The Urine Biomarker PUR-4 Is Positively Associated with the Amount of Gleason 4 in Human Prostate Cancers
Abstract
1. Introduction
2. Materials and Methods
2.1. Urine RNA Extraction and PUR Signatures
2.2. Biopsy Tissue Analyses
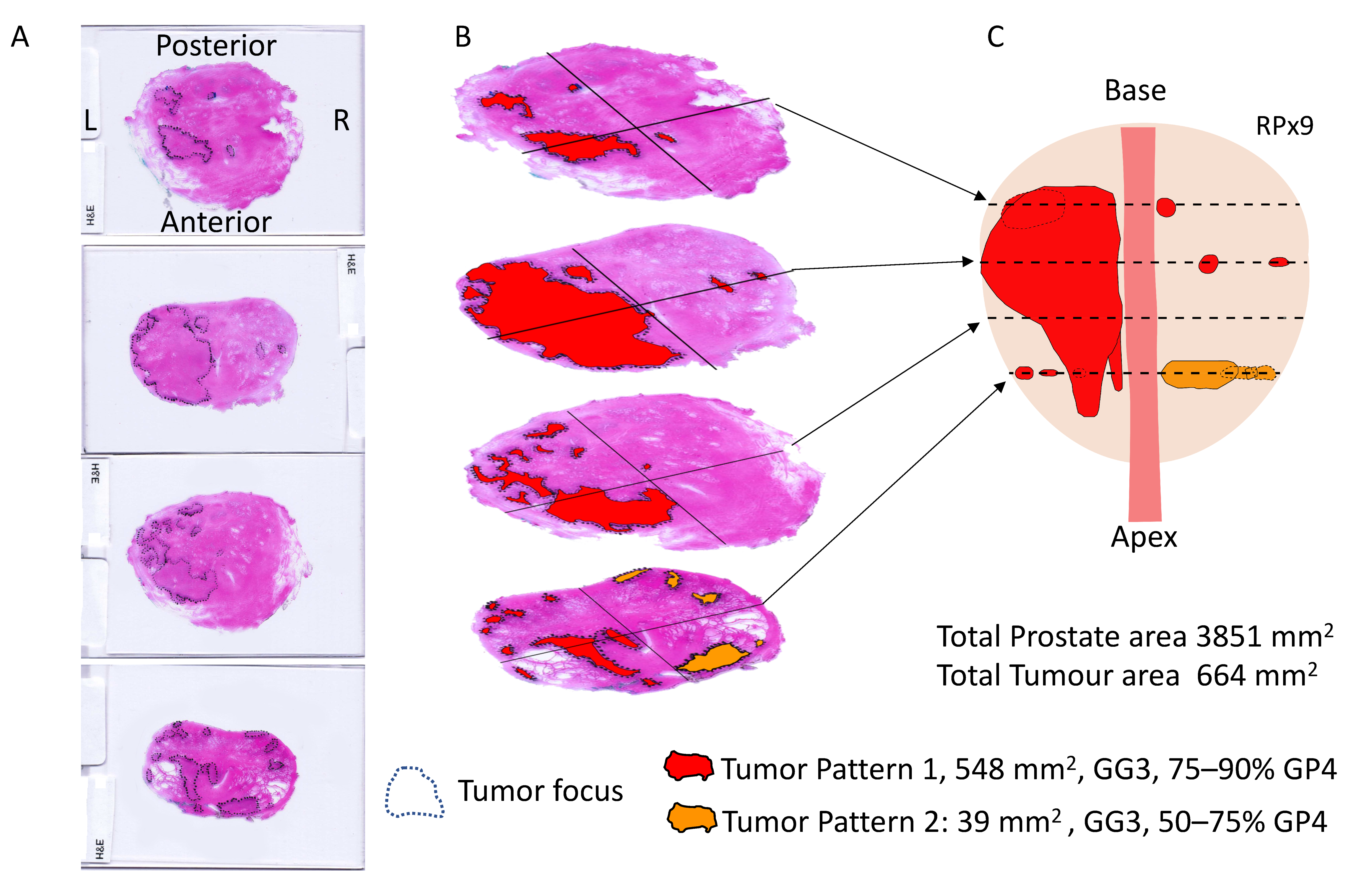
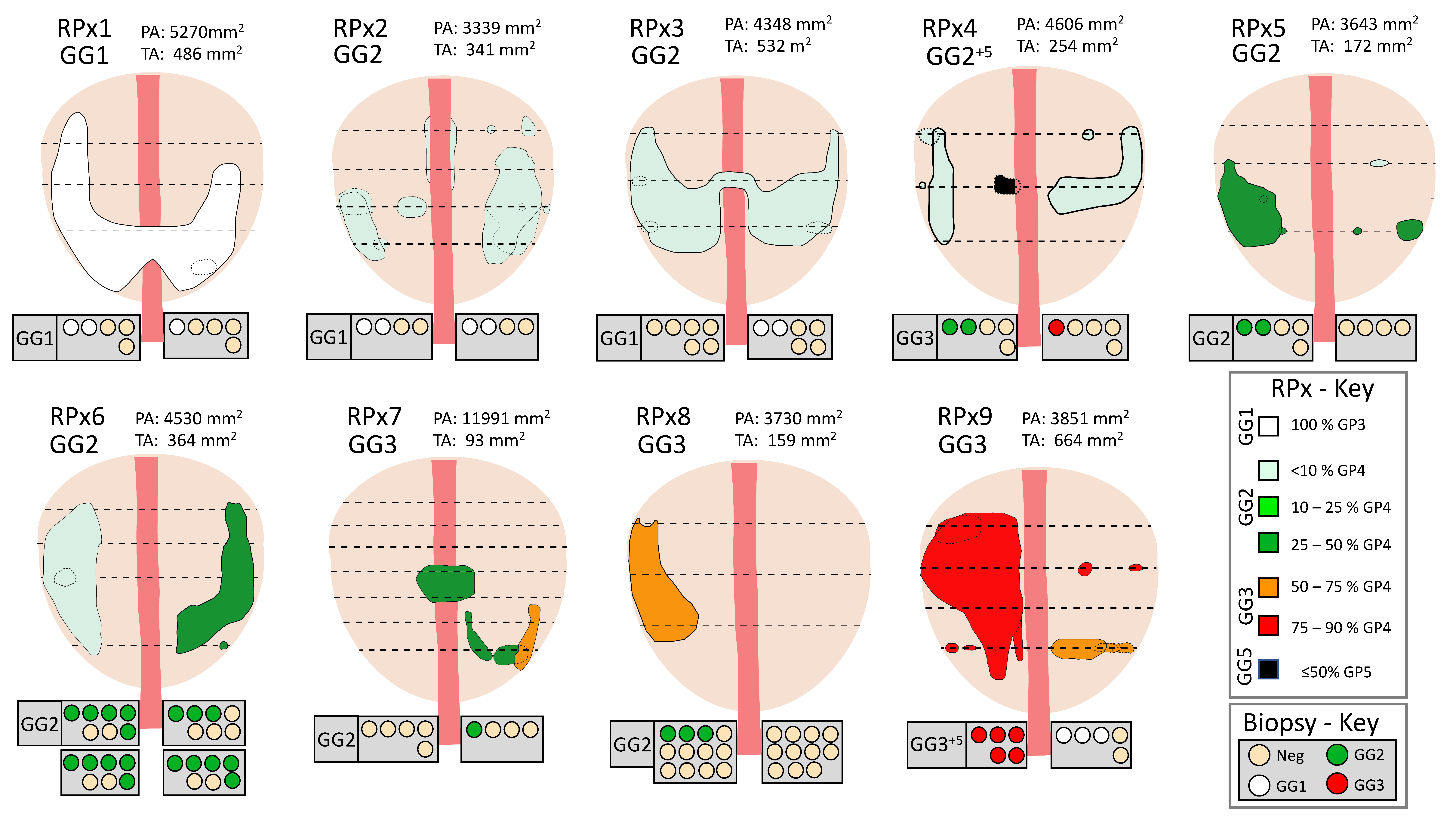
2.3. Radical Prostatectomy Analyses
2.4. Statistical Analyses
3. Results
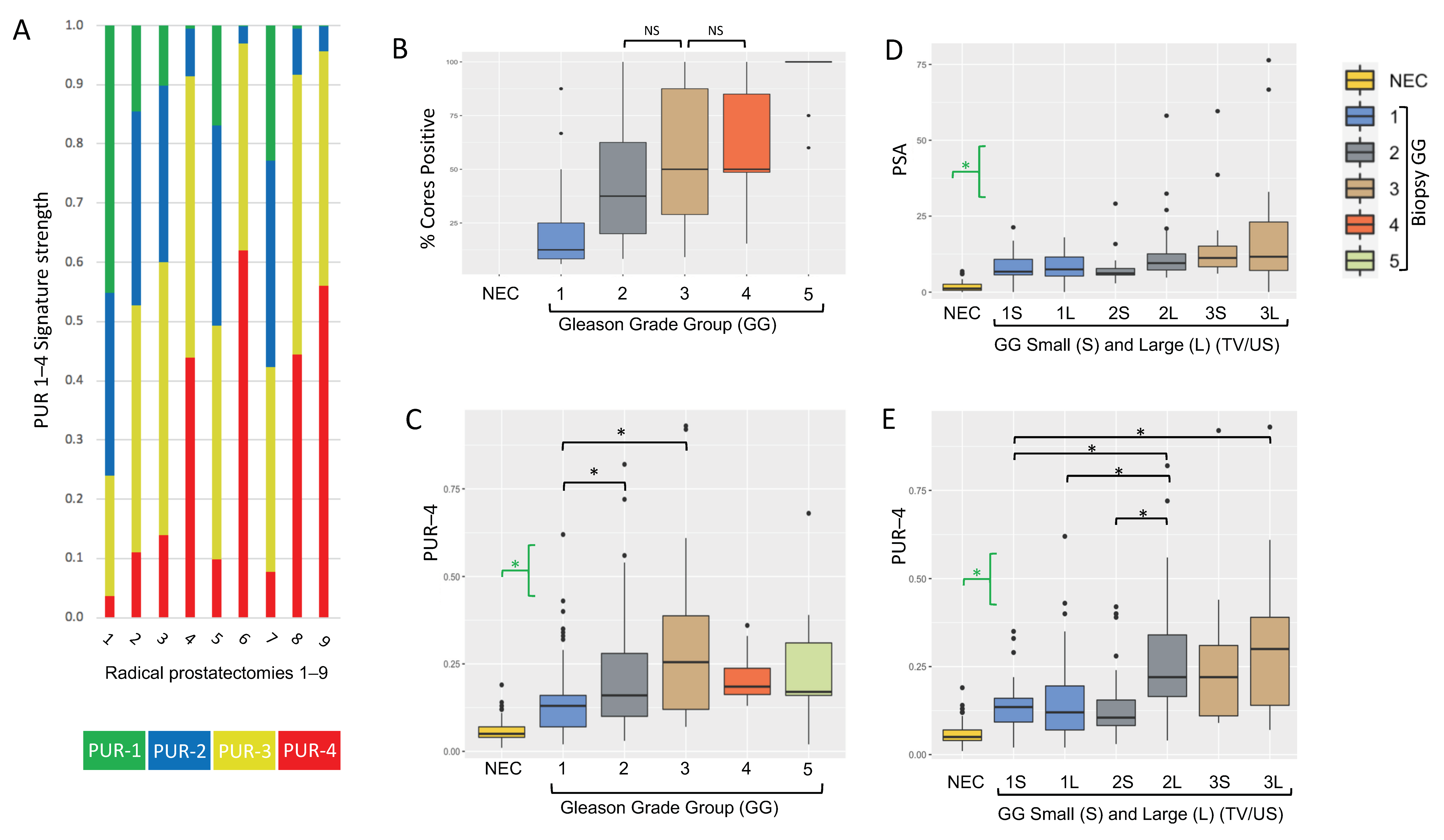
3.1. Analyses of Prostate Needle Biopsy Tissue
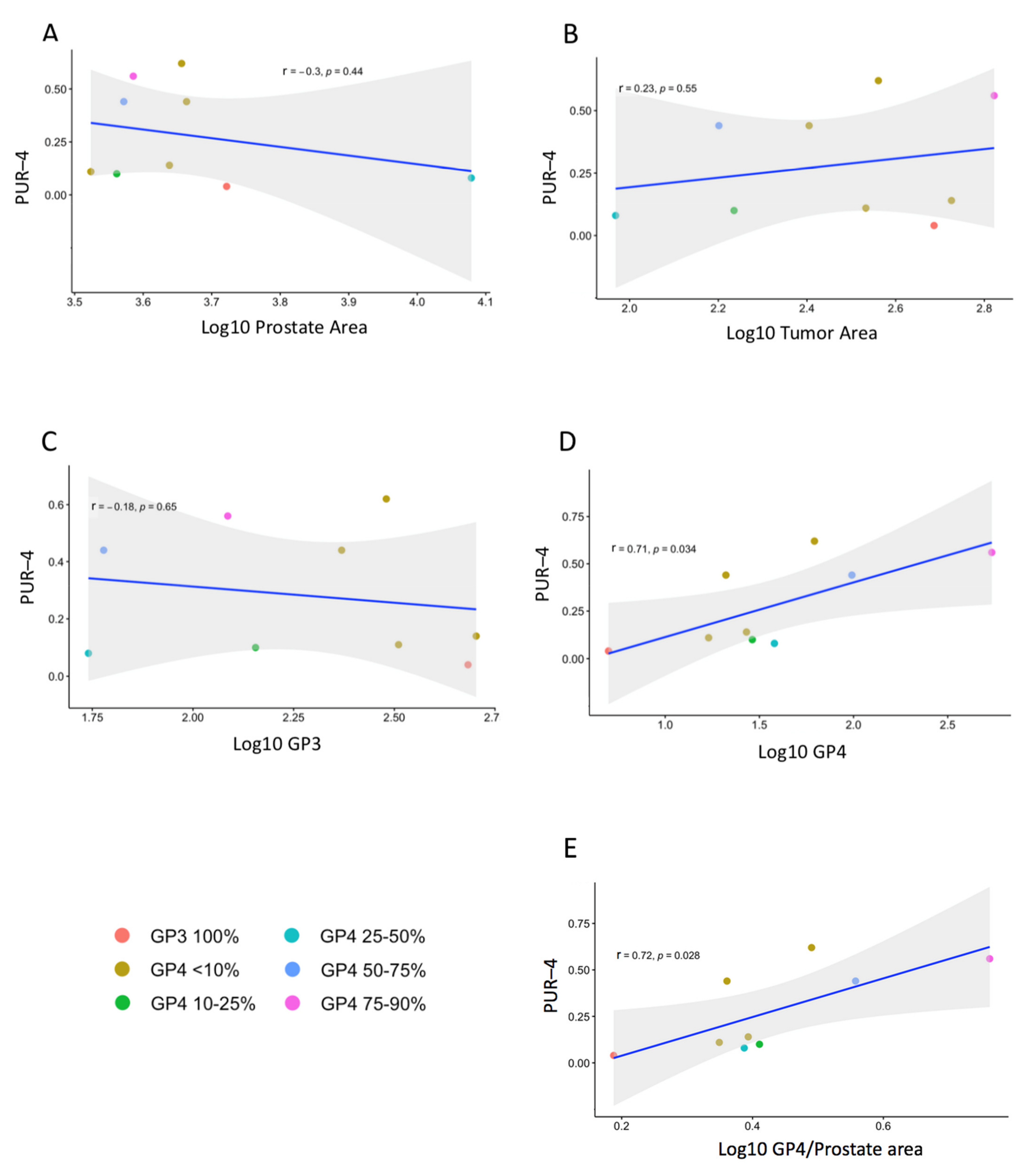
3.2. Prostatectomy Analyses
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Stark, J.R.; Perner, S.; Stampfer, M.J.; Sinnott, J.A.; Finn, S.; Eisenstein, A.S.; Ma, J.; Fiorentino, M.; Kurth, T.; Loda, M.; et al. Gleason score and lethal prostate cancer: Does 3 + 4 = 4 + 3? J. Clin. Oncol. 2009, 27, 3459–3464. [Google Scholar] [CrossRef] [PubMed]
- Choy, B.; Pearce, S.M.; Anderson, B.B.; Shalhav, A.L.; Zagaja, G.; Eggener, S.E.; Paner, G.P. Prognostic Significance of Percentage and Architectural Types of Contemporary Gleason Pattern 4 Prostate Cancer in Radical Prostatectomy. Am. J. Surg. Pathol. 2016, 40, 1400–1406. [Google Scholar] [CrossRef]
- Spalding, A.C.; Daignault, S.; Sandler, H.M.; Shah, R.B.; Pan, C.C.; Ray, M.E. Percent positive biopsy cores as a prognostic factor for prostate cancer treated with external beam radiation. Urology 2007, 69, 936–940. [Google Scholar] [CrossRef] [PubMed]
