The Genome of the “Sea Vomit” Didemnum vexillum
Abstract
1. Introduction
2. Materials and Methods
2.1. DNA and RNA Sequencing
2.1.1. DNA Extraction
2.1.2. Partial Degradation of Genomic DNA
2.1.3. RNA Extraction
2.1.4. Genome Sequencing
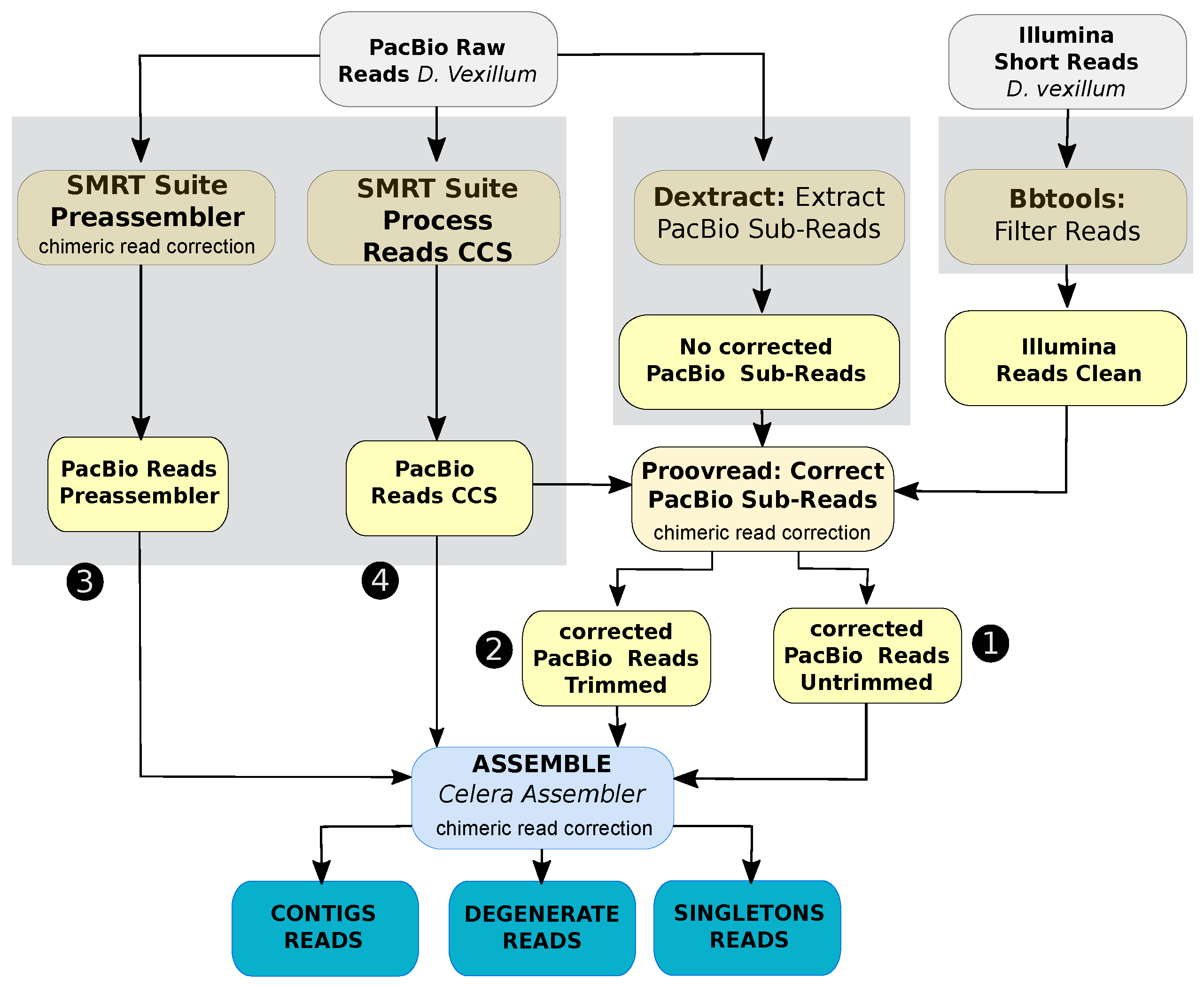
2.2. Assembly of D. vexillum Genome
2.2.1. Data Preprocessing and Pre-Assembling
2.2.2. Contig-Level Assembly
2.2.3. Genome Scaffolding
2.2.4. Assembly Polishing
2.2.5. Assessment of Genome Assembly Quality
2.3. Transcriptome Data Assembly
2.4. Genome Annotation
2.5. Identification of Contamination
2.6. Annotation of Non-Coding RNAs
2.7. Computational Identification of miRNAs
2.8. Mitochondrial Genes
2.9. Genome Size and GC Content Estimation
2.10. Functional Annotation of Protein Coding Genes
2.10.1. Protein Enrichment Analysis
2.10.2. Interaction Analysis of Proteins
2.10.3. Annotation of Homeobox Proteins
2.10.4. Detection of Orthologous Proteins Involved in Skeletogenesis
2.10.5. Gene Phylogenies
2.11. Genome Browser Construction
3. Results
3.1. Assembly of the D. vexillum Genome
3.2. Transcriptome Sequencing and Assembly
3.3. Genome Annotation
3.3.1. Detection and Analysis of Repetitive Regions
3.3.2. Annotation of Protein-Coding Genes
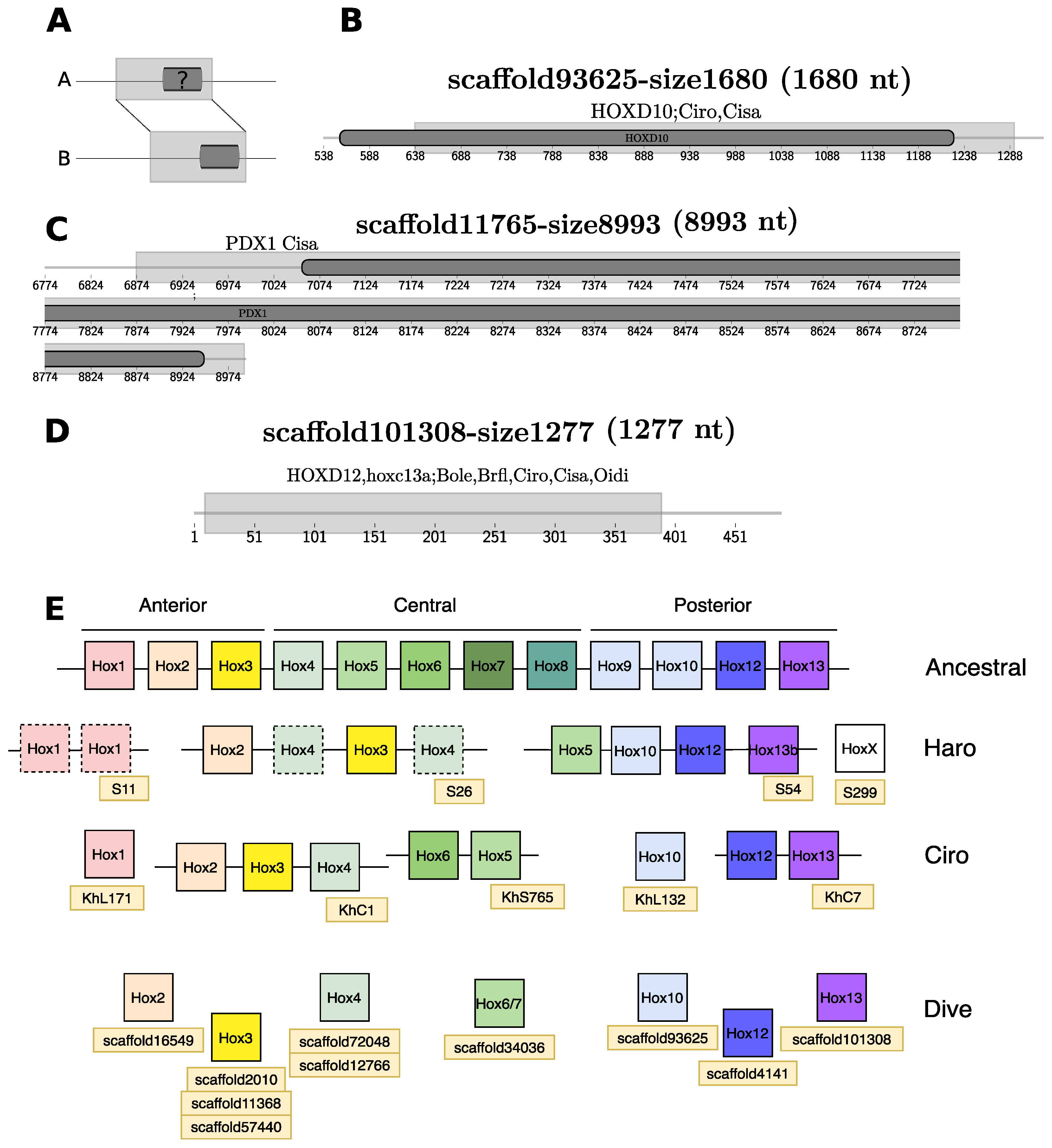
3.3.3. Homeobox Transcription Factors
3.3.4. Annotation of Non-Coding RNAs
3.3.5. Mitochondrial Genome
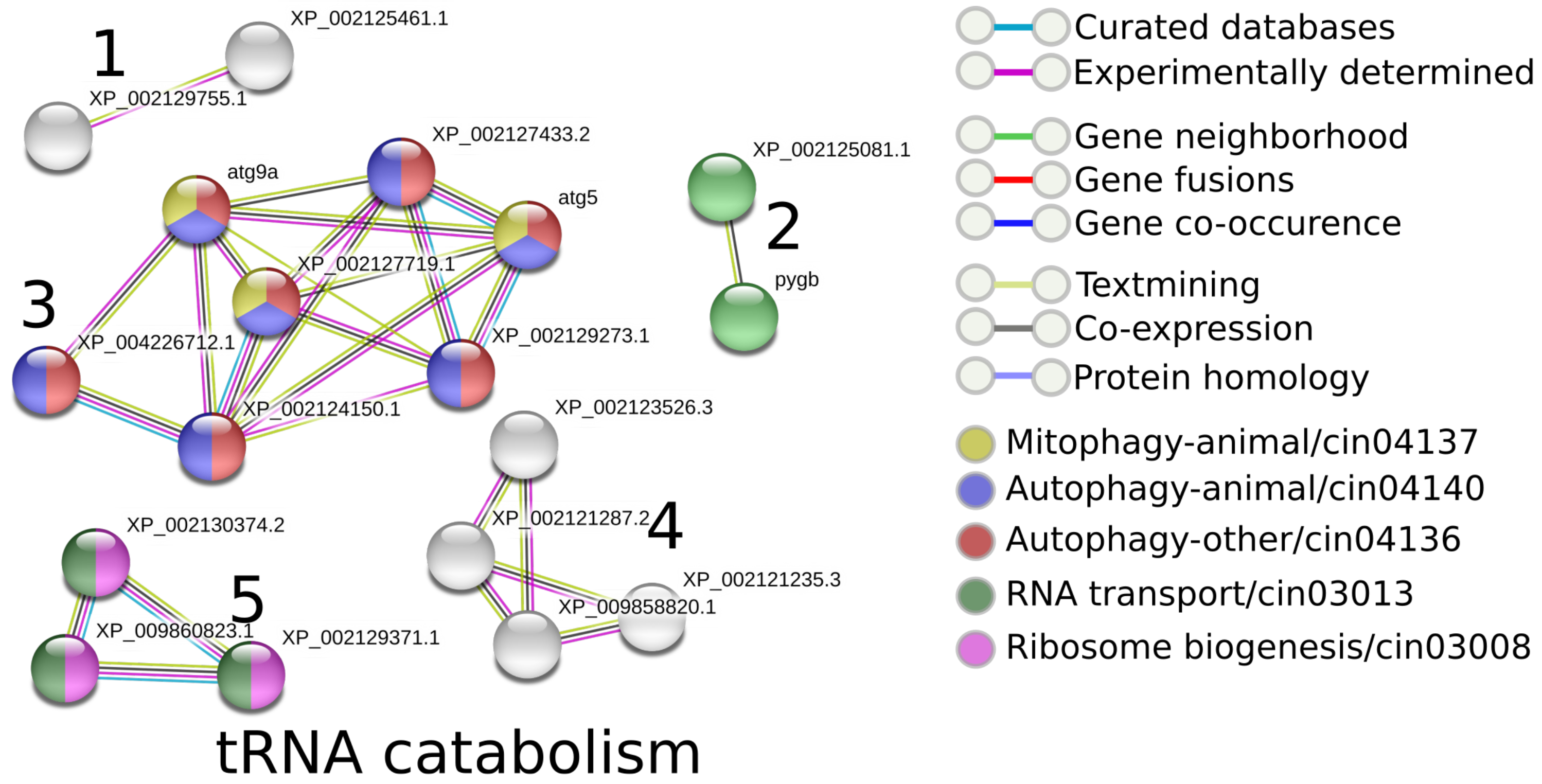
3.3.6. Functional Annotation and Comparison of Proteins across the Tunicates
3.4. Genome Browser and Analysis of Genomic Coordinates
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| BUSCO | Benchmarking Universal Single-Copy Orthologs |
| CM | covariance model |
| Hh | Hedgehog |
| HMM | Hidden Markov Models |
| HOX | homeobox |
| lncRNA | long non-coding RNA |
| ML | Maximum Likelihood |
| miRNA | microRNA |
| misc RNAs | miscellaneous RNAs |
| ncRNA | non-coding RNA |
| RMST | Rhabdomyosarcoma 2-associated transcript |
| rRNA | ribosomal RNA |
| snRNA | small nuclear RNA |
| snoRNA | small nucleolar RNA |
| tRNA | transfer RNA |
References
- Kott, P. A complex didemnid ascidian from Whangamata, New Zealand. J. Mar. Biol. Assoc. UK 2002, 82, 625–628. [Google Scholar] [CrossRef][Green Version]
- Lambert, G. Adventures of a sea squirt sleuth: Unraveling the identity of Didemnum vexillum, a global ascidian invader. Aquat. Invasions 2009, 4, 5–28. [Google Scholar] [CrossRef]
- Valentine, P.C.; Carman, M.R.; Blackwood, D.S.; Heffron, E.J. Ecological observations on the colonial ascidian Didemnum sp. in a New England tide pool habitat. J. Exp. Mar. Biol. Ecol. 2007, 342, 109–121. [Google Scholar] [CrossRef]
