Application of Domain- and Genotype-Specific Models to Infer Post-Transcriptional Regulation of Segmentation Gene Expression in Drosophila
Abstract
:1. Introduction
2. Materials and Methods
2.1. Model and Data
2.2. Model Fitting and Hypotheses
2.3. Parameter Optimization
3. Results
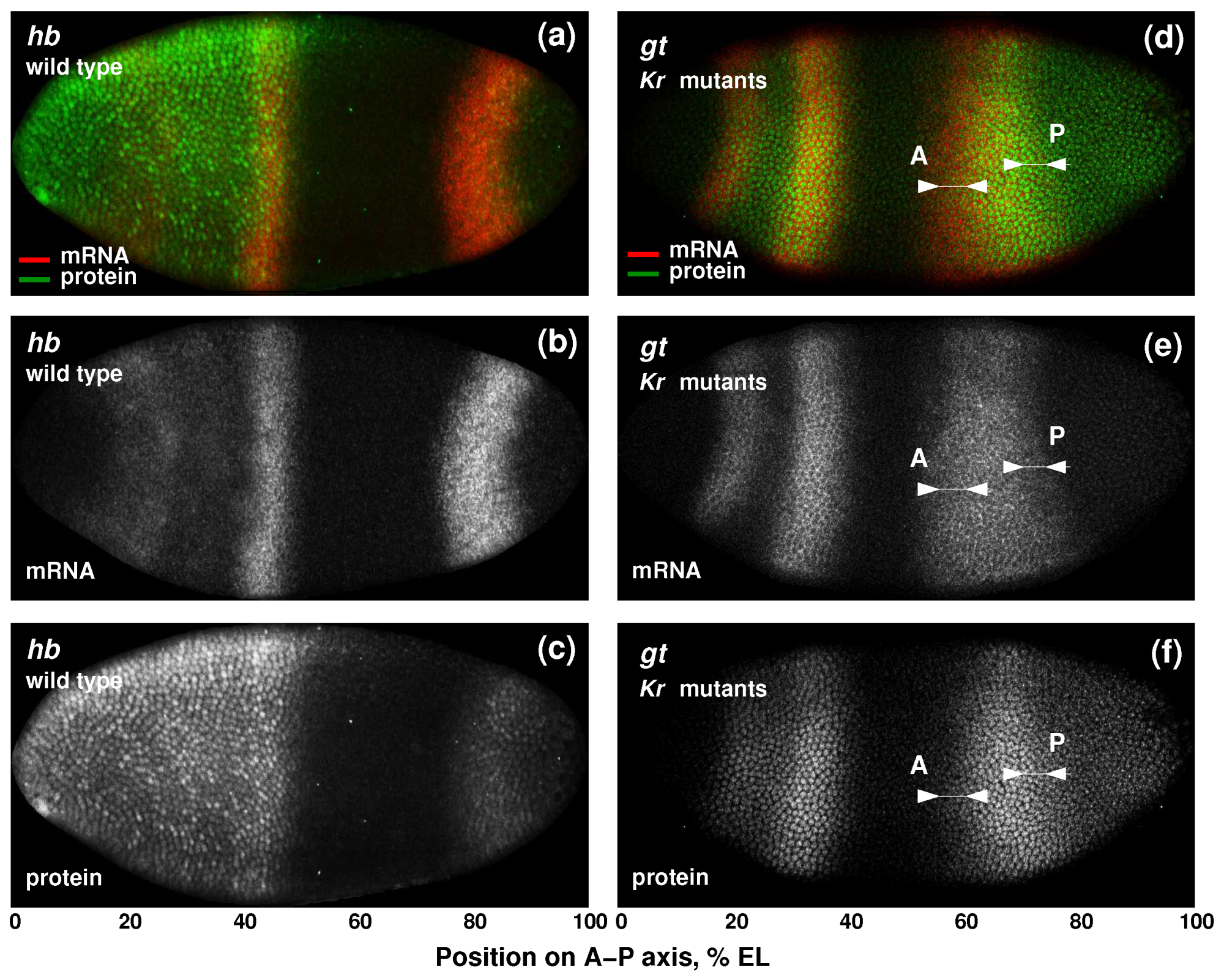
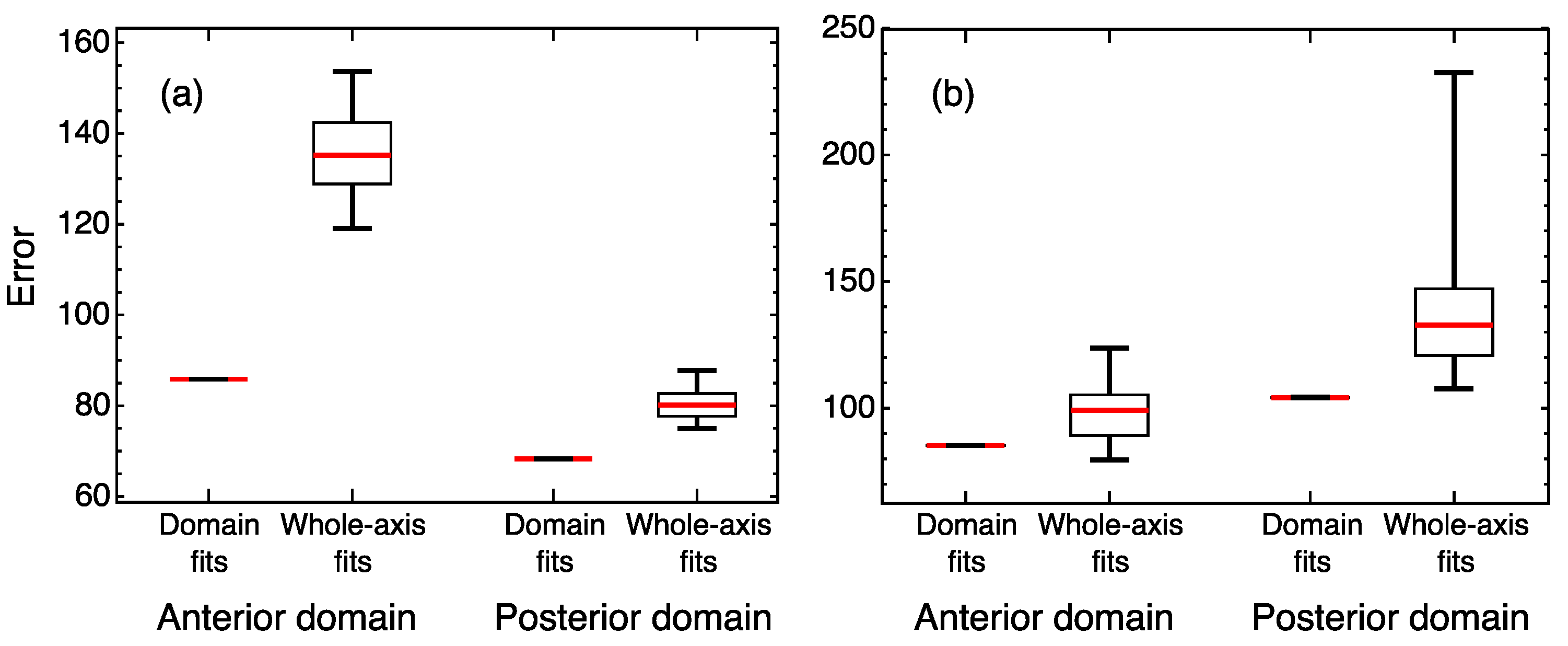
3.1. Position-Specific Models for Hunchback Expression in Wild Type Embryos
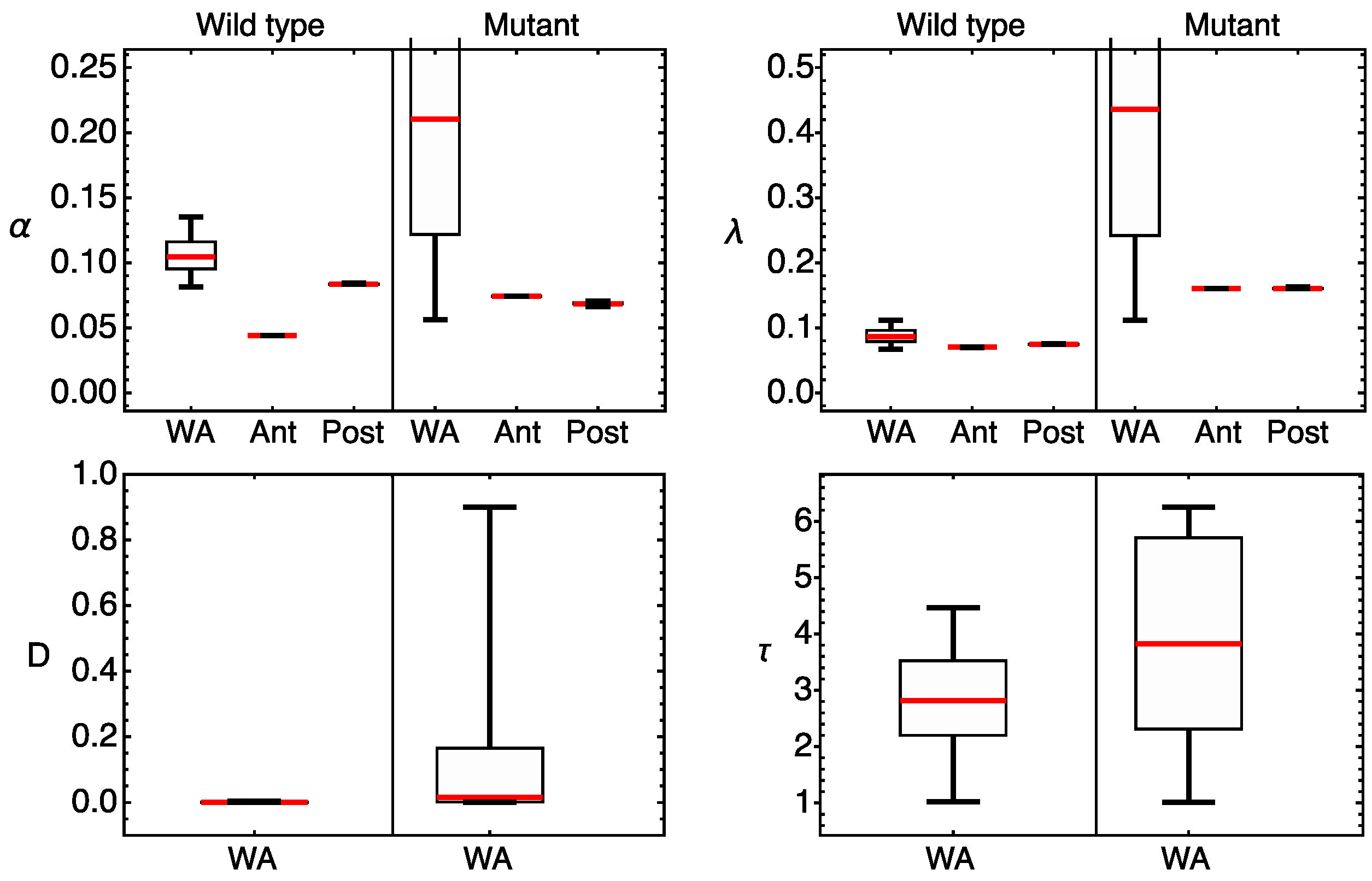
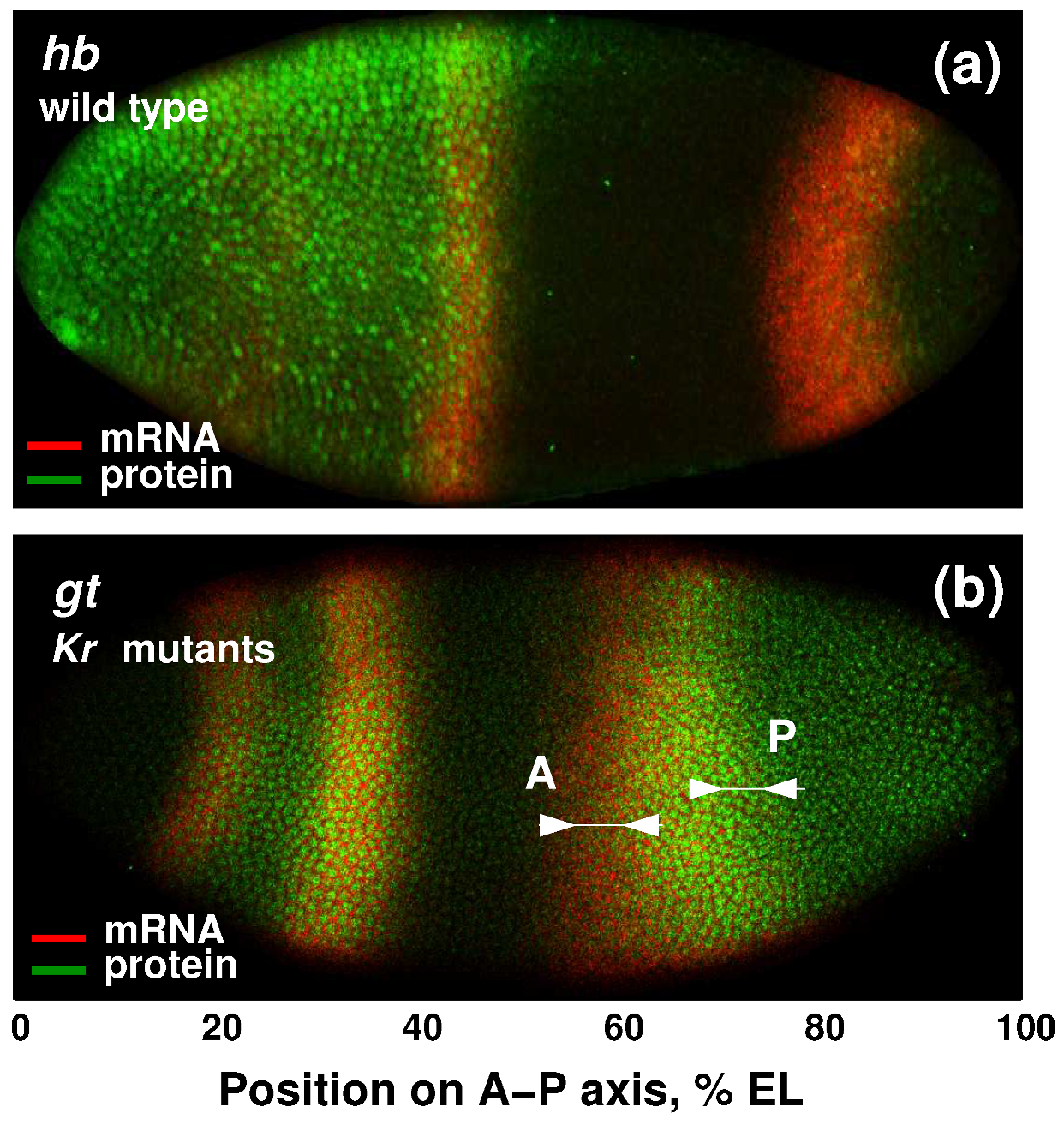
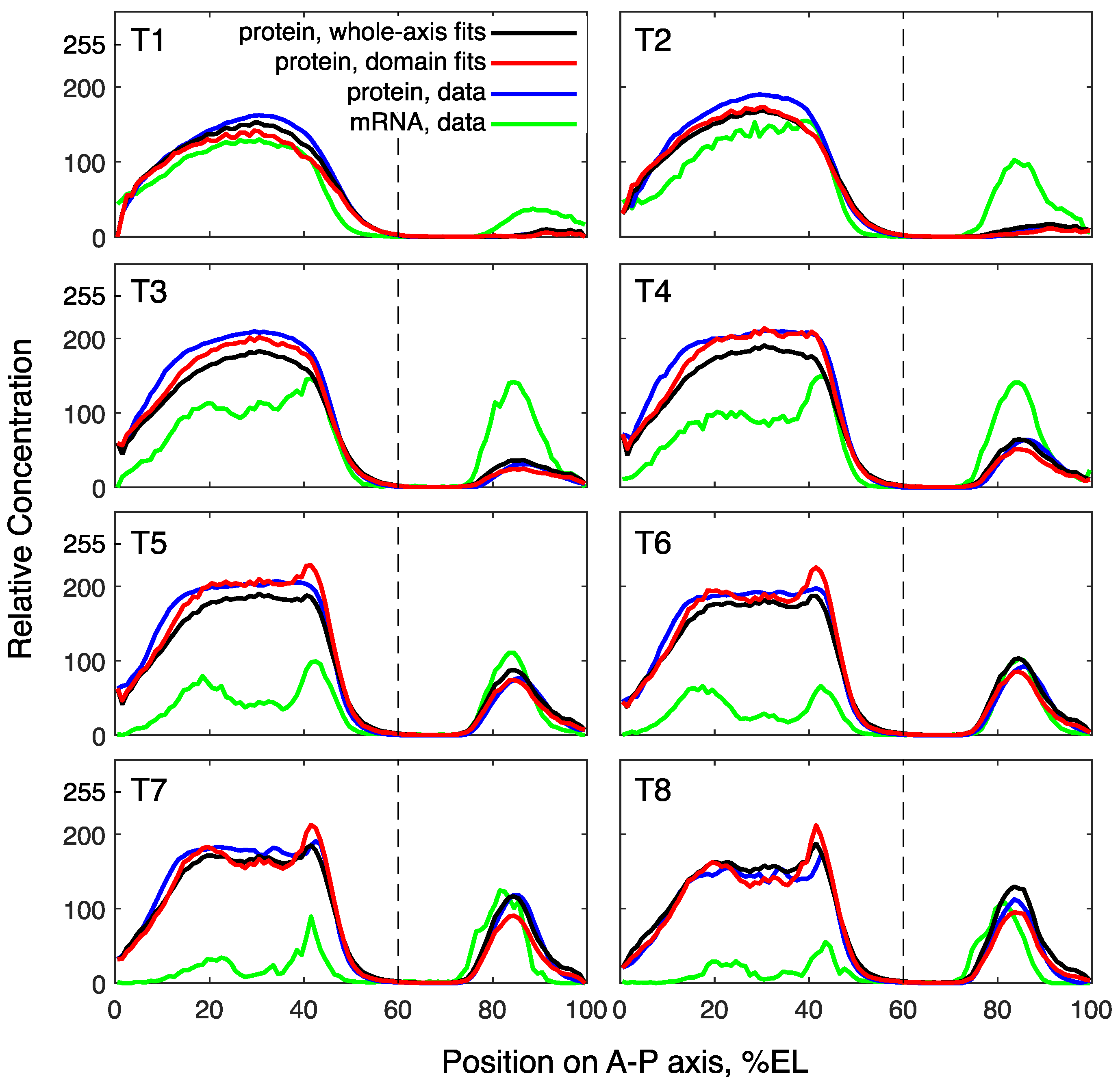
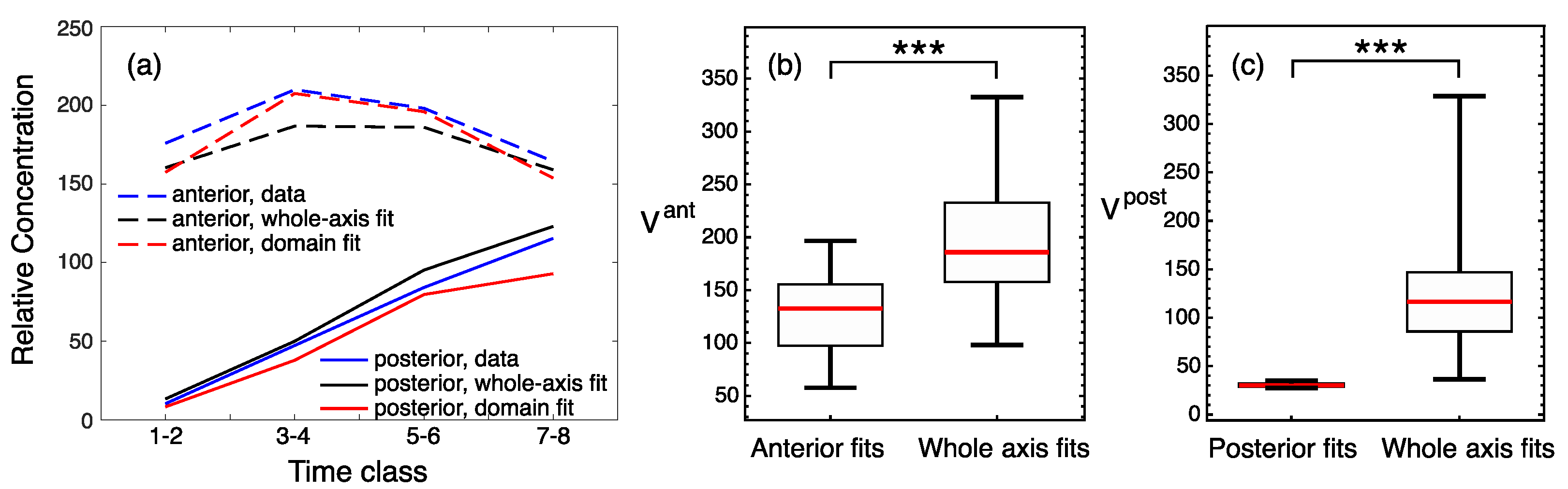
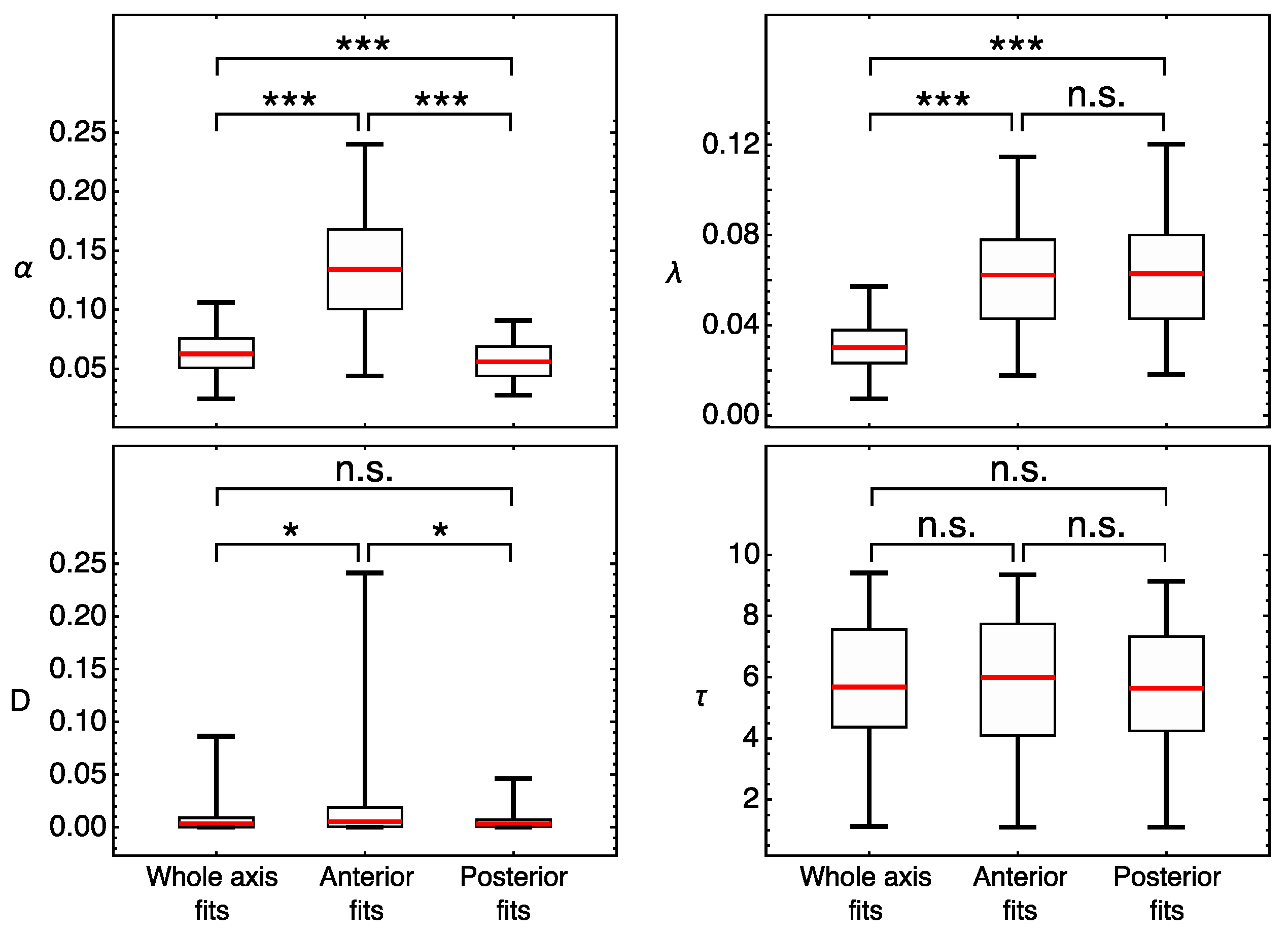
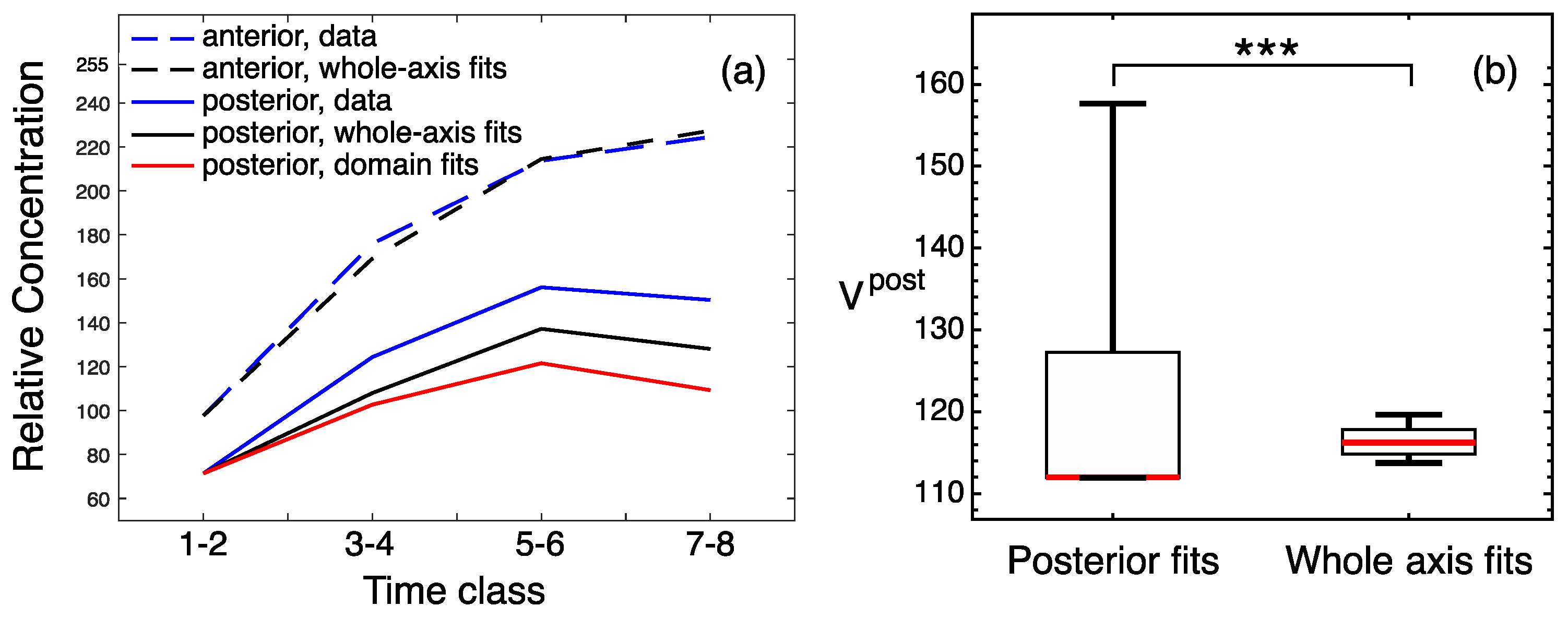
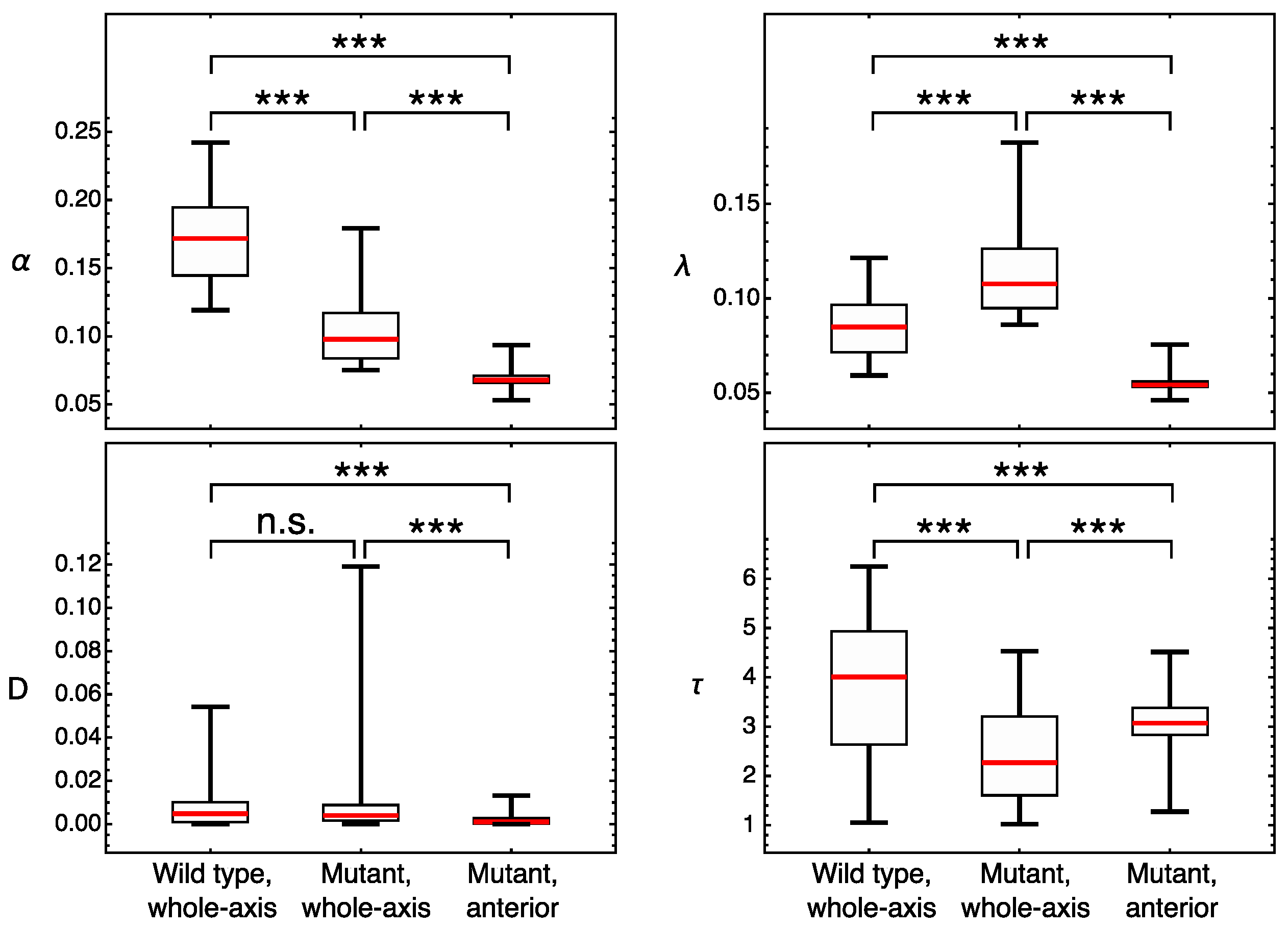
3.2. Modeling Gt Protein Dynamics in Wild-Type Embryos and Kruppel Mutants
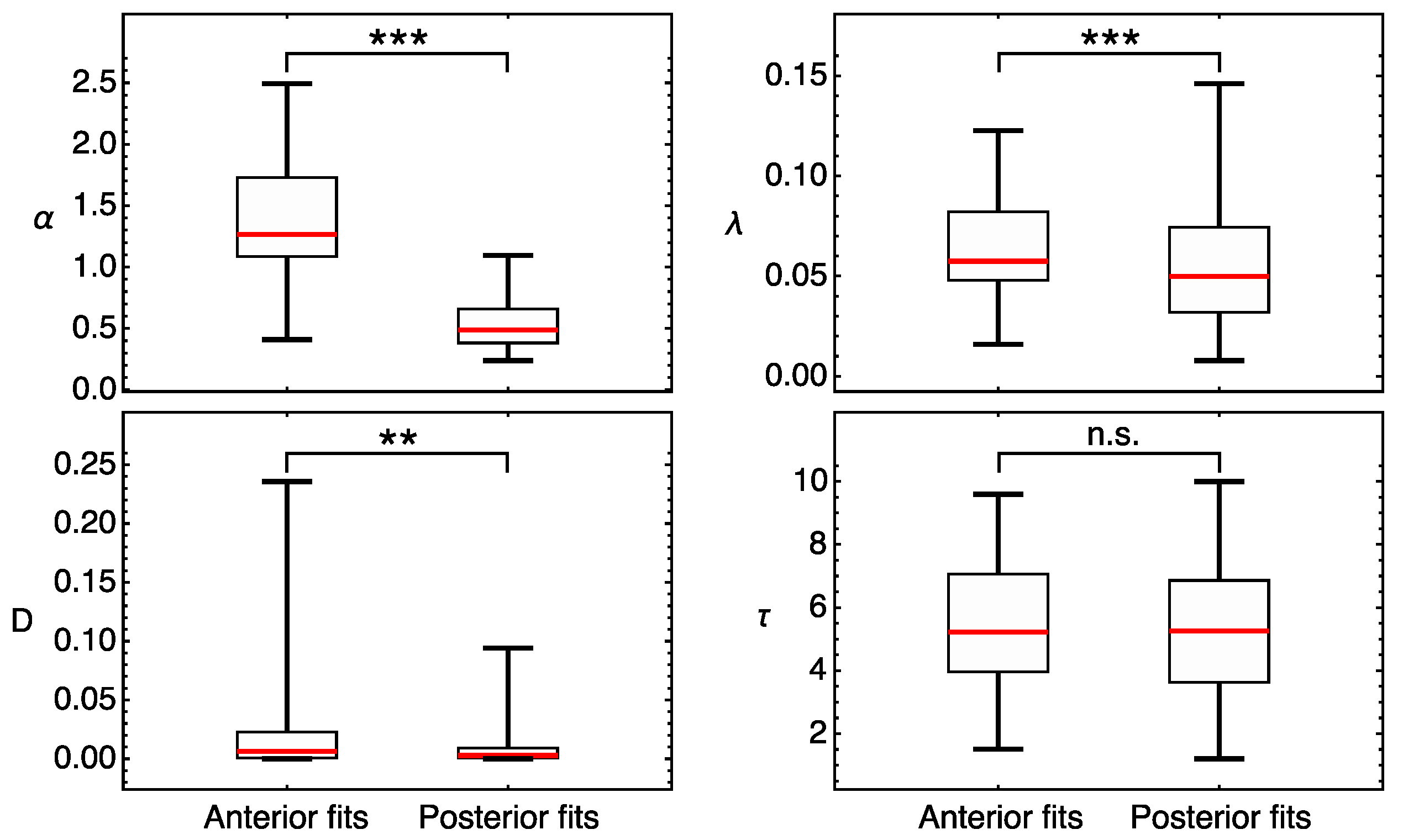
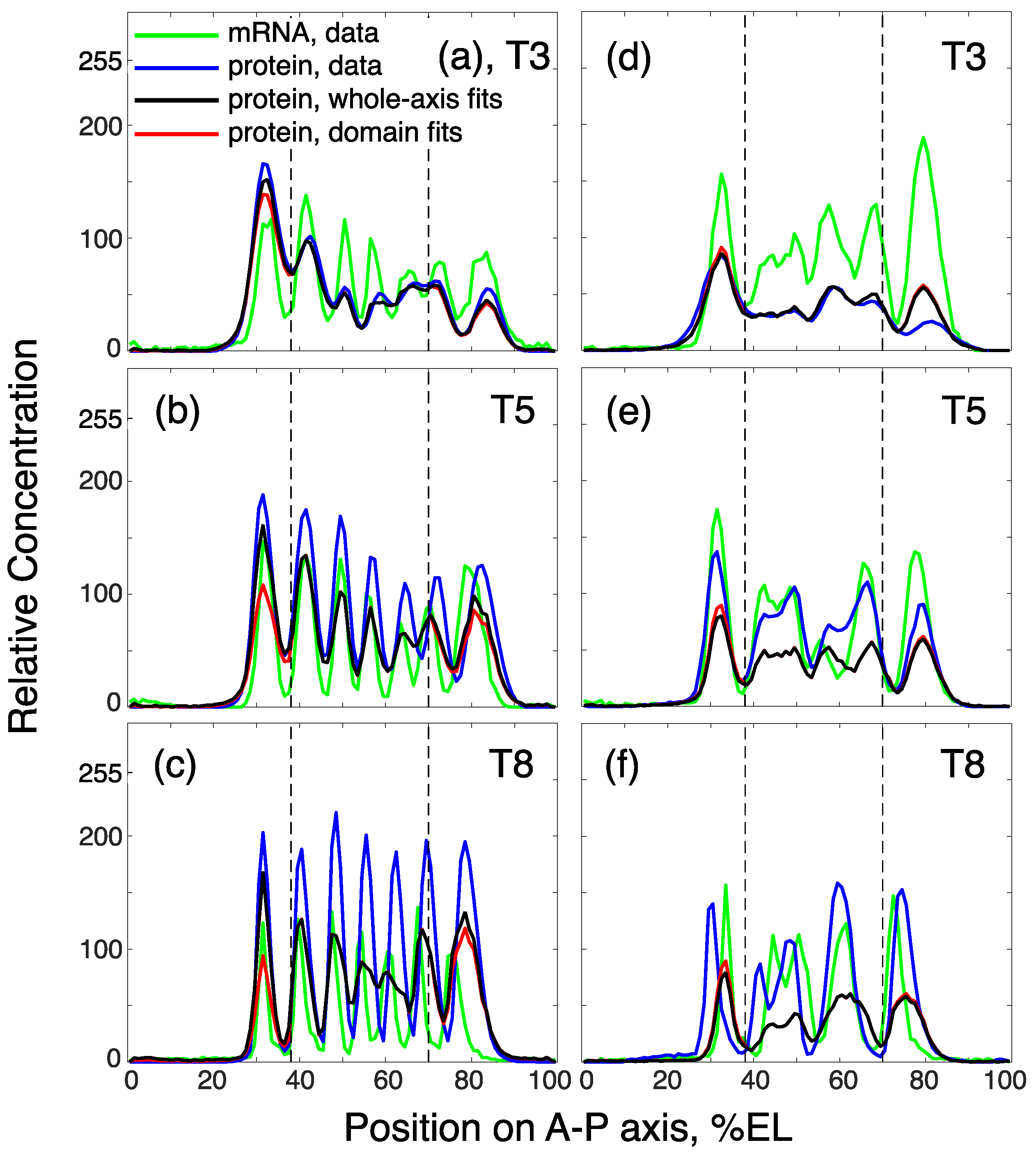
3.3. Model Fails to Reproduce Full Dynamics of Even-Skipped Expression
4. Discussion
4.1. Model Predicts Position-Specific Regulation of Hb Protein Production
4.2. Genotype-Specific Modeling of Gt Protein Dynamics
4.3. Dynamic Sculpting of Eve Pattern: Modeling Failures and Perspectives
4.4. Limitations of the Modeling Approach
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
References
- Schwanhäusser, B.; Busse, D.; Li, N.; Dittmar, G.; Schuchhardt, J.; Wolf, J.; Chen, W.; Selbach, M. Global quantification of mammalian gene expression control. Nature 2011, 473, 337–342. [Google Scholar] [CrossRef] [Green Version]
- Li, J.J.; Bickel, P.J.; Biggin, M.D. System wide analyses have underestimated protein abundances and the importance of transcription in mammals. PeerJ 2014, 2, e270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koussounadis, A.; Langdon, S.P.; Um, I.H.; Harrison, D.J.; Smith, V.A. Relationship between differentially expressed mRNA and mRNA-protein correlations in a xenograft model system. Sci. Rep. 2015, 5, 10775. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, Z.; Teo, G.; Krueger, S.; Rock, T.M.; Koh, H.W.; Choi, H.; Vogel, C. Differential dynamics of the mammalian mRNA and protein expression response to misfolding stress. Mol. Syst. Biol. 2016, 12, 855. [Google Scholar] [CrossRef]