- Cooperberg, M.R.; Freedland, S.J.; Pasta, D.J.; Elkin, E.P.; Presti, J.C.; Amling, C.L.; Terris, M.K.; Aronson, W.J.; Kane, C.J.; Carroll, P.R. Multiinstitutional validation of the UCSF cancer of the prostate risk assessment for prediction of recurrence after radical prostatectomy. Cancer 2006, 107, 2384–2391. [Google Scholar] [CrossRef]
- Arora, R.; Koch, M.O.; Eble, J.N.; Ulbright, T.M.; Li, L.; Cheng, L. Heterogeneity of Gleason grade in multifocal adenocarcinoma of the prostate. Cancer 2004, 100, 2362–2366. [Google Scholar] [CrossRef] [PubMed]
- Le, J.D.; Tan, N.; Shkolyar, E.; Lu, D.Y.; Kwan, L.; Marks, L.S.; Huang, J.; Margolis, D.J.A.; Raman, S.S.; Reiter, R.E. Multifocality and prostate cancer detection by multiparametric magnetic resonance imaging: Correlation with whole-mount histopathology. Eur. Urol. 2015, 67, 569–576. [Google Scholar] [CrossRef] [PubMed]
- Cooper, C.S.; Eeles, R.; Wedge, D.C.; Van Loo, P.; Gundem, G.; Alexandrov, L.B.; Kremeyer, B.; Butler, A.; Lynch, A.G.; Camacho, N.; et al. Analysis of the genetic phylogeny of multifocal prostate cancer identifies multiple independent clonal expansions in neoplastic and morphologically normal prostate tissue. Nat. Genet. 2015, 47, 367–372. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, P.-N.; Violette, P.; Chan, S.; Tanguay, S.; Kassouf, W.; Aprikian, A.; Chen, J.Z. A Panel of TMPRSS2:ERG Fusion Transcript Markers for Urine-Based Prostate Cancer Detection with High Specificity and Sensitivity. Eur. Urol. 2010, 59, 407–414. [Google Scholar] [CrossRef] [PubMed]