- Ordóñez, V.; Pascual, M.; Fernández-Tejedor, M.; Pineda, M.C.; Tagliapietra, D.; Turon, X. Ongoing expansion of the worldwide invader Didemnum vexillum (Ascidiacea) in the Mediterranean Sea: High plasticity of its biological cycle promotes establishment in warm waters. Biol. Invasions 2015, 17, 2075–2085. [Google Scholar] [CrossRef]
- Bullard, S.G.; Sedlack, B.; Reinhardt, J.F.; Litty, C.; Gareau, K.; Whitlatch, R.B. Fragmentation of colonial ascidians: Differences in reattachment capability among species. J. Exp. Mar. Biol. Ecol. 2007, 342, 166–168. [Google Scholar] [CrossRef]
- Cottier-Cook, E.J.; Minchin, D.; Geisler, R.; Graham, J.; Mogg, A.; Sayer, M.D.; Matejusova, I. Biosecurity implications of the highly invasive carpet sea-squirt Didemnum vexillum Kott, 2002 for a protected area of global significance. Manag. Biol. Invasions 2019, 10, 311–323. [Google Scholar] [CrossRef]
- Bullard, S.G.; Lambert, G.; Carman, M.R.; Byrnes, J.; Whitlatch, R.B.; Ruiz, G.; Miller, R.J.; Harris, L.; Valentine, P.C.; Collie, J.S.; et al. The colonial ascidian Didemnum sp. A: Current distribution, basic biology and potential threat to marine communities of the northeast and west coasts of North America. J. Exp. Mar. Biol. Ecol. 2007, 342, 99–108. [Google Scholar] [CrossRef]
- Brunetti, R.; Gissi, C.; Pennati, R.; Caicci, F.; Gasparini, F.; Manni, L. Morphological evidence that the molecularly determined Ciona intestinalis type A and type B are different species: Ciona robusta and Ciona intestinalis. J. Zool. Syst. Evol. Res. 2015, 53, 186–193. [Google Scholar] [CrossRef]
- Dehal, P.; Satou, Y.; Campbell, R.K.; Chapman, J.; Degnan, B.; De Tomaso, A.; Davidson, B.; Di Gregorio, A.; Gelpke, M.; Goodstein, D.M.; et al. The draft genome of Ciona intestinalis: Insights into chordate and vertebrate origins. Science 2002, 298, 2157–2167. [Google Scholar] [CrossRef]
- Small, K.S.; Brudno, M.; Hill, M.M.; Sidow, A. A haplome alignment and reference sequence of the highly polymorphic Ciona savignyi genome. Genome Biol. 2007, 8, R41. [Google Scholar] [CrossRef] [PubMed]
- Hill, M.M.; Broman, K.W.; Stupka, E.; Smith, W.C.; Jiang, D.; Sidow, A. The C. savignyi genetic map and its integration with the reference sequence facilitates insights into chordate genome evolution. Genome Res. 2008, 18, 1369–1379. [Google Scholar] [CrossRef] [PubMed]
- Satou, Y.; Nakamura, R.; Yu, D.; Yoshida, R.; Hamada, M.; Fujie, M.; Hisata, K.; Takeda, H.; Satoh, N. A Nearly Complete Genome of Ciona intestinalis Type A (C. robusta) Reveals the Contribution of Inversion to Chromosomal Evolution in the Genus Ciona. Genome Biol. Evol. 2019, 11, 3144–3157. [Google Scholar] [CrossRef]
