruzynski, A.; Bartholomae, C.C.; Nowrouzi, A.; Arens, A.; Howe, S.J.; Recchia, A.; Cattoglio, C.; Wang, W.; et al. Comprehensive genomic access to vector integration in clinical gene therapy. Nat. Med. 2009, 15, 1431–1436. [Google Scholar] [CrossRef]
- Paruzynski, A.; Arens, A.; Gabriel, R.; Bartholomae, C.C.; Scholz, S.; Wang, W.; Wolf, S.; Glimm, H.; Schmidt, M.; von Kalle, C. Genome-wide high-throughput integrome analyses by nrLAM-PCR and next-generation sequencing. Nat. Protocol 2010, 5, 1379–1395. [Google Scholar] [CrossRef]
- Patro, S.C.; Brandt, L.D.; Bale, M.J.; Halvas, E.K.; Joseph, K.W.; Shao, W.; Wu, X.; Guo, S.; Murrell, B.; Wiegand, A.; et al. Combined HIV-1 sequence and integration site analysis informs viral dynamics and allows reconstruction of replicating viral ancestors. Proc. Natl. Acad. Sci. USA 2019, 116, 25891–25899. [Google Scholar] [CrossRef]
- Lee, G.Q.; Bangsberg, D.R.; Mo, T.; Lachowski, C.; Brumme, C.J.; Zhang, W.; Lima, V.D.; Boum, Y.; Mwebesa, B.B.; Muzoora, C.; et al. Prevalence and clinical impacts of HIV-1 intersubtype recombinants in Uganda revealed by near-full-genome population and deep sequencing approaches. AIDS 2017, 31, 2345–2354. [Google Scholar] [CrossRef]
- Wagner, T.A.; McLaughlin, S.; Garg, K.; Cheung, C.Y.; Larsen, B.B.; Styrchak, S.; Huang, H.C.; Edlefsen, P.T.; Mullins, J.I.; Frenkel, L.M. HIV latency. Proliferation of cells with HIV integrated into cancer genes contributes to persistent infection. Science 2014, 345, 570–573. [Google Scholar] [CrossRef]
- Serrao, E.; Cherepanov, P.; Engelman, A.N. Amplification, Next-generation Sequencing, and Genomic DNA Mapping of Retroviral Integration Sites. J. Vis. Exp. 2016, 53840. [Google Scholar] [CrossRef]
- Rouzioux, C.; Avettand-Fenoel, V. Total HIV DNA: A global marker of HIV persistence. Retrovirology 2018, 15, 30. [Google Scholar] [CrossRef]
- Rutsaert, S.; De Spiegelaere, W.; Van Hecke, C.; De Scheerder, M.A.; Kiselinova, M.; Vervisch, K.; Trypsteen, W.; Vandekerckhove, L. In-depth validation of total HIV-1 DNA assays for quantification of various HIV-1 subtypes. Sci. Rep. 2018, 8, 17274. [Google Scholar] [CrossRef]
- Thomas, J.; Ruggiero, A.; Procopio, F.A.; Pantaleo, G.; Paxton, W.A.; Pollakis, G. Comparative analysis and generation of a robust HIV-1 DNA quantification assay. J. Virol. Methods 2018, 263, 24–31. [Google Scholar] [CrossRef]
- Brussel, A.; Delelis, O.; Sonigo, P. Alu-LTR real-time nested PCR assay for quantifying integrated HIV-1 DNA. Methods Mol. Biol. 2005, 304, 139–154. [Google Scholar] [CrossRef]
- Agosto, L.M.; Yu, J.J.; Dai, J.; Kaletsky, R.; Monie, D.; O’Doherty, U. HIV-1 integrates into resting CD4+ T cells even at low inoculums as demonstrated with an improved assay for HIV-1 integration. Virology 2007, 368, 60–72. [Google Scholar] [CrossRef]
- Liszewski, M.K.; Yu, J.J.; O’Doherty, U. Detecting HIV-1 integration by repetitive-sampling Alu-gag PCR. Methods 2009, 47, 254–260. [Google Scholar] [CrossRef]
- Brady, T.; Kelly, B.J.; Male, F.; Roth, S.; Bailey, A.; Malani, N.; Gijsbers, R.; O’Doherty, U.; Bushman, F.D. Quantitation of HIV DNA integration: Effects of differential integration site distributions on Alu-PCR assays. J. Virol. Methods 2013, 189, 53–57. [Google Scholar] [CrossRef]