- Groskopf, J.; Aubin, S.M.J.; Deras, I.L.; Blase, A.; Bodrug, S.; Clark, C.; Brentano, S.; Mathis, J.; Pham, J.; Meyer, T.; et al. APTIMA PCA3 molecular urine test: Development of a method to aid in the diagnosis of prostate cancer. Clin. Chem. 2006, 52, 1089–1095. [Google Scholar] [CrossRef] [PubMed]
- Tomlins, S.A.; Day, J.R.; Lonigro, R.J.; Hovelson, D.H.; Siddiqui, J.; Kunju, L.P.; Dunn, R.L.; Meyer, S.; Hodge, P.; Groskopf, J.; et al. Urine TMPRSS2:ERG Plus PCA3 for Individualized Prostate Cancer Risk Assessment. Eur. Urol. 2016, 70, 45–53. [Google Scholar] [CrossRef] [PubMed]
- McKiernan, J.; Donovan, M.J.; Margolis, E.; Partin, A.; Carter, B.; Brown, G.; Torkler, P.; Noerholm, M.; Skog, J.; Shore, N.; et al. A Prospective Adaptive Utility Trial to Validate Performance of a Novel Urine Exosome Gene Expression Assay to Predict High-grade Prostate Cancer in Patients with Prostate-specific Antigen 2–10 ng/mL at Initial Biopsy. Eur. Urol. 2018, 74, 731–738. [Google Scholar] [CrossRef] [PubMed]
- Connell, S.P.; Hanna, M.; McCarthy, F.; Hurst, R.; Webb, M.; Curley, H.; Walker, H.; Mills, R.; Ball, R.Y.; Sanda, M.G.; et al. A Four-Group Urine Risk Classifier for Predicting Outcome in Prostate Cancer Patients. BJU Int. 2019, 124, 609–620. [Google Scholar] [CrossRef] [PubMed]
- Cohen, J. Statistical Power Analysis for the Behavioral Sciences; Academic Press: Cambridge, MA, USA, 2013. [Google Scholar]