- Wei, J.; Zhang, J.; Lu, Q.; Ren, P.; Guo, X.; Wang, J.; Li, X.; Chang, Y.; Duan, S.; Wang, S.; et al. Genomic basis of environmental adaptation in the leathery sea squirt (Styela clava). Mol. Ecol. Resour. 2020, 20, 1414–1431. [Google Scholar] [CrossRef]
- Colombera, D.; Fenaux, R. Chromosome form and Number in the Larvacea. Boll. Zool. 1973, 40, 347–353. [Google Scholar] [CrossRef]
- Seo, H.C.; Kube, M.; Edvardsen, R.B.; Jensen, M.F.; Beck, A.; Spriet, E.; Gorsky, G.; Thompson, E.M.; Lehrach, H.; Reinhardt, R.; et al. Miniature genome in the marine chordate Oikopleura dioica. Science 2001, 294, 2506. [Google Scholar] [CrossRef]
- Denoeud, F.; Henriet, S.; Mungpakdee, S.; Aury, J.M.; Da Silva, C.; Brinkmann, H.; Mikhaleva, J.; Olsen, L.C.; Jubin, C.; Cañestro, C.; et al. Plasticity of animal genome architecture unmasked by rapid evolution of a pelagic tunicate. Science 2010, 330, 1381–1385. [Google Scholar] [CrossRef]
- Wang, K.; Tomura, R.; Chen, W.; Kiyooka, M.; Ishizaki, H.; Aizu, T.; Minakuchi, Y.; Seki, M.; Suzuki, Y.; Omotezako, T.; et al. A genome database for a Japanese population of the larvacean Oikopleura dioica. Dev. Growth Differ. 2020, 62, 450–461. [Google Scholar] [CrossRef]
- Bliznina, A.; Masunaga, A.; Mansfield, M.J.; Tan, Y.; Liu, A.W.; West, C.; Rustagi, T.; Chien, H.C.; Kumar, S.; Pichon, J.; et al. Telomere-to-telomere assembly of the genome of an individual Oikopleura dioica from Okinawa using Nanopore-based sequencing. BMC Genom. 2021, 22, 222. [Google Scholar] [CrossRef] [PubMed]
- Jue, N.K.; Batta-Lona, P.G.; Trusiak, S.; Obergfell, C.; Bucklin, A.; O’Neill, M.J.; O’Neill, R.J. Rapid Evolutionary Rates and Unique Genomic Signatures Discovered in the First Reference Genome for the Southern Ocean Salp, Salpa thompsoni (Urochordata, Thaliacea). Genome Biol. Evol. 2016, 8, 3171–3186. [Google Scholar] [CrossRef]
- Wang, K.; Dantec, C.; Lemaire, P.; Onuma, T.A.; Nishida, H. Genome-wide survey of miRNAs and their evolutionary history in the ascidian, Halocynthia roretzi. BMC Genom. 2017, 18, 314. [Google Scholar] [CrossRef]
- Stolfi, A.; Lowe, E.K.; Racioppi, C.; Ristoratore, F.; Brown, C.T.; Swalla, B.J.; Christiaen, L. Divergent mechanisms regulate conserved cardiopharyngeal development and gene expression in distantly related ascidians. eLife 2014, 3, e03728. [Google Scholar] [CrossRef]
- DeBiasse, M.B.; Colgan, W.N.; Harris, L.; Davidson, B.; Ryan, J.F. Inferring Tunicate Relationships and the Evolution of the Tunicate Hox Cluster with the Genome of Corella inflata. Genome Biol. Evol. 2020, 12, 948–964. [Google Scholar] [CrossRef]
- Brozovic, M.; Dantec, C.; Dardaillon, J.; Dauga, D.; Faure, E.; Gineste, M.; Louis, A.; Naville, M.; Nitta, K.R.; Piette, J.; et al. ANISEED 2017: Extending the integrated ascidian database to the exploration and evolutionary comparison of genome-scale datasets. Nucleic Acids Res. 2017, 46, D718–D725. [Google Scholar] [CrossRef]