- Liu, Y.; Beyer, A.; Aebersold, R. On the Dependency of Cellular Protein Levels on mRNA Abundance. Cell 2016, 165, 535–550. [Google Scholar] [CrossRef] [Green Version]
- Alli Shaik, A.; Wee, S.; Li, R.H.; Li, Z.; Carney, T.J.; Mathavan, S.; Gunaratne, J. Functional mapping of the zebrafish early embryo proteome and transcriptome. J. Proteome Res. 2014, 13, 5536–5550. [Google Scholar] [CrossRef] [PubMed]
- Grün, D.; Kirchner, M.; Thierfelder, N.; Stoeckius, M.; Selbach, M.; Rajewsky, N. Conservation of mRNA and protein expression during development of C. Elegans. Cell Rep. 2014, 6, 565–577. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Casas-Vila, N.; Bluhm, A.; Sayols, S.; Dinges, N.; Dejung, M.; Altenhein, T.; Kappei, D.; Altenhein, B.; Roignant, J.Y.; Butter, F. The developmental proteome of Drosophila Melanogaster. Genome Res. 2017, 27, 1273–1285. [Google Scholar] [CrossRef] [Green Version]
- Gao, Y.; Liu, X.; Tang, B.; Li, C.; Kou, Z.; Li, L.; Liu, W.; Wu, Y.; Kou, X.; Li, J.; et al. Protein Expression Landscape of Mouse Embryos during Pre-implantation Development. Cell Rep. 2017, 21, 3957–3969. [Google Scholar] [CrossRef] [Green Version]
- Becker, K.; Bluhm, A.; Casas-Vila, N.; Dinges, N.; Dejung, M.; Sayols, S.; Kreutz, C.; Roignant, J.Y.; Butter, F.; Legewie, S. Quantifying post-transcriptional regulation in the development of Drosophila Melanogaster. Nat. Commun. 2018, 9, 4970. [Google Scholar] [CrossRef]
- Smits, A.H.; Lindeboom, R.G.; Perino, M.; van Heeringen, S.J.; Veenstra, G.J.; Vermeulen, M. Global absolute quantification reveals tight regulation of protein expression in single Xenopus Eggs. Nucleic Acids Res. 2014, 42, 9880–9891. [Google Scholar] [CrossRef] [Green Version]
- Vogel, C.; Marcotte, E.M. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat. Rev. Genet. 2012, 13, 227–232. [Google Scholar] [CrossRef]
- Mair, F.; Erickson, J.R.; Voillet, V.; Simoni, Y.; Bi, T.; Tyznik, A.J.; Martin, J.; Gottardo, R.; Newell, E.W.; Prlic, M. A Targeted Multi-omic Analysis Approach Measures Protein Expression and Low-Abundance Transcripts on the Single-Cell Level. Cell Rep. 2020, 31, 107499. [Google Scholar] [CrossRef]