- Evans, S.M.; Bandarage, V.P.; Kronborg, C.; Earnest, A.; Millar, J.; Clouston, D. Gleason group concordance between biopsy and radical prostatectomy specimens: A cohort study from Prostate Cancer Outcome Registry-Victoria. Prostate Int. 2016, 4, 145–151. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.D.; Mahal, B.A.; Muralidhar, V.; Nezolosky, M.D.; Vastola, M.E.; Labe, S.A.; Boldbaatar, N.; King, M.T.; Martin, N.E.; Orio, P.F., 3rd; et al. Risk of Upgrading and Upstaging Among 10,000 Patients with Gleason 3 + 4 Favorable Intermediate-risk Prostate Cancer. Eur. Urol. Focus 2019, 5, 69–76. [Google Scholar] [CrossRef]
- Epstein, J.I.; Feng, Z.; Trock, B.J.; Pierorazio, P.M. Upgrading and Downgrading of Prostate Cancer from Biopsy to Radical Prostatectomy: Incidence and Predictive Factors Using the Modified Gleason Grading System and Factoring in Tertiary Grades. Eur. Urol. 2012, 61, 1019–1024. [Google Scholar] [CrossRef] [PubMed]
- Kane, C.J.; Eggener, S.E.; Shindel, A.W.; Andriole, G.L. Variability in Outcomes for Patients with Intermediate-risk Prostate Cancer (Gleason Score 7, International Society of Urological Pathology Gleason Group 2–3) and Implications for Risk Stratification: A Systematic Review. Eur. Urol. Focus 2017, 3, 487–497. [Google Scholar] [CrossRef] [PubMed]
- Sanda, M.G.; Cadeddu, J.A.; Kirkby, E.; Chen, R.C.; Crispino, T.; Fontanarosa, J.; Freedland, S.J.; Greene, K.; Klotz, L.H.; Makarov, D.V.; et al. Clinically Localized Prostate Cancer: AUA/ASTRO/SUO Guideline. Part I: Risk Stratification, Shared Decision Making, and Care Options. J. Urol. 2017, 199, 683–690. [Google Scholar] [CrossRef]