- Voskoboynik, A.; Neff, N.F.; Sahoo, D.; Newman, A.M.; Pushkarev, D.; Koh, W.; Passarelli, B.; Fan, H.C.; Mantalas, G.L.; Palmeri, K.J.; et al. The genome sequence of the colonial chordate, Botryllus schlosseri. Elife 2013, 2, e00569. [Google Scholar] [CrossRef]
- Blanchoud, S.; Rutherford, K.; Zondag, L.; Gemmell, N.J.; Wilson, M.J. De novo draft assembly of the Botrylloides leachii genome provides further insight into tunicate evolution. Sci. Rep. 2018, 8, 5518. [Google Scholar] [CrossRef]
- Velandia-Huerto, C.A.; Gittenberger, A.; Brown, F.D.; Stadler, P.F.; Bermúdez-Santana, C.I. Automated detection of ncRNAs in the draft genome sequence of a basal chordate: The Carpet Sea Squirt Didemnum vexillum. BMC Genom. 2016, 17, 591. [Google Scholar] [CrossRef]
- Sasakura, Y.; Ogura, Y.; Treen, N.; Yokomori, R.; Park, S.J.; Nakai, K.; Saiga, H.; Sakuma, T.; Yamamoto, T.; Fujiwara, S.; et al. Transcriptional regulation of a horizontally transferred gene from bacterium to chordate. Proc. R. Soc. B Biol. Sci. 2016, 283, 20161712. [Google Scholar] [CrossRef]
- Delsuc, F.; Brinkmann, H.; Chourrout, D.; Philippe, H. Tunicates and not cephalochordates are the closest living relatives of vertebrates. Nature 2006, 439, 965–968. [Google Scholar] [CrossRef]
- Putnam, N.H.; Butts, T.; Ferrier, D.E.; Furlong, R.F.; Hellsten, U.; Kawashima, T.; Robinson-Rechavi, M.; Shoguchi, E.; Terry, A.; Yu, J.K.; et al. The amphioxus genome and the evolution of the chordate karyotype. Nature 2008, 453, 1064–1071. [Google Scholar] [CrossRef] [PubMed]
- Berná, L.; Alvarez-Valin, F. Evolutionary Genomics of Fast Evolving Tunicates. Genome Biol. Evol. 2014, 6, 1724–1738. [Google Scholar] [CrossRef]
- Wang, S.; Harting, J.; Tseng, E.; Baybayan, P. Getting the Most Out of Your PacBio Libraries with Size Selection; Technical Report; PacBio: Menlo Park, CA, USA, 2019. [Google Scholar]
- Myers, G. DEXTRACTOR. 2015. Available online: https://github.com/thegenemyers/DEXTRACTOR (accessed on 26 July 2019).
- Förster, F.; Schultz, J.; Hedrich, R.; Hackl, T. proovread: Large-scale high-accuracy PacBio correction through iterative short read consensus. Bioinformatics 2014, 30, 3004–3011. [Google Scholar] [CrossRef]
- Myers, E.W.; Sutton, G.G.; Delcher, A.L.; Dew, I.M.; Fasulo, D.P.; Flanigan, M.J.; Kravitz, S.A.; Mobarry, C.M.; Reinert, K.H.J.; Remington, K.A.; et al. A whole-genome assembly of Drosophila. Science 2000, 287, 2196–2204. [Google Scholar] [CrossRef] [PubMed]
