Origin of Pathogens of Grapevine Crown Gall Disease in Hokkaido in Japan as Characterized by Molecular Epidemiology of Allorhizobium vitis Strains
Abstract
:1. Introduction
2. Materials and Methods
2.1. Multi-Locus Sequence Analysis (MLSA)
2.2. Logistic Regression
2.3. Odds Ratio
3. Results
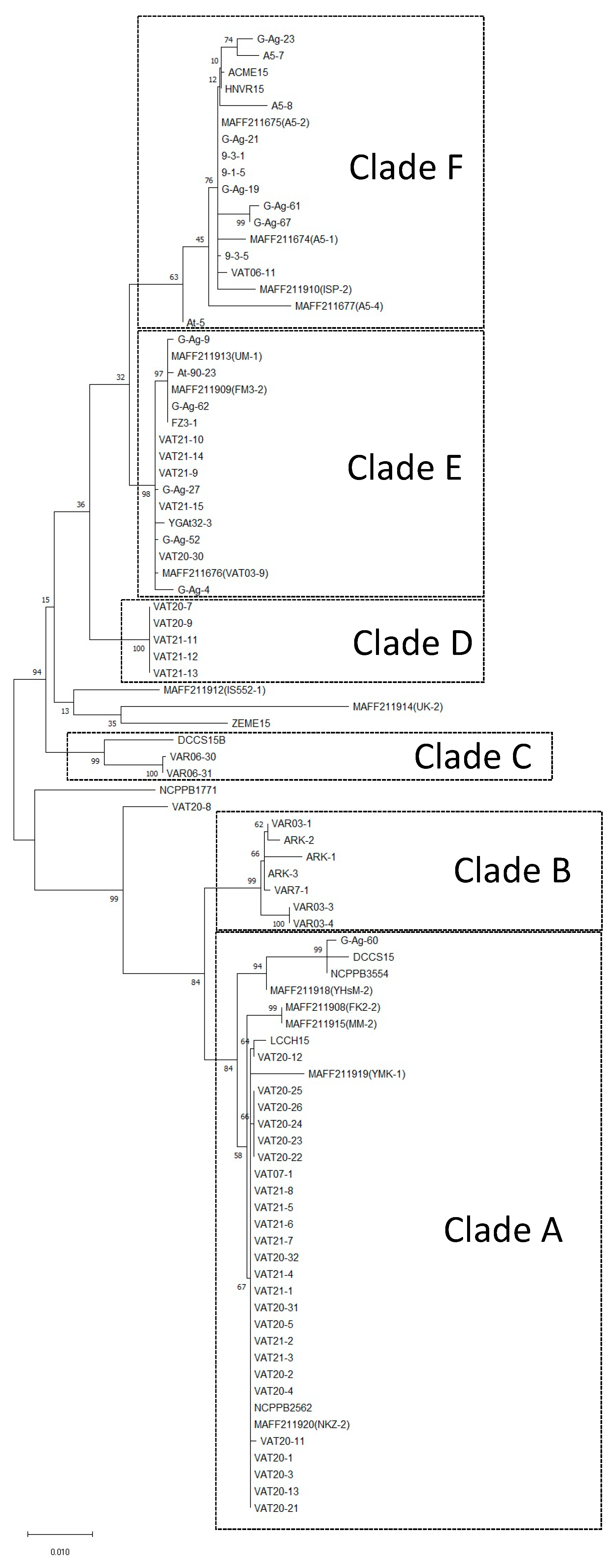
3.1. Multi-Locus Sequence Analysis (MLSA)
3.2. Logistic Regression
3.3. Odds Ratio
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mousavi, S.A.; Willems, A.; Nesme, X.; de Lajudie, P.; Lindstrom, K. Revised phylogeny of Rhizobiaceae: Proposal of the delineation of Pararhizobium gen. nov., and 13 new species combinations. Syst. Appl. Microbiol. 2015, 38, 84–90. [Google Scholar] [CrossRef]
- Burr, T.J.; Otten, L. Crown gall of grape: Biology and disease management. Annu. Rev. Phytopathol. 1999, 37, 53–80. [Google Scholar] [CrossRef]
- Chilton, M.D.; Drummond, M.H.; Merlo, D.J.; Sciaky, D.; Montoya, A.L.; Gordon, M.P.; Nester, W. Stable incorporation of plasmid DNA into higher plant cells: The molecular basis of crown gall tumorigenesis. Cell 1997, 11, 263–271. [Google Scholar] [CrossRef]
- Gelvin, S.B. Traversing the cell: Agrobacterium T-DNA’s journey to the host genome. Front. Plant Sci. 2012, 3, 52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pitzschke, A.; Hirt, H. New insights into an old story: Agrobacterium-induced tumor formation in plants by plant transformation. EMBO J. 2010, 29, 1021–1032. [Google Scholar] [CrossRef]
- Morris, R. Genes specifying auxin and cytokinin biosynthesis in phytopathogens. Annu. Rev. Plant Physiol. 1986, 37, 509–538. [Google Scholar] [CrossRef]
- Burr, T.J.; Bazzi, C.; Süle, S.; Otten, L. Crown gall of grape: Biology of Agrobacterium vitis and the development of disease control strategies. Plant Dis. 1998, 82, 1288–1297. [Google Scholar] [CrossRef] [Green Version]
- Wächter, R.; Langhans, M.; Aloni, R.; Götz, S.; Weilmünster, A.; Koops, A.; Temguia, L.; Mistrik, I.; Pavlovkin, J.; Rascher, U.; et al. Vascularization, high-volume solution flow, and localized roles for enzymes of sucrose metabolism during tumorigenesis by Agrobacterium tumefaciens. Plant Physiol. 2003, 133, 1024–1037. [Google Scholar] [CrossRef] [Green Version]
- Gohlke, J.; Deeken, R. Plant responses to Agrobacterium tumefaciens and crown gall development. Front. Plant Sci. 2014, 5, 155. [Google Scholar] [CrossRef] [Green Version]
- Kawaguchi, A. Studies on the diagnosis and biological control of grapevine crown gall and phylogenetic analysis of tumorigenic Rhizobium vitis. J. Gen. Plant Pathol. 2009, 75, 462–463. [Google Scholar] [CrossRef]