- Zumsteg, Z.S.; Zelefsky, M.J. Short-term androgen deprivation therapy for patients with intermediate-risk prostate cancer undergoing dose-escalated radiotherapy: The standard of care? Lancet Oncol. 2012, 13, e259–e269. [Google Scholar] [CrossRef]
- Gosselaar, C.; Kranse, R.; Roobol, M.J.; Roemeling, S.; Schröder, F.H. The interobserver variability of digital rectal examination in a large randomized trial for the screening of prostate cancer. Prostate 2008, 68, 985–993. [Google Scholar] [CrossRef]
- Ankerst, D.P.; Miyamoto, R.; Nair, P.V.; Pollock, B.H.; Thompson, I.M.; Parekh, D.J. Yearly prostate specific antigen and digital rectal examination fluctuations in a screened population. J. Urol. 2009, 181, 2071–2076. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Naji, L.; Randhawa, H.; Sohani, Z.; Dennis, B.; Lautenbach, D.; Kavanagh, O.; Bawor, M.; Banfield, L.; Profetto, J. Digital Rectal Examination for Prostate Cancer Screening in Primary Care: A Systematic Review and Meta-Analysis. Ann. Fam. Med. 2018, 16, 149–154. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, H.U.; El-Shater Bosaily, A.; Brown, L.C.; Gabe, R.; Kaplan, R.; Parmar, M.K.; Collaco-Moraes, Y.; Ward, K.; Hindley, R.G.; Freeman, A.; et al. Diagnostic accuracy of multi-parametric MRI and TRUS biopsy in prostate cancer (PROMIS): A paired validating confirmatory study. Lancet 2017, 25, 815–822. [Google Scholar] [CrossRef]
- Eldred-Evans, D.; Burak, P.; Connor, M.J.; Day, E.; Evans, M.; Fiorentino, F.; Gammon, M.; Hosking-Jervis, F.; Klimowska-Nassar, N.; McGuire, W.; et al. Population-Based Prostate Cancer Screening with Magnetic Resonance Imaging or Ultrasonography. JAMA Oncol. 2021, 7, 395–402. [Google Scholar] [CrossRef] [PubMed]