- Pryszcz, L.P.; Gabaldón, T. Redundans: An assembly pipeline for highly heterozygous genomes. Nucleic Acids Res. 2016, 44, e113. [Google Scholar] [CrossRef]
- Salmela, L.; Rivals, E. LoRDEC: Accurate and efficient long read error correction. Bioinformatics 2014, 30, 3506–3514. [Google Scholar] [CrossRef]
- PacBio. Unanimity. 2015. Available online: https://github.com/PacificBiosciences/ccs (accessed on 26 July 2019).
- Myers, E. Daligner. 2016. Available online: https://github.com/thegenemyers/DALIGNER (accessed on 26 July 2019).
- Tischler, G.; Myers, E.W. Non hybrid long read consensus using local de Bruijn graph assembly. bioRxiv 2017, 106252. [Google Scholar] [CrossRef]
- Boetzer, M.; Pirovano, W. SSPACE-LongRead: Scaffolding bacterial draft genomes using long read sequence information. BMC Bioinform. 2014, 15, 211. [Google Scholar] [CrossRef]
- PacBio. QUIVER. 2016. Available online: https://github.com/PacificBiosciences/GenomicConsensus (accessed on 26 July 2019).
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef]
- Bushnell, B. BBmap. 2016. Available online: https://sourceforge.net/projects/bbmap (accessed on 26 July 2019).
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644. [Google Scholar] [CrossRef]
- Wu, T.D.; Reeder, J.; Lawrence, M.; Becker, G.; Brauer, M.J. GMAP and GSNAP for Genomic Sequence Alignment: Enhancements to Speed, Accuracy, and Functionality. In Statistical Genomics: Methods and Protocols; Mathé, E., Davis, S., Eds.; Springer: New York, NY, USA, 2016; pp. 283–334. [Google Scholar] [CrossRef]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M.; et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013, 8, 1494–1512. [Google Scholar] [CrossRef]
- Cantarel, B.L.; Korf, I.; Robb, S.M.; Parra, G.; Ross, E.; Moore, B.; Holt, C.; Alvarado, A.S.; Yandell, M. MAKER: An easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res. 2008, 18, 188–196. [Google Scholar] [CrossRef]
- Stanke, M.; Schöffmann, O.; Morgenstern, B.; Waack, S. Gene prediction in eukaryotes with a generalized hidden Markov model that uses hints from external sources. BMC Bioinform. 2006, 7, 62. [Google Scholar] [CrossRef] [PubMed]
- Smit, A.F.A.; Hubley, R.; Green, P. RepeatMasker Open-4.0. 2013–2015. 2015. Available online: http://www.repeatmasker.org (accessed on 26 July 2019).
- Jurka, J.; Kapitonov, V.V.; Pavlicek, A.; Klonowski, P.; Kohany, O.; Walichiewicz, J. Repbase Update, a database of eukaryotic repetitive elements. Cytogen. Genome Res. 2005, 110, 462–467. [Google Scholar] [CrossRef] [PubMed]
- Korf, I. Gene finding in novel genomes. BMC Bioinform. 2004, 5, 59. [Google Scholar] [CrossRef]
- Smit, A.F.; Hubley, R. RepeatModeler Open-1.0. 2008. Available online: http://www.repeatmasker.org (accessed on 26 July 2019).