- Kawaguchi, A.; Inoue, K.; Nasu, H. Inhibition of crown gall formation by Agrobacterium radiobacter biovar 3 strains isolated from grapevine. J. Gen. Plant Pathol. 2005, 71, 422–430. [Google Scholar] [CrossRef]
- Kawaguchi, A.; Inoue, K.; Nasu, H. Biological control of grapevine crown gall by nonpathogenic Agrobacterium vitis strain VAR03-1. J. Gen. Plant Pathol. 2007, 73, 133–138. [Google Scholar] [CrossRef]
- Kawaguchi, A.; Inoue, K.; Ichinose, Y. Biological control of crown gall of grapevine, rose, and tomato by nonpathogenic Agrobacterium vitis strain VAR03-1. Phytopathology 2008, 98, 1218–1225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kawaguchi, A.; Kondo, K.; Inoue, K. Biological control of apple crown gall by nonpathogenic Rhizobium vitis strain VAR03-1. J. Gen. Plant Pathol. 2012, 78, 287–293. [Google Scholar] [CrossRef]
- Saito, K.; Watanabe, M.; Matsui, H.; Yamamoto, M.; Ichinose, Y.; Toyoda, K.; Kawaguchi, A.; Noutoshi, Y. Characterization of the suppressive effects of the biological control strain VAR03-1 of Rhizobium vitis on the virulence of tumorigenic R. vitis. J. Gen. Plant Pathol. 2018, 84, 58–64. [Google Scholar] [CrossRef]
- Kawaguchi, A.; Inoue, K. New antagonistic strains of non-pathogenic Agrobacterium vitis to control grapevine crown gall. J. Phytopathol. 2012, 160, 509–518. [Google Scholar] [CrossRef]
- Kawaguchi, A. Biological control of crown gall on grapevine and root colonization by nonpathogenic Rhizobium vitis strain ARK-1. Microbes Environ. 2013, 28, 306–311. [Google Scholar] [CrossRef] [Green Version]
- Kawaguchi, A. Reduction in pathogen populations at grapevine wound sites is associated with the mechanism underlying the biological control of crown gall by Rhizobium vitis strain ARK-1. Microbes Environ. 2014, 29, 296–302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kawaguchi, A. Biological control agent Agrobacterium vitis strain ARK-1 suppresses expression of the virD2 and virE2 genes in tumorigenic A. vitis. Eur. J. Plant Pathol. 2015, 143, 789–799. [Google Scholar] [CrossRef]
- Kawaguchi, A.; Inoue, K.; Tanina, K.; Nita, M. Biological control for grapevine crown gall using nonpathogenic Rhizobium vitis strain ARK-1. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2017, 93, 547–560. [Google Scholar] [CrossRef] [Green Version]