- Walz, J. The “PROMIS” of Magnetic Resonance Imaging Cost Effectiveness in Prostate Cancer Diagnosis? Eur. Urol. 2018, 73, 31–32. [Google Scholar] [CrossRef] [PubMed]
| NEC | GG1 | GG2 | GG3 | GG4 | GG5 | |
|---|---|---|---|---|---|---|
| Total (%) | 62 (22.4%) | 93 (33.6%) | 61 (22.0%) | 38 (13.7%) | 12 (4.3%) | 11 (4.0%) |
| Age in years | 66 (17) | 67 (6.5) | 68 (9) | 73 (8.25) | 71 (12.25) | 69 (8.5) |
| PSA ng/mL | 1.2 (2) | 7.3 (5.5) | 7.8 (4.5) | 11.35 (11.03) | 13.45 (12.7) | 18.7 (45.95) |
| Prostate volume US | NA | 23 (7.5) | 42.43 (24.7) | 49.03 (28.4) | 50.39 (22.9) | 55.93 (24.97) |
| PSAD | NA | 0.292 (0.31) | 0.179 (0.12) | 0.256 (0.22) | 0.287 (0.38) | 0.31 (0.56) |
| Biopsy Cores taken | NA | 11 (4) | 9 (2) | 8 (6) | 5.5 (5.25) | 4 (6.5) |
| Biopsy Cores positive | NA | 1 (1) | 3 (3) | 3.5 (2.75) | 3.5 (3.5) | 4 (5) |
| % biopsy cores positive | NA | 12.5 (16.67) | 37.5 (42.5) | 50 (58.57) | 50 (36.39) | 100 (0) |
| % biopsy cores positive/US | NA | 0.003 (0.004) | 0.001 (0.001) | 0.001 (0.001) | 0.001 (0.001) | 0.001 (0.002) |
| PUR-1 | 0.358 (0.206) | 0.119 (0.165) | 0.078 (0.145) | 0.03 (0.119) | 0.059 (0.04) | 0.07 (0.059) |
| PUR-2 | 0.321 (0.036) | 0.302 (0.078) | 0.275 (0.151) | 0.187 (0.217) | 0.248 (0.07) | 0.201 (0.124) |
| PUR-3 | 0.256 (0.129) | 0.442 (0.15) | 0.452 (0.116) | 0.475 (0.119) | 0.506 (0.032) | 0.489 (0.073) |