- Akam, M. The molecular basis for metameric pattern in the Drosophila Embryo. Development 1987, 101, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Jaeger, J. The gap gene network. Cell. Mol. Life Sci. CMLS 2011, 68, 243–274. [Google Scholar] [CrossRef] [Green Version]
- Jaeger, J.; Reinitz, J. Drosophila Blastoderm Patterning. Curr. Opin. Genet. Dev. 2012, 22, 533–541. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frasch, M.; Hoey, T.; Rushlow, C.; Doyle, H.; Levine, M. Characterization and localization of the Even-Skipped Protein Drosophila. EMBO J. 1987, 6, 749–759. [Google Scholar] [CrossRef] [PubMed]
- Surkova, S.; Kosman, D.; Kozlov, K.N.; Myasnikova, E.; Samsonova, A.A.; Spirov, A.; Vanario-Alonso, C.E.; Samsonova, M.G.; Reinitz, J. Characterization of the Drosophila Segm. Determ. Morphome. Dev. Biol. 2008, 313, 844–862. [Google Scholar] [CrossRef] [Green Version]
- Loncar, D.; Singer, S.J. Cell membrane formation during the cellularization of the syncytial blastoderm of Drosophila. Proc. Natl. Acad. Sci. USA 1995, 92, 2199–2203. [Google Scholar] [CrossRef] [Green Version]
- Jaeger, J.; Surkova, S.; Blagov, M.; Janssens, H.; Kosman, D.; Kozlov, K.N.; Myasnikova, E.; Vanario-Alonso, C.E.; Samsonova, M.G.; Sharp, D.H.; et al. Dynamic control of positional information in the early Drosophila Embryo. Nature 2004, 430, 368–371. [Google Scholar] [CrossRef]
- Jaeger, J.; Blagov, M.; Kosman, D.; Kozlov, K.N.; Myasnikova, E.; Surkova, S.; Vanario-Alonso, C.E.; Samsonova, M.G.; Sharp, D.H.; Reinitz, J. Dynamical analysis of regulatory interactions in the gap gene system of Drosophila Melanogaster. Genetics 2004, 167, 1721–1737. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perkins, T.J.; Jaeger, J.; Reinitz, J.; Glass, L. Reverse engineering the gap gene network of Drosophila Melanogaster. PLoS Comput. Biol. 2006, 2, e51. [Google Scholar] [CrossRef]
- Surkova, S.; Spirov, A.V.; Gursky, V.V.; Janssens, H.; Kim, A.R.; Radulescu, O.; Vanario-Alonso, C.E.; Sharp, D.H.; Samsonova, M.G.; Reinitz, J. Canalization of gene expression in the Drosophila Blastoderm Gap Gene Cross Regul. PLoS Biol. 2009, 7, e1000049. [Google Scholar] [CrossRef] [Green Version]