- Kawaguchi, A.; Nita, M.; Ishii, T.; Watanabe, M.; Noutoshi, Y. Biological control agent Rhizobium (=Agrobacterium) vitis strain ARK-1 suppresses expression of the essential and non-essential vir genes of tumorigenic R. vitis. BMC Res. Notes. 2019, 12, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Wong, A.T.; Kawaguchi, A.; Nita, M. Efficacy of a biological control agent Rhizobium vitis ARK-1 against Virginia R. vitis isolates, and relative relationship among Japanese and Virginia R. vitis isolates. Crop Prot. 2021, 146, 105685. [Google Scholar] [CrossRef]
- Burr, T.J.; Katz, B.H. Grapevine cuttings as potential sites of survival and means of dissemination of Agrobacterium tumefaciens. Plant Dis. 1984, 68, 976–978. [Google Scholar] [CrossRef]
- Kawaguchi, A. Genetic diversity of Rhizobium vitis strains in Japan based on multilocus sequence analysis of pyrG, recA and rpoD. J. Gen. Plant Pathol. 2011, 77, 299–303. [Google Scholar] [CrossRef]
- Sawada, H.; Ieki, H. Phenotypic characteristics of the genus Agrobacterium. Annu. Phytopathol. Soc. Jpn. 1992, 58, 37–45. [Google Scholar] [CrossRef]
- Sawada, H.; Ieki, H.; Takikawa, Y. Identification of grapevine crown gall bacteria isolated in Japan. Annu. Phytopathol. Soc. Jpn. 1990, 56, 199–206. [Google Scholar] [CrossRef] [Green Version]
- Sawada, H.; Imada, J.; Ieki, H. Serogroups of Agrobacterium tumefaciens biovar 3 determined using somatic antigens. Annu. Phytopathol. Soc. Jpn. 1992, 58, 52–57. [Google Scholar] [CrossRef] [Green Version]
- Kawaguchi, A.; Sawada, H.; Inoue, K.; Nasu, H. Multiplex PCR for the identification of Agrobacterium biover 3 strainsI. J. Gen. Plant Pathol. 2005, 71, 54–59. [Google Scholar] [CrossRef]
- Kawaguchi, A.; Sawada, H.; Ichinose, Y. Phylogenetic and serological analyses reveal genetic diversity of Agrobacterium vitis strains in Japan. Plant Pathol. 2008, 57, 747–753. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547. [Google Scholar] [CrossRef]
- Kawaguchi, A.; Tanina, K.; Takehara, T. Molecular epidemiology of Pseudomonas syringae pv. syringae strains isolated from barley and wheat infected with bacterial black node. J. Gen. Plant Pathol. 2017, 83, 162–168. [Google Scholar] [CrossRef]
- Kawaguchi, A.; Inoue, K.; Tanina, K. Evaluation of the nonpathogenic Agrobacterium vitis strain ARK-1 for crown gall control in diverse plant species. Plant Dis. 2015, 99, 409–414. [Google Scholar] [CrossRef] [Green Version]
- Stover, E.W.; Swarz, H.J.; Burr, T.J. Eondophytic Agrobacterium in crown gall-resistant and susceptible Vitis genetypes. Vitis 1997, 36, 21–26. [Google Scholar]
- Misawa, T. A field survey of crown gall of grapevine (Vitis vinifera) caused by Agrobacterium vitis in Hokkaido. Annu. Rept. Plant Prot. N. Jpn. 2004, 55, 82–83. (In Japanese) [Google Scholar]