- De Spiegelaere, W.; Malatinkova, E.; Lynch, L.; Van Nieuwerburgh, F.; Messiaen, P.; O’Doherty, U.; Vandekerckhove, L. Quantification of Integrated HIV DNA by Repetitive-Sampling Alu-HIV PCR on the Basis of Poisson Statistics. Clin. Chem. 2014, 60, 886–895. [Google Scholar] [CrossRef]
- Eriksson, S.; Graf, E.H.; Dahl, V.; Strain, M.C.; Yukl, S.A.; Lysenko, E.S.; Bosch, R.J.; Lai, J.; Chioma, S.; Emad, F.; et al. Comparative analysis of measures of viral reservoirs in HIV-1 eradication studies. PLoS Pathog. 2013, 9, e1003174. [Google Scholar] [CrossRef]
- Kiselinova, M.; De Spiegelaere, W.; Buzon, M.J.; Malatinkova, E.; Lichterfeld, M.; Vandekerckhove, L. Integrated and Total HIV-1 DNA Predict Ex Vivo Viral Outgrowth. PLoS Pathog. 2016, 12, e1005472. [Google Scholar] [CrossRef][Green Version]
- O’Doherty, U.; Swiggard, W.J.; Jeyakumar, D.; McGain, D.; Malim, M.H. A sensitive, quantitative assay for human immunodeficiency virus type 1 integration. J. Virol. 2002, 76, 10942–10950. [Google Scholar] [CrossRef]
- Yu, J.J.; Wu, T.L.; Liszewski, M.K.; Dai, J.; Swiggard, W.J.; Baytop, C.; Frank, I.; Levine, B.L.; Yang, W.; Theodosopoulos, T.; et al. A more precise HIV integration assay designed to detect small differences finds lower levels of integrated DNA in HAART treated patients. Virology 2008, 379, 78–86. [Google Scholar] [CrossRef] [PubMed]
- Mexas, A.M.; Graf, E.H.; Pace, M.J.; Yu, J.J.; Papasavvas, E.; Azzoni, L.; Busch, M.P.; Di Mascio, M.; Foulkes, A.S.; Migueles, S.A.; et al. Concurrent measures of total and integrated HIV DNA monitor reservoirs and ongoing replication in eradication trials. Aids 2012, 26, 2295–2306. [Google Scholar] [CrossRef] [PubMed]
- Malatinkova, E.; De Spiegelaere, W.; Bonczkowski, P.; Kiselinova, M.; Vervisch, K.; Trypsteen, W.; Johnson, M.; Verhofstede, C.; de Looze, D.; Murray, C.; et al. Impact of a decade of successful antiretroviral therapy initiated at HIV-1 seroconversion on blood and rectal reservoirs. eLife 2015, 4, e09115. [Google Scholar] [CrossRef] [PubMed]
- Ruggiero, A.; De Spiegelaere, W.; Cozzi-Lepri, A.; Kiselinova, M.; Pollakis, G.; Beloukas, A.; Vandekerckhove, L.; Strain, M.; Richman, D.; Phillips, A.; et al. During Stably Suppressive Antiretroviral Therapy Integrated HIV-1 DNA Load in Peripheral Blood is Associated with the Frequency of CD8 Cells Expressing HLA-DR/DP/DQ. EBioMedicine 2015, 2, 1153–1159. [Google Scholar] [CrossRef]
- Ganor, Y.; Real, F.; Sennepin, A.; Dutertre, C.A.; Prevedel, L.; Xu, L.; Tudor, D.; Charmeteau, B.; Couedel-Courteille, A.; Marion, S.; et al. HIV-1 reservoirs in urethral macrophages of patients under suppressive antiretroviral therapy. Nat. Microbiol. 2019, 4, 633–644. [Google Scholar] [CrossRef]
- Tremeaux, P.; Lenfant, T.; Boufassa, F.; Essat, A.; Melard, A.; Gousset, M.; Delelis, O.; Viard, J.P.; Bary, M.; Goujard, C.; et al. Increasing contribution of integrated forms to total HIV DNA in blood during HIV disease progression from primary infection. Ebiomedicine 2019, 41, 455–464. [Google Scholar] [CrossRef]
- Rasmussen, T.A.; Tolstrup, M.; Brinkmann, C.R.; Olesen, R.; Erikstrup, C.; Solomon, A.; Winckelmann, A.; Palmer, S.; Dinarello, C.; Buzon, M.; et al. Panobinostat, a histone deacetylase inhibitor, for latent-virus reactivation in HIV-infected patients on suppressive antiretroviral therapy: A phase 1/2, single group, clinical trial. Lancet HIV 2014, 1, e13–e21. [Google Scholar] [CrossRef]