- Surkova, S.; Spirov, A.V.; Gursky, V.V.; Janssens, H.; Kim, A.R.; Radulescu, O.; Vanario-Alonso, C.E.; Sharp, D.H.; Samsonova, M.G.; Reinitz, J. Canalization of gene expression and domain shifts in the Drosophila Blastoderm by Dynamical Attractors. PLoS Comput. Biol. 2009, 5, e1000303. [Google Scholar] [CrossRef] [Green Version]
- Crombach, A.; Wotton, K.R.; Cicin-Sain, D.; Ashyraliyev, M.; Jaeger, J. Efficient reverse-engineering of a developmental gene regulatory network. PLoS Comput. Biol. 2012, 8, e1002589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kozlov, K.N.; Surkova, S.; Myasnikova, E.; Reinitz, J.; Samsonova, M.G. Modeling of gap gene expression in Drosophila Kruppel Mutants. PLoS Comput. Biol. 2012, 8, e1002635. [Google Scholar] [CrossRef]
- Clark, E. Dynamic patterning by the Drosophila Pair-Rule Netw. Reconciles Long-Germ Short-Germ Segmentation. PLoS Biol. 2017, 15, e2002439. [Google Scholar] [CrossRef] [Green Version]
- Becker, K.; Balsa-Canto, E.; Cicin-Sain, D.; Hoermann, A.; Janssens, H.; Banga, J.R.; Jaeger, J. Reverse-Engineering Post-Transcriptional Regulation of Gap Genes in Drosophila Melanogaster. PLoS Comput. Biol. 2013, 9, e1003281. [Google Scholar] [CrossRef] [Green Version]
- Wharton, R.P.; Struhl, G. RNA regulatory elements mediate control of Drosophila Body Pattern Posterior Morphogen Nanos. Cell 1991, 67, 955–967. [Google Scholar] [CrossRef]
- Rivera-Pomar, R.; Niessing, D.; Schmidt-Ott, U.; Gehring, W.; Jäckle, H. RNA binding and translational suppression by Bicoid. Nature 1996, 379, 746–749. [Google Scholar] [CrossRef]
- Cho, P.F.; Gamberi, C.; Cho-Park, Y.A.; Cho-Park, I.B.; Lasko, P.; Sonenberg, N. Cap-dependent translational inhibition establishes two opposing morphogen gradients in Drosophila Embryos. Curr. Biol. 2006, 16, 2035–2041. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lasko, P. Posttranscriptional regulation in Drosophila Oocytes Early Embryos. Wiley Interdiscip. Rev. RNA 2011, 2, 408–416. [Google Scholar] [CrossRef]
- Surkova, S.; Sokolkova, A.; Kozlov, K.; Nuzhdin, S.V.; Samsonova, M. Quantitative analysis reveals genotype- and domain- specific differences between mRNA and protein expression of segmentation genes in Drosophila. Dev. Biol. 2019, 448, 48–58. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, T.A.; Koceja, C.; Eisa-Beygi, S.; Kleinstiver, B.P.; Kumar, S.N.; Lin, C.W.; Li, K.; Prabhudesai, S.; Joung, J.K.; Ramchandran, R. Temporal and Spatial Post-Transcriptional Regulation of Zebrafish Tie1 MRNA Long Noncoding RNA During Brain Vascular Assembly. Arter. Thromb Vasc Biol. 2018, 38, 1562–1575. [Google Scholar] [CrossRef] [Green Version]