- Misawa, T. Infection and disease development of crown gall in pathogen-free grapevine seedlings replanted in infested vineyard. Annu. Rept. Plant Prot. N. Jpn. 2004, 55, 84–86. (In Japanese) [Google Scholar]
- Gan, H.M.; Szegedi, E.; Fersi, R.; Chebil, S.; Kovács, L.; Kawaguchi, A.; Hudson, A.O.; Burr, T.J.; Savka, M.A. Insight into the microbial co-occurrence and diversity of 73 grapevine (Vitis vinifera) crown galls collected across the northern hemisphere. Front. Microbiol. 2019, 13, 1896. [Google Scholar] [CrossRef] [Green Version]
- Kuzmanović, N.; Biondi, E.; Bertaccini, A.; Obradović, A. Genetic relatedness and recombination analysis of Allorhizobium vitis strains associated with grapevine crown gall outbreaks in Europe. J. Appl. Microbiol. 2015, 119, 786–796. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Noutoshi, Y.; Toyoda, A.; Ishii, T.; Saito, K.; Watanabe, M.; Kawaguchi, A. Complete genome sequence data of tumorigenic Rhizobium vitis strain VAT03-9, a causal agent of grapevine crown gall disease. Mol. Plant Microbe Interact. 2020, 33, 1280–1282. [Google Scholar] [CrossRef]
- Noutoshi, Y.; Toyoda, A.; Ishii, T.; Saito, K.; Watanabe, M.; Kawaguchi, A. Complete genome sequence data of nonpathogenic and nonantagonistic strain of Rhizobium vitis VAR06-30 isolated from grapevine rhizosphere. Mol. Plant Microbe Interact. 2020, 33, 1283–1285. [Google Scholar] [CrossRef]
- Noutoshi, Y.; Toyoda, A.; Ishii, T.; Saito, K.; Watanabe, M.; Kawaguchi, A. Complete genome sequence data of nonpathogenic Rhizobium vitis strain VAT03-1, a biological control agent for grapevine crown gall disease. Mol. Plant Microbe Interact. 2020, 33, 1451–1453. [Google Scholar] [CrossRef]
| Strains (Former Name) | Ti or N a | Cultivar b | Location of Vineyard | Prefcture/State | Country | Isolated Year | Genetic Group c |
|---|---|---|---|---|---|---|---|
| MAFF663001 (G-Ag-27) | Ti | Kyoho | Matsumoto | Nagano | Japan | Before 2000 | E |
| MAFF212292 (YGAt32-3) | Ti | Garnet A | Yamanashi | Yamanashi | Japan | Before 2000 | E |
| MAFF663017 (G-Ag-4) | Ti | Kyoho | Shimane | Shimane | Japan | Before 2000 | E |
| MAFF663004 (G-Ag-9) | Ti | Kyoho | Yokota | Shimane | Japan | Before 2000 | E |
| MAFF211676 (VAT03-9) | Ti | Seto Giants | Asaguchi | Okayama | Japan | 2000 to 2009 | E |
| MAFF211944 (G-Ag-62) | Ti | Kyoho | Sagae | Yamagata | Japan | Before 2000 | E |
| MAFF211889 (G-Ag-52) | Ti | Kyoho | Hanamaki | Iwate | Japan | Before 2000 | E |
| At-90-23 | Ti | Kyoho | Shimane | Shimane | Japan | Before 2000 | E |
| FZ-3-1 | Ti | Zweigeltrebe | Furano | Hokkaido | Japan | 2000 to 2009 | E |