- Lomsadze, A.; Ter-Hovhannisyan, V.; Chernoff, Y.O.; Borodovsky, M. Gene identification in novel eukaryotic genomes by self-training algorithm. Nucleic Acids Res. 2005, 33, 6494–6506. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Lechner, M.; Findeiß, S.; Steiner, L.; Marz, M.; Stadler, P.F.; Prohaska, S.J. Proteinortho: Detection of (Co-)Orthologs in Large-Scale Analysis. BMC Bioinform. 2011, 12, 124. [Google Scholar] [CrossRef] [PubMed]
- Huerta-Cepas, J.; Szklarczyk, D.; Heller, D.; Hernández-Plaza, A.; Forslund, S.K.; Cook, H.; Mende, D.R.; Letunic, I.; Rattei, T.; Jensen, L.J.; et al. eggNOG 5.0: A hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 2018, 47, D309–D314. [Google Scholar] [CrossRef]
- Huerta-Cepas, J.; Forslund, K.; Coelho, L.P.; Szklarczyk, D.; Jensen, L.J.; von Mering, C.; Bork, P. Fast Genome-Wide Functional Annotation through Orthology Assignment by eggNOG-Mapper. Mol. Biol. Evol. 2017, 34, 2115–2122. [Google Scholar] [CrossRef]
- Yazbeck, A.M.; Stadler, P.F.; Tout, K.; Fallmann, J. Automatic Curation of Large Comparative Animal MicroRNA Data Sets. Bioinformatics 2019, 35, 4553–4559. [Google Scholar] [CrossRef]
- The RNAcentral Consortium. RNAcentral: A hub of information for non-coding RNA sequences. Nucleic Acids Res. 2018, 47, D221–D229. [Google Scholar] [CrossRef]
- Smith, K.F.; Abbott, C.L.; Saito, Y.; Fidler, A.E. Comparison of whole mitochondrial genome sequences from two clades of the invasive ascidian, Didemnum vexillum. Mar. Genom. 2015, 19, 75–83. [Google Scholar] [CrossRef]
- Darling, A.E.; Mau, B.; Perna, N.T. progressiveMauve: Multiple Genome Alignment with Gene Gain, Loss and Rearrangement. PLoS ONE 2010, 5, e11147. [Google Scholar] [CrossRef] [PubMed]
- Nordberg, H.; Cantor, M.; Dusheyko, S.; Hua, S.; Poliakov, A.; Shabalov, I.; Smirnova, T.; Grigoriev, I.V.; Dubchak, I. The genome portal of the Department of Energy Joint Genome Institute: 2014 updates. Nucleic Acids Res. 2013, 42, D26–D31. [Google Scholar] [CrossRef]
- Danks, G.; Campsteijn, C.; Parida, M.; Butcher, S.; Doddapaneni, H.; Fu, B.; Petrin, R.; Metpally, R.; Lenhard, B.; Wincker, P.; et al. OikoBase: A genomics and developmental transcriptomics resource for the urochordate Oikopleura dioica. Nucleic Acids Res. 2012, 41, D845–D853. [Google Scholar] [CrossRef] [PubMed]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2014, 12, 59–60. [Google Scholar] [CrossRef]
- Klopfenstein, D.V.; Zhang, L.; Pedersen, B.S.; Ramírez, F.; Warwick Vesztrocy, A.; Naldi, A.; Mungall, C.J.; Yunes, J.M.; Botvinnik, O.; Weigel, M.; et al. GOATOOLS: A Python library for Gene Ontology analyses. Sci. Rep. 2018, 8, 10872. [Google Scholar] [CrossRef]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016. [Google Scholar]
- Wickham, H. Tidyverse: Easily Install and Load the ‘Tidyverse’. R Package Version 1.2.1; 2017. Available online: https://cran.r-project.org/web/packages/tidyverse/index.html (accessed on 3 December 2021).
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2019. [Google Scholar]
- Supek, F.; Bošnjak, M.; Škunca, N.; Šmuc, T. REVIGO Summarizes and Visualizes Long Lists of Gene Ontology Terms. PLoS ONE 2011, 6, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2018, 47, D607–D613. [Google Scholar] [CrossRef] [PubMed]
- Yates, B.; Braschi, B.; Gray, K.A.; Seal, R.L.; Tweedie, S.; Bruford, E.A. Genenames.org: The HGNC and VGNC resources in 2017. Nucleic Acids Res. 2016, 45, D619–D625. [Google Scholar] [CrossRef]
- Yates, A.D.; Achuthan, P.; Akanni, W.; Allen, J.; Allen, J.; Alvarez-Jarreta, J.; Amode, M.R.; Armean, I.M.; Azov, A.G.; Bennett, R.; et al. Ensembl 2020. Nucleic Acids Res. 2019, 48, D682–D688. [Google Scholar] [CrossRef]
- Sekigami, Y.; Kobayashi, T.; Omi, A.; Nishitsuji, K.; Ikuta, T.; Fujiyama, A.; Satoh, N.; Saiga, H. Hox gene cluster of the ascidian, Halocynthia roretzi, reveals multiple ancient steps of cluster disintegration during ascidian evolution. Zool. Lett. 2017, 3, 17. [Google Scholar] [CrossRef]
- Sekigami, Y.; Kobayashi, T.; Omi, A.; Nishitsuji, K.; Ikuta, T.; Fujiyama, A.; Satoh, N.; Saiga, H. Note to: Hox gene cluster of the ascidian, Halocynthia roretzi, reveals multiple ancient steps of cluster disintegration during ascidian evolution. Zool. Lett. 2019, 5, 8. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Y.F.; Butts, T.; Holland, P.W.H. HomeoDB: A database of homeobox gene diversity. Evol. Dev. 2008, 10, 516–518. [Google Scholar] [CrossRef]