- Maier, T.; Schmidt, A.; Güell, M.; Kühner, S.; Gavin, A.C.; Aebersold, R.; Serrano, L. Quantification of mRNA and protein and integration with protein turnover in a bacterium. Mol. Syst. Biol. 2011, 7, 511. [Google Scholar] [CrossRef]
- Jovanovic, M.; Rooney, M.; Mertins, P.; Przybylski, D.; Chevrier, N.; Satija, R.; Rodriguez, E.H.; Fields, A.P.; Schwartz, S.; Raychowdhury, R.; et al. Dynamic profiling of the protein life cycle in response to pathogens. Science 2015, 347, 1259038. [Google Scholar] [CrossRef] [Green Version]
- Eldon, E.D.; Pirrotta, V. Interactions of the Drosophila gap gene giant with maternal and zygotic pattern-forming genes. Development 1991, 111, 367–378. [Google Scholar] [CrossRef] [PubMed]
- Kraut, R.; Levine, M. Spatial regulation of the gap gene Giant Drosophila development. Development 1991, 111, 601–609. [Google Scholar] [CrossRef] [PubMed]
- Surkova, S.; Golubkova, E.; Panok, L.; Mamon, L.; Reinitz, J.; Samsonova, M. Quantitative dynamics and increased variability of segmentation gene expression in the Drosophila Krüppel Knirps Mutants. Dev. Biol. 2013, 376, 99–112. [Google Scholar] [CrossRef] [Green Version]
- Sonoda, J.; Wharton, R. Recruitment of Nanos to Hunchback MRNA Pumilio. Genes Dev. 1999, 13, 2704–2712. [Google Scholar] [CrossRef] [Green Version]
- Chagnovich, D.; Lehmann, R. Poly(A)-independent regulation of maternal hunchback translation in the Drosophila Embryo. Proc. Natl. Acad. Sci. USA 2001, 98, 11359–11364. [Google Scholar] [CrossRef] [Green Version]
- Vinter, D.J.; Hoppe, C.; Minchington, T.G.; Sutcliffe, C.; Ashe, H.L. Dynamics of hunchback translation in real-time and at single-mRNA resolution in the Drosophila Embryo. Development 2021, 148, dev196121. [Google Scholar] [CrossRef] [PubMed]
- Wieschaus, E.; Nüsslein-Volhard, C.; Kluding, H. Krüppel, a gene whose activity is required early in the zygotic genome for normal embryonic segmentation. Dev. Biol. 1984, 104, 172–186. [Google Scholar] [CrossRef]