| VAT20-30 | Ti | Pinot Noir | Chitose | Hokkaido | Japan | After 2020 | E |
| VAT21-9 | Ti | Zweigeltrebe | Yoichi | Hokkaido | Japan | After 2020 | E |
| VAT21-10 | Ti | Kerner | Yoichi | Hokkaido | Japan | After 2020 | E |
| VAT21-14 | Ti | Cambell Early | Yoichi | Hokkaido | Japan | After 2020 | E |
| VAT21-15 | Ti | Cambell Early | Yoichi | Hokkaido | Japan | After 2020 | E |
| ACME15 | Ti | Merlot | Winchester | Virginia | USA | 2010 to 2019 | E |
| HNVR15 | Ti | Viognier | Gordonsville | Virginia | USA | 2010 to 2019 | E |
| MAFF211909 (FM-3-2) | Ti | Müller-Thurgau | Furano | Hokkaido | Japan | 2000 to 2009 | E |
| MAFF211913 (UM-1) | Ti | Müller-Thurgau | Urausu | Hokkaido | Japan | 2000 to 2009 | E |
| MAFF211301 (At-5) | Ti | Kyoho | Shimane | Shimane | Japan | Before 2000 | F |
| MAFF211674 (A5-1) | Ti | Kyoho | Yokote | Akita | Japan | 2000 to 2009 | F |
| MAFF211675 (A5-2) | Ti | Kyoho | Yokote | Akita | Japan | 2000 to 2009 | F |
| MAFF211677 (A5-4) | Ti | Kyoho | Yokote | Akita | Japan | 2000 to 2009 | F |
| A5-7 | Ti | Kyoho | Yokote | Akita | Japan | 2000 to 2009 | F |
| MAFF211302 (A5-8) | Ti | Kyoho | Yokote | Akita | Japan | 2000 to 2009 | F |
| VAT06-11 | Ti | Aurora Black | Ukan | Okayama | Japan | 2000 to 2009 | F |
| MAFF663006 (G-Ag-19) | Ti | Rizamat | Shiojiri | Nagano | Japan | Before 2000 | F |
| MAFF663007 (G-Ag-21) | Ti | Kyoho | Shiojiri | Nagano | Japan | Before 2000 | F |
| MAFF663008 (G-Ag-23) | Ti | Kyoho | Shiojiri | Nagano | Japan | Before 2000 | F |
| 9-1-5 | Ti | Kyoho | Hanamaki | Akita | Japan | 2000 to 2009 | F |
| 9-3-1 | Ti | Kyoho | Hanamaki | Akita | Japan | 2000 to 2009 | F |
| 9-3-5 | Ti | Kyoho | Hanamaki | Akita | Japan | 2000 to 2009 | F |
| MAFF211943 (G-Ag-61) | Ti | Beniizu | Sannohe | Aomori | Japan | Before 2000 | F |
| MAFF211949 (G-Ag-67) | Ti | Kyoho | Yokote | Akita | Japan | Before 2000 | F |
| MAFF211910 (ISP-2) | Ti | Pinot Noir | Ikeda | Hokkaido | Japan | 2000 to 2009 | F |
| MAFF212306 (VAR03-1) | N | Seto Giants | Okayama | Okayama | Japan | 2000 to 2009 | B |
| ARK-1 | N | Pione | Okayama | Okayama | Japan | 2000 to 2009 | B |
| MAFF212307 (VAR03-3) | N | Seto Giants | Okayama | Okayama | Japan | 2000 to 2009 | B |
| MAFF212308 (VAR03-4) | N | Seto Giants | Okayama | Okayama | Japan | 2000 to 2009 | B |
| ARK-2 | N | Pione | Okayama | Okayama | Japan | 2000 to 2009 | B |
| ARK-3 | N | Pione | Okayama | Okayama | Japan | 2000 to 2009 | B |
| MAFF212313 (VAR7-1) | N | Seto Giants | Okayama | Okayama | Japan | 2000 to 2009 | B |
| VAR06-30 | N | Aurora Black | Ukan | Okayama | Japan | 2000 to 2009 | C |