- Jacobson, J.M.; Routy, J.P.; Welles, S.; DeBenedette, M.; Tcherepanova, I.; Angel, J.B.; Asmuth, D.M.; Stein, D.K.; Baril, J.G.; McKellar, M.; et al. Dendritic Cell Immunotherapy for HIV-1 Infection Using Autologous HIV-1 RNA: A Randomized, Double-Blind, Placebo-Controlled Clinical Trial. J. Acquir. Immune Defic. Syndr. 2016, 72, 31–38. [Google Scholar] [CrossRef]
- Tapia, G.; Hojen, J.F.; Okvist, M.; Olesen, R.; Leth, S.; Nissen, S.K.; VanBelzen, D.J.; O’Doherty, U.; Mork, A.; Krogsgaard, K.; et al. Sequential Vacc-4x and romidepsin during combination antiretroviral therapy (cART): Immune responses to Vacc-4x regions on p24 and changes in HIV reservoirs. J. Infect. 2017, 75, 555–571. [Google Scholar] [CrossRef]
- Vibholm, L.; Schleimann, M.H.; Hojen, J.F.; Benfield, T.; Offersen, R.; Rasmussen, K.; Olesen, R.; Dige, A.; Agnholt, J.; Grau, J.; et al. Short-Course Toll-Like Receptor 9 Agonist Treatment Impacts Innate Immunity and Plasma Viremia in Individuals With Human Immunodeficiency Virus Infection. Clin. Infect. Dis. 2017, 64, 1686–1695. [Google Scholar] [CrossRef]
- Moron-Lopez, S.; Navarro, J.; Jimenez, M.; Rutsaert, S.; Urrea, V.; Puertas, M.C.; Torrella, A.; De Clercq, L.; Ribas, B.P.; Galvez, C.; et al. Switching From a Protease Inhibitor-based Regimen to a Dolutegravir-based Regimen: A Randomized Clinical Trial to Determine the Effect on Peripheral Blood and Ileum Biopsies From Antiretroviral Therapy-suppressed Human Immunodeficiency Virus-infected Individuals. Clin. Infect. Dis. 2019, 69, 1320–1328. [Google Scholar] [CrossRef]
- Pinzone, M.R.; O’Doherty, U. Measuring integrated HIV DNA ex vivo and in vitro provides insights about how reservoirs are formed and maintained. Retrovirology 2018, 15, 22. [Google Scholar] [CrossRef]
- Avettand-Fenoel, V.; Chaix, M.L.; Blanche, S.; Burgard, M.; Floch, C.; Toure, K.; Allemon, M.C.; Warszawski, J.; Rouzioux, C.; The French Pediatric Cohort Study ANRS—CO 01 Group. LTR real-time PCR for HIV-1 DNA quantitation in blood cells for early diagnosis in infants born to seropositive mothers treated in HAART area (ANRS CO 01). J. Med. Virol. 2009, 81, 217–223. [Google Scholar] [CrossRef]
- Avettand-Fènoël, V.; Hocqueloux, L.; Ghosn, J.; Cheret, A.; Frange, P.; Melard, A.; Viard, J.P.; Rouzioux, C. Total HIV-1 DNA, a Marker of Viral Reservoir Dynamics with Clinical Implications. Clin. Microbiol. Rev. 2016, 29, 859–880. [Google Scholar] [CrossRef]
- Ruggiero, A.; Cozzi-Lepri, A.; Beloukas, A.; Richman, D.; Khoo, S.; Phillips, A.; Geretti, A.M.; Group, E.S. Factors Associated with Persistence of Plasma HIV-1 RNA during Long-term Continuously Suppressive Firstline Antiretroviral Therapy. Open Forum Infect. Dis. 2018, 5, ofy032. [Google Scholar] [CrossRef]
- Symons, J.; Chopra, A.; Malatinkova, E.; De Spiegelaere, W.; Leary, S.; Cooper, D.; Abana, C.O.; Rhodes, A.; Rezaei, S.D.; Vandekerckhove, L.; et al. HIV integration sites in latently infected cell lines: Evidence of ongoing replication. Retrovirology 2017, 14, 2. [Google Scholar] [CrossRef]