- Choi, H.M.; Beck, V.A.; Pierce, N.A. Next-generation in situ hybridization chain reaction: Higher gain, lower cost, greater durability. ACS Nano 2014, 8, 4284–4294. [Google Scholar] [CrossRef]
- Nagaso, H.; Murata, T.; Day, N.; Yokoyama, K.K. Simultaneous detection of RNA and protein by in situ hybridization and immunological staining. J. Histochem. Cytochem. 2001, 49, 1177–1182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Surkova, S.; Myasnikova, E.; Janssens, H.; Kozlov, K.N.; Samsonova, A.A.; Reinitz, J.; Samsonova, M. Pipeline for acquisition of quantitative data on segmentation gene expression from confocal images. Fly 2008, 2, 58–66. [Google Scholar] [CrossRef] [Green Version]
- Wu, X.; Vasisht, V.; Kosman, D.; Reinitz, J.; Small, S. Thoracic patterning by the Drosophila Gap Gene Hunchback. Dev. Biol. 2001, 237, 79–92. [Google Scholar] [CrossRef] [Green Version]
- Perry, M.W.; Bothma, J.P.; Luu, R.D.; Levine, M. Precision of Hunchback Expression in the Drosophila Embryo. Curr. Biol. 2012, 22, 2247–2252. [Google Scholar] [CrossRef] [Green Version]
- Mohler, J.; Eldon, E.D.; Pirrotta, V. A novel spatial transcription pattern associated with the segmentation gene, giant, of Drosophila. EMBO J. 1989, 8, 1539–1548. [Google Scholar] [CrossRef]
- Janssens, H.; Hou, S.; Jaeger, J.; Kim, A.R.; Myasnikova, E.; Sharp, D.; Reinitz, J. Quantitative and predictive model of transcriptional control of the Drosophila melanogaster even skipped gene. Nat. Genet. 2006, 38, 1159–1165. [Google Scholar] [CrossRef]
- Liu, Y.; Barr, K.; Reinitz, J. Fully interpretable deep learning model of transcriptional control. Bioinformatics 2020, 36 (Suppl. 1), i499–i507. [Google Scholar] [CrossRef] [PubMed]
- Lasko, P. mRNA localization and translational control in Drosophila Oogenesis. Cold Spring Harb. Perspect. Biol. 2012, 4, a012294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McKee, A.E.; Minet, E.; Stern, C.; Riahi, S.; Stiles, C.D.; Silver, P.A. A genome-wide in situ hybridization map of RNA-binding proteins reveals anatomically restricted expression in the developing mouse brain. BMC Dev. Biol. 2005, 5, 14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Forinash, K.D.; McGivern, J.; Fritz, B.; Dorey, K.; Sheets, M.D. Spatially restricted translation of the xCR1 mRNA in Xenopus Embryos. Mol. Cell. Biol. 2009, 29, 3791–3802. [Google Scholar] [CrossRef] [Green Version]