| VAR06-31 | N | Aurora Black | Ukan | Okayama | Japan | 2000 to 2009 | C |
| DCCS15B | N | Cabernet Sauvignon | Etlan | Virginia | USA | 2010 to 2019 | C |
| NCPPB3554T | Ti | Unknown | Unknown | Unknown | Australia | Before 2000 | A |
| DCCS15 | Ti | Cabernet Sauvignon | Etlan | Virginia | USA | 2010 to 2019 | A |
| MAFF211942 (G-Ag-60) | Ti | Cambell Early | Nanbu | Aomori | Japan | 2000 to 2009 | A |
| MAFF211918 (YHsM-2) | Ti | Müller-Thurgau | Yoichi | Hokkaido | Japan | 2000 to 2009 | A |
| VAT07-1 | Ti | Aurora Black | Asaguchi | Okayama | Japan | 2000 to 2009 | A |
| NCPPB2562 | Ti | Unknown | Unknown | Unknown | Greece | Before 2000 | A |
| MAFF211919 (YMK-1) | Ti | Kerner | Yoichi | Hokkaido | Japan | 2000 to 2009 | A |
| MAFF211920 (NKZ-2) | Ti | Zweigeltrebe | Niki | Hokkaido | Japan | 2000 to 2009 | A |
| MAFF211908 (FK-2-2) | Ti | Kerner | Furano | Hokkaido | Japan | 2000 to 2009 | A |
| MAFF211915 (MM-2) | Ti | Müller-Thurgau | Mikasa | Hokkaido | Japan | 2000 to 2009 | A |
| LCCH15 | Ti | Chardonnay | Charlottesville | Virginia | USA | 2010 to 2019 | A |
| VAT20-1 | Ti | Zweigeltrebe | Urausu | Hokkaido | Japan | After 2020 | A |
| VAT20-2 | Ti | Zweigeltrebe | Urausu | Hokkaido | Japan | After 2020 | A |
| VAT20-3 | Ti | Kerner | Urausu | Hokkaido | Japan | After 2020 | A |
| VAT21-4 | Ti | Kerner | Urausu | Hokkaido | Japan | After 2020 | A |
| VAT21-5 | Ti | Kerner | Urausu | Hokkaido | Japan | After 2020 | A |
| VAT20-11 | Ti | Zweigeltrebe | Yoichi | Hokkaido | Japan | After 2020 | A |
| VAT20-12 | Ti | Kerner | Yoichi | Hokkaido | Japan | After 2020 | A |
| VAT20-13 | Ti | Zweigeltrebe | Yoichi | Hokkaido | Japan | After 2020 | A |
| VAT20-21 | Ti | Kerner | Niseko | Hokkaido | Japan | After 2020 | A |
| VAT20-22 | Ti | Kerner | Niseko | Hokkaido | Japan | After 2020 | A |
| VAT20-23 | Ti | Kerner | Niseko | Hokkaido | Japan | After 2020 | A |
| VAT20-24 | Ti | Kerner | Niseko | Hokkaido | Japan | After 2020 | A |
| VAT20-25 | Ti | Kerner | Niseko | Hokkaido | Japan | After 2020 | A |
| VAT20-26 | Ti | Kerner | Niseko | Hokkaido | Japan | After 2020 | A |
| VAT20-31 | Ti | Pinot Noir | Chitose | Hokkaido | Japan | After 2020 | A |
| VAT20-32 | Ti | Pinot Noir | Chitose | Hokkaido | Japan | After 2020 | A |
| VAT21-1 | Ti | Zweigeltrebe | Yoichi | Hokkaido | Japan | After 2020 | A |
| VAT21-2 | Ti | Zweigeltrebe | Yoichi | Hokkaido | Japan | After 2020 | A |
| VAT21-3 | Ti | Zweigeltrebe | Yoichi | Hokkaido | Japan | After 2020 | A |
| VAT21-4 | Ti | Zweigeltrebe | Yoichi | Hokkaido | Japan | After 2020 | A |
| VAT21-5 | Ti | Zweigeltrebe | Yoichi | Hokkaido | Japan | After 2020 | A |