- Harris, R.S. Improved Pairwise Alignment of Genomic DNA. Ph.D. Thesis, Pennsylvania State University, University Park, PA, USA, 2007. [Google Scholar]
- You, L.; Chi, J.; Huang, S.; Yu, T.; Huang, G.; Feng, Y.; Sang, X.; Gao, X.; Li, T.; Yue, Z.; et al. LanceletDB: An integrated genome database for lancelet, comparing domain types and combination in orthologues among lancelet and other species. Database 2019, 2019, baz056. [Google Scholar] [CrossRef]
- Hecht, J.; Stricker, S.; Wiecha, U.; Stiege, A.; Panopoulou, G.; Podsiadlowski, L.; Poustka, A.J.; Dieterich, C.; Ehrich, S.; Suvorova, J.; et al. Evolution of a Core Gene Network for Skeletogenesis in Chordates. PLoS Genet. 2008, 4, e1000025. [Google Scholar] [CrossRef]
- Eddy, S.R. Accelerated Profile HMM Searches. PLoS Comp. Biol. 2011, 7, e1002195. [Google Scholar] [CrossRef]
- Nah, G.S.S.; Tay, B.H.; Brenner, S.; Osato, M.; Venkatesh, B. Characterization of the Runx Gene Family in a Jawless Vertebrate, the Japanese Lamprey (Lethenteron japonicum). PLoS ONE 2014, 9, e113445. [Google Scholar] [CrossRef] [PubMed]
- Huerta-Cepas, J.; Serra, F.; Bork, P. ETE 3: Reconstruction, Analysis, and Visualization of Phylogenomic Data. Mol. Biol. Evol. 2016, 33, 1635–1638. [Google Scholar] [CrossRef]
- Hoff, K.J. MakeHub: Fully automated generation of UCSC Genome Browser Assembly Hubs. Genom. Proteom. Bioinform. 2019, 17, 546–549. [Google Scholar] [CrossRef] [PubMed]
- Raney, B.J.; Dreszer, T.R.; Barber, G.P.; Clawson, H.; Fujita, P.A.; Wang, T.; Nguyen, N.; Paten, B.; Zweig, A.S.; Karolchik, D.; et al. Track data hubs enable visualization of user-defined genome-wide annotations on the UCSC Genome Browser. Bioinformatics 2013, 30, 1003–1005. [Google Scholar] [CrossRef]
- Marz, M.; Kirsten, T.; Stadler, P.F. Evolution of Spliceosomal snRNA Genes in Metazoan Animals. J. Mol. Evol. 2008, 67, 594–607. [Google Scholar] [CrossRef]
- Menzel, P.; Gorodkin, J.; Stadler, P.F. The Tedious Task of Finding Homologous Non-coding RNA Genes. RNA 2009, 15, 2075–2082. [Google Scholar] [CrossRef]
- Velandia-Huerto, C.A.; Brown, F.D.; Gittenberger, A.; Stadler, P.F.; Bermúdez-Santana, C.I. Nonprotein-Coding RNAs as Regulators of Development in Tunicates. In Marine Organisms as Model Systems in Biology and Medicine; Kloc, M., Kubiak, J., Eds.; Springer: Cham, Switzerland, 2018; Volume 65, pp. 197–225. [Google Scholar] [CrossRef]
- Chodroff, R.A.; Goodstadt, L.; Sirey, T.M.; Oliver, P.L.; Davies, K.E.; Green, E.D.; Molnár, Z.; Ponting, C.P. Long noncoding RNA genes: Conservation of sequence and brain expression among diverse amniotes. Genome Biol. 2010, 11, R72. [Google Scholar] [CrossRef]
- Obradovic Wagner, D.; Aspenberg, P. Where did bone come from? Acta Orthop. 2011, 82, 393–398. [Google Scholar] [CrossRef]
- Guth, S.I.E.; Wegner, M. Having it both ways: Sox protein function between conservation and innovation. Cell. Mol. Life Sci. 2008, 65, 3000–3018. [Google Scholar] [CrossRef]
- Takatori, N.; Satou, Y.; Satoh, N. Expression of hedgehog genes in Ciona intestinalis embryos. Mech. Dev. 2002, 116, 235–238. [Google Scholar] [CrossRef]
- Shimeld, S.M. The evolution of the hedgehog gene family in chordates: Insights from amphioxus hedgehog. Dev. Genes Evol. 1999, 209, 40–47. [Google Scholar] [CrossRef]
- Ingham, P.W.; McMahon, A.P. Hedgehog signaling in animal development: Paradigms and principles. Genes Dev. 2001, 15, 3059–3087. [Google Scholar] [CrossRef]
- Stolfi, A.; Sasakura, Y.; Chalopin, D.; Satou, Y.; Christiaen, L.; Dantec, C.; Endo, T.; Naville, M.; Nishida, H.; Swalla, B.J.; et al. Guidelines for the nomenclature of genetic elements in tunicate genomes. Genesis 2015, 53, 1–14. [Google Scholar] [CrossRef]
- Hirose, E. Acid Containers and Cellular Networks in the Ascidian Tunic with Special Remarks on Ascidian Phylogeny. Zool. Sci. 2001, 18, 723–731. [Google Scholar] [CrossRef]
- Shapiro, R.; Klein, R.S. The Deamination of Cytidine and Cytosine by Acidic Buffer Solutions. Mutagenic Implications. Biochemistry 1966, 5, 2358–2362. [Google Scholar] [CrossRef]
- Lindahl, T.; Nyberg, B. Heat-induced deamination of cytosine residues in deoxyribonucleic acid. Biochemistry 1974, 13, 3405–3410. [Google Scholar] [CrossRef]