- Kaschula, R.; Pinho, S.; Alonso, C.R. MicroRNA-dependent regulation of Hox gene expression sculpts fine-grain morphological patterns in a Drosophila Appendage. Development 2018, 145, dev161133. [Google Scholar] [CrossRef] [Green Version]
- Chua, B.A.; Van Der Werf, I.; Jamieson, C.; Signer, R. Post-Transcriptional Regulation of Homeostatic, Stressed, and Malignant Stem Cells. Cell Stem Cell 2020, 26, 138–159. [Google Scholar] [CrossRef] [PubMed]
- Pultz, M.A.; Westendorf, L.; Gale, S.D.; Hawkins, K.; Lynch, J.; Pitt, J.N.; Reeves, N.L.; Yao, J.C.; Small, S.; Desplan, C.; et al. A major role for zygotic Hunchback Patterning Nasonia Embryo. Development 2005, 132, 3705–3715. [Google Scholar] [CrossRef] [Green Version]
- Perry, M.W.; Boettiger, A.N.; Levine, M. Multiple enhancers ensure precision of gap gene-expression patterns in the Drosophila Embryo. Proc. Natl. Acad. Sci. USA 2011, 108, 13570–13575. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Menon, K.P.; Andrews, S.; Murthy, M.; Gavis, E.R.; Zinn, K. The translational repressors Nanos and Pumilio have divergent effects on presynaptic terminal growth and postsynaptic glutamate receptor subunit composition. J. Neurosci. 2009, 29, 5558–5572. [Google Scholar] [CrossRef]
- Miles, W.O.; Korenjak, M.; Griffiths, L.M.; Dyer, M.A.; Provero, P.; Dyson, N.J. Post-transcriptional gene expression control by NANOS is up-regulated and functionally important in pRb-deficient cells. EMBO J. 2014, 33, 2201–2215. [Google Scholar] [CrossRef] [Green Version]
- Olesnicky, E.C.; Bhogal, B.; Gavis, E.R. Combinatorial use of translational co-factors for cell type-specific regulation during neuronal morphogenesis in Drosophila. Dev. Biol. 2012, 365, 208–218. [Google Scholar] [CrossRef] [Green Version]
- Tanenbaum, M.E.; Gilbert, L.A.; Qi, L.S.; Weissman, J.S.; Vale, R.D. A protein-tagging system for signal amplification in gene expression and fluorescence imaging. Cell 2014, 159, 635–646. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pichon, X.; Bastide, A.; Safieddine, A.; Chouaib, R.; Samacoits, A.; Basyuk, E.; Peter, M.; Mueller, F.; Bertrand, E. Visualization of single endogenous polysomes reveals the dynamics of translation in live human cells. J. Cell Biol. 2016, 214, 769–781. [Google Scholar] [CrossRef] [Green Version]