- Van der Sluis, R.M.; van Montfort, T.; Centlivre, M.; Schopman, N.C.; Cornelissen, M.; Sanders, R.W.; Berkhout, B.; Jeeninga, R.E.; Paxton, W.A.; Pollakis, G. Quantitation of HIV-1 DNA with a sensitive TaqMan assay that has broad subtype specificity. J. Virol. Methods 2013, 187, 94–102. [Google Scholar] [CrossRef]
- Carstensen, B. Comparing methods of measurement: Extending the LoA by regression. Stat. Med. 2010, 29, 401–410. [Google Scholar] [CrossRef]
- Ruggiero, A.; Malatinkova, E.; Rutsaert, S.; Paxton, W.A.; Vandekerckhove, L.; Spiegelaere, W.D. Utility of integrated HIV-1 DNA quantification in cure studies. Future Virol. 2017, 12, 215–225. [Google Scholar] [CrossRef]
- Rainwater-Lovett, K.; Ziemniak, C.; Watson, D.; Luzuriaga, K.; Siberry, G.; Petru, A.; Chen, Y.; Uprety, P.; McManus, M.; Ho, Y.C.; et al. Paucity of Intact Non-Induced Provirus with Early, Long-Term Antiretroviral Therapy of Perinatal HIV Infection. PloS ONE 2017, 12, e0170548. [Google Scholar] [CrossRef]
- Pollack, R.A.; Jones, R.B.; Pertea, M.; Bruner, K.M.; Martin, A.R.; Thomas, A.S.; Capoferri, A.A.; Beg, S.A.; Huang, S.H.; Karandish, S.; et al. Defective HIV-1 Proviruses Are Expressed and Can Be Recognized by Cytotoxic T Lymphocytes, which Shape the Proviral Landscape. Cell Host Microbe 2017, 21, 494–506. [Google Scholar] [CrossRef]
- Maldarelli, F.; Wu, X.; Su, L.; Simonetti, F.R.; Shao, W.; Hill, S.; Spindler, J.; Ferris, A.L.; Mellors, J.W.; Kearney, M.F.; et al. HIV latency. Specific HIV integration sites are linked to clonal expansion and persistence of infected cells. Science 2014, 345, 179–183. [Google Scholar] [CrossRef]
- Murray, J.M.; Zaunders, J.J.; McBride, K.L.; Xu, Y.; Bailey, M.; Suzuki, K.; Cooper, D.A.; Emery, S.; Kelleher, A.D.; Koelsch, K.K.; et al. HIV DNA subspecies persist in both activated and resting memory CD4+ T cells during antiretroviral therapy. J. Virol. 2014, 88, 3516–3526. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Malatinkova, E.; Thomas, J.; De Spiegelaere, W.; Rutsaert, S.; Geretti, A.M.; Pollakis, G.; Paxton, W.A.; Vandekerckhove, L.; Ruggiero, A. Measuring Proviral HIV-1 DNA: Hurdles and Improvements to an Assay Monitoring Integration Events Utilising Human Alu Repeat Sequences. Life 2021, 11, 1410. https://doi.org/10.3390/life11121410
Malatinkova E, Thomas J, De Spiegelaere W, Rutsaert S, Geretti AM, Pollakis G, Paxton WA, Vandekerckhove L, Ruggiero A. Measuring Proviral HIV-1 DNA: Hurdles and Improvements to an Assay Monitoring Integration Events Utilising Human Alu Repeat Sequences. Life. 2021; 11(12):1410. https://doi.org/10.3390/life11121410
Chicago/Turabian StyleMalatinkova, Eva, Jordan Thomas, Ward De Spiegelaere, Sofie Rutsaert, Anna Maria Geretti, Georgios Pollakis, William A. Paxton, Linos Vandekerckhove, and Alessandra Ruggiero. 2021. "Measuring Proviral HIV-1 DNA: Hurdles and Improvements to an Assay Monitoring Integration Events Utilising Human Alu Repeat Sequences" Life 11, no. 12: 1410. https://doi.org/10.3390/life11121410
APA StyleMalatinkova, E., Thomas, J., De Spiegelaere, W., Rutsaert, S., Geretti, A. M., Pollakis, G., Paxton, W. A., Vandekerckhove, L., & Ruggiero, A. (2021). Measuring Proviral HIV-1 DNA: Hurdles and Improvements to an Assay Monitoring Integration Events Utilising Human Alu Repeat Sequences. Life, 11(12), 1410. https://doi.org/10.3390/life11121410