- Schwartz, D.C.; Cantor, C.R. Separation of yeast chromosome-sized DNAs by pulsed field gradient gel electrophoresis. Cell 1984, 37, 67–75. [Google Scholar] [CrossRef]
- Vogelstein, B.; Gillespie, D. Preparative and analytical purification of DNA from agarose. Proc. Natl. Acad. Sci. USA 1979, 76, 615–619. [Google Scholar] [CrossRef]
- Zhang, H.-B.; Zhao, X.; Ding, X.; Paterson, A.H.; Wing, R.A. Preparation of megabase-size DNA from plant nuclei. Plant J. 1995, 7, 175–184. [Google Scholar] [CrossRef]
- Rinkevich, B.; Fidler, A.E. Initiating laboratory culturing of the invasive ascidian Didemnum vexillum. Manag. Biol. Invasions 2014, 5, 55–62. [Google Scholar] [CrossRef][Green Version]
- Casso, M.; Tagliapietra, D.; Turon, X.; Pascula, M. High fusibility and chimera prevalence in an invasive colonial ascidian. Sci. Rep. 2019, 9, 15673. [Google Scholar] [CrossRef] [PubMed]
- Watts, A.M.; Hopkins, G.A.; Goldstien, S.J. Chimerism and population dieback alter genetic inference related to invasion pathways and connectivity of biofouling populations on artificial substrata. Ecol. Evol. 2019, 9, 3089–3104. [Google Scholar] [CrossRef]
- Rinkevich, B. Natural chimerism in colonial urochordates. J. Exp. Mar. Biol. Ecol. 2005, 322, 93–109. [Google Scholar] [CrossRef]
- Tsagkogeorga, G.; Cahais, V.; Galtier, N. The population genomics of a fast evolver: High levels of diversity, functional constraint, and molecular adaptation in the tunicate Ciona intestinalis. Genome Biol. Evol. 2012, 4, 852–861. [Google Scholar] [CrossRef]
- Gittenberger, A.; Rensing, M.; Niemantsverdriet, P.; Schrieken, N.; D’Hont, A.; Stegenga, H. Soorteninventarisatie oesterputten en oesterpercelen, 2015. I.o.v. Bureau Risicobeoordeling & onderzoeksprogrammering, Nederlandse Voedsel- en Warenautoriteit, Ministerie van Economische Zaken. Technical Report 2015_19, GiMaRIS, 2015. Available online: https://www.nvwa.nl/documenten (accessed on 3 December 2021).
- Gittenberger, A.; Wesdorp, K.; Rensing, M. Biofouling as a transport vector of non-native marine species in the Dutch Delta, along the North Sea coast and in the Wadden Sea. Commissioned by Office for Risk Assessment and Research, the Netherlands Food and Consumer Product Safety Authority. Technical Report 2017_09, GiMaRIS, 2017. Available online: https://www.nvwa.nl/documenten (accessed on 3 December 2021).



| Assembly | Estimated Size (kb) | Number Contigs (c)/ Scaffolds (s) | L50 | N50 | GC Content | IUPAC | Putative Gene Number | Putative Protein Number |
|---|---|---|---|---|---|---|---|---|
| Draft [26] | 542,259 | 882,106 (c) | 152,090 | 918 | N/A | N/A | ||
| This work | 517,553 | 109,769 (s) | 25,281 | 6539 | 0.0155 | 62,194 | 64,424 |
| ncRNA Family | Homology | Mapped | Final |
|---|---|---|---|
| Cis-Reg | 3 (333) | 0 | 3 (333) |
| miRNAs | 248 (2065) | 17 (20) | 235 (1582) |
| misc RNAs | 1 (1) | 1 (1) | 2 (2) |
| lncRNAs | 2 (8) | 0 | 2 (8) |
| Ribozyme | 3 (11) | 0 | 3 (11) |
| rRNAs | 4 (84) | 0 | 4 (84) |
| snoRNAs | 6 (9) | 6 (9) | 12 (18) |
| snRNAs | 9 (87) | 2 (34) | 9 (115) |
| tRNAs | 23 (2724) | NA | 23 (2724) |
| • mt-tRNAs | 0 | 21 | 21 |
| • mt-rRNAs | 0 | 2 | 2 |
| Total | 277 (5322) | 26 (64) | 271 (4877) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Parra-Rincón, E.; Velandia-Huerto, C.A.; Gittenberger, A.; Fallmann, J.; Gatter, T.; Brown, F.D.; Stadler, P.F.; Bermúdez-Santana, C.I. The Genome of the “Sea Vomit” Didemnum vexillum. Life 2021, 11, 1377. https://doi.org/10.3390/life11121377
Parra-Rincón E, Velandia-Huerto CA, Gittenberger A, Fallmann J, Gatter T, Brown FD, Stadler PF, Bermúdez-Santana CI. The Genome of the “Sea Vomit” Didemnum vexillum. Life. 2021; 11(12):1377. https://doi.org/10.3390/life11121377
Chicago/Turabian StyleParra-Rincón, Ernesto, Cristian A. Velandia-Huerto, Adriaan Gittenberger, Jörg Fallmann, Thomas Gatter, Federico D. Brown, Peter F. Stadler, and Clara I. Bermúdez-Santana. 2021. "The Genome of the “Sea Vomit” Didemnum vexillum" Life 11, no. 12: 1377. https://doi.org/10.3390/life11121377
APA StyleParra-Rincón, E., Velandia-Huerto, C. A., Gittenberger, A., Fallmann, J., Gatter, T., Brown, F. D., Stadler, P. F., & Bermúdez-Santana, C. I. (2021). The Genome of the “Sea Vomit” Didemnum vexillum. Life, 11(12), 1377. https://doi.org/10.3390/life11121377