- Dufourt, J.; Bellec, M.; Trullo, A.; Dejean, M.; De Rossi, S.; Favard, C.; Lagha, M. Imaging translation dynamics in live embryos reveals spatial heterogeneities. Science 2021, 372, 840–844. [Google Scholar] [CrossRef]
- Berrocal, A.; Lammers, N.C.; Garcia, H.G.; Eisen, M.B. Kinetic sculpting of the seven stripes of the Drosophila even-skipped gene. eLife 2020, 9, e61635. [Google Scholar] [CrossRef] [PubMed]
- Buchler, N.E.; Gerland, U.; Hwa, T. Nonlinear protein degradation and the function of genetic circuits. Proc. Natl. Acad. Sci. USA 2005, 102, 9559–9564. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.; Hansen, D.; Eck, E.; Kim, Y.J.; Turner, M.; Alamos, S.; Garcia, H. Real-time single-cell characterization of the eukaryotic transcription cycle reveals correlations between RNA initiation, elongation, and cleavage. PLoS Comput. Biol. 2021, 17, e1008999. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Duk, M.A.; Gursky, V.V.; Samsonova, M.G.; Surkova, S.Y. Application of Domain- and Genotype-Specific Models to Infer Post-Transcriptional Regulation of Segmentation Gene Expression in Drosophila. Life 2021, 11, 1232. https://doi.org/10.3390/life11111232
Duk MA, Gursky VV, Samsonova MG, Surkova SY. Application of Domain- and Genotype-Specific Models to Infer Post-Transcriptional Regulation of Segmentation Gene Expression in Drosophila. Life. 2021; 11(11):1232. https://doi.org/10.3390/life11111232
Chicago/Turabian StyleDuk, Maria A., Vitaly V. Gursky, Maria G. Samsonova, and Svetlana Yu. Surkova. 2021. "Application of Domain- and Genotype-Specific Models to Infer Post-Transcriptional Regulation of Segmentation Gene Expression in Drosophila" Life 11, no. 11: 1232. https://doi.org/10.3390/life11111232
APA StyleDuk, M. A., Gursky, V. V., Samsonova, M. G., & Surkova, S. Y. (2021). Application of Domain- and Genotype-Specific Models to Infer Post-Transcriptional Regulation of Segmentation Gene Expression in Drosophila. Life, 11(11), 1232. https://doi.org/10.3390/life11111232