| VAT21-6 | Ti | Zweigeltrebe | Yoichi | Hokkaido | Japan | After 2020 | A |
| VAT21-7 | Ti | Zweigeltrebe | Yoichi | Hokkaido | Japan | After 2020 | A |
| VAT21-8 | Ti | Zweigeltrebe | Yoichi | Hokkaido | Japan | After 2020 | A |
| VAT20-7 | Ti | Zweigeltrebe | Urausu | Hokkaido | Japan | After 2020 | D |
| VAT20-9 | Ti | Zweigeltrebe | Urausu | Hokkaido | Japan | After 2020 | D |
| VAT21-11 | Ti | Zweigeltrebe | Urausu | Hokkaido | Japan | After 2020 | D |
| VAT21-12 | Ti | Zweigeltrebe | Urausu | Hokkaido | Japan | After 2020 | D |
| VAT21-13 | Ti | Kerner | Urausu | Hokkaido | Japan | After 2020 | D |
| VAT20-8 | Ti | Zweigeltrebe | Urausu | Hokkaido | Japan | After 2020 | nc |
| ZEME15 | Ti | Merlot | Hamilton | Virginia | USA | 2010 to 2019 | nc |
| MAFF211912 (IS552-1) | Ti | Pinot Noir | Ikeda | Hokkaido | Japan | 2000 to 2009 | nc |
| MAFF211914 (UK-2) | Ti | Kerner | Urausu | Hokkaido | Japan | 2000 to 2009 | nc |
| NCPPB1771 | Ti | Unknown | Unknown | Unknown | Iran | Before 2000 | nc |
| Variable | Parameter Estimate | Standard Error | z Value | p Value b | |
|---|---|---|---|---|---|
| Category | Factor | ||||
| Location of vineyard | Yoichi | 0.971 | 0.683 | 1.422 | 0.155 |
| Prefecture/state | Hokkaido | 2.027 | 0.577 | 3.512 | 4.5 × 10−4 |
| y-Intercept | −1.819 | 0.440 | −4.133 | 3.6 × 10−5 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kawaguchi, A.; Sone, T.; Ochi, S.; Matsushita, Y.; Noutoshi, Y.; Nita, M. Origin of Pathogens of Grapevine Crown Gall Disease in Hokkaido in Japan as Characterized by Molecular Epidemiology of Allorhizobium vitis Strains. Life 2021, 11, 1265. https://doi.org/10.3390/life11111265
Kawaguchi A, Sone T, Ochi S, Matsushita Y, Noutoshi Y, Nita M. Origin of Pathogens of Grapevine Crown Gall Disease in Hokkaido in Japan as Characterized by Molecular Epidemiology of Allorhizobium vitis Strains. Life. 2021; 11(11):1265. https://doi.org/10.3390/life11111265
Chicago/Turabian StyleKawaguchi, Akira, Teruo Sone, Sunao Ochi, Yosuke Matsushita, Yoshiteru Noutoshi, and Mizuho Nita. 2021. "Origin of Pathogens of Grapevine Crown Gall Disease in Hokkaido in Japan as Characterized by Molecular Epidemiology of Allorhizobium vitis Strains" Life 11, no. 11: 1265. https://doi.org/10.3390/life11111265
APA StyleKawaguchi, A., Sone, T., Ochi, S., Matsushita, Y., Noutoshi, Y., & Nita, M. (2021). Origin of Pathogens of Grapevine Crown Gall Disease in Hokkaido in Japan as Characterized by Molecular Epidemiology of Allorhizobium vitis Strains. Life, 11(11), 1265. https://doi.org/10.3390/life11111265