| PUR-4 | 0.049 (0.036) | 0.126 (0.09) | 0.162 (0.18) | 0.255 (0.269) | 0.188 (0.075) | 0.171 (0.146) |
| GG | Majority GG, %GP4 | Prostate Area (mm2) | Tumor Area (mm2) | GP3 Area | GP4 Area | Age | PSA | PUR-4 | |
|---|---|---|---|---|---|---|---|---|---|
| RPx 1 | GG1 | GG1+4 | 5269.9 | 486.4 | 481.6 | 4.8 | 54 | 5.5 | 0.04 |
| RPx 2 | GG2 | GG2, <10% GP4 | 3339.0 | 341.2 | 324.1 | 17.1 | 64 | 15 | 0.11 |
| RPx 3 | GG2 | GG2, <10% GP4 | 4347.6 | 531.8 | 505.2 | 26.6 | 52 | 5.8 | 0.14 |
| RPx 4 | GG2+5 | GG2, <10% GP4 | 4606.0 | 254.5 | 233.9 | 20.5 | 70 | 8.4 | 0.44 |
| RPx 5 | GG2 | GG2, 10–25% GP4 | 3643.2 | 172.1 | 143.3 | 28.8 | 66 | 5.2 | 0.10 |
| RPx 6 | GG2 | GG2, <10% GP4 | 4529.7 | 364.0 | 302.0 | 62.0 | 68 | 6.7 | 0.62 |
| RPx 7 | GG3 | GG2, 25–50% GP4 | 11,991.1 | 92.9 | 54.8 | 38.0 | 67 | 10.3 | 0.08 |
| RPx 8 | GG3 | GG3, 50–75% GP4 | 3729.6 | 158.7 | 60.3 | 98.4 | 65 | 7.4 | 0.44 |
| RPx 9 | GG3 | GG3, 75–90% GP4 | 3851.4 | 664.4 | 122.2 | 542.2 | 75 | 2.9 | 0.56 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ball, R.Y.; Cardenas, R.; Winterbone, M.S.; Hanna, M.Y.; Parker, C.; Hurst, R.; Brewer, D.S.; D’Sa, L.; Mills, R.; Cooper, C.S.; et al. The Urine Biomarker PUR-4 Is Positively Associated with the Amount of Gleason 4 in Human Prostate Cancers. Life 2021, 11, 1172. https://doi.org/10.3390/life11111172
Ball RY, Cardenas R, Winterbone MS, Hanna MY, Parker C, Hurst R, Brewer DS, D’Sa L, Mills R, Cooper CS, et al. The Urine Biomarker PUR-4 Is Positively Associated with the Amount of Gleason 4 in Human Prostate Cancers. Life. 2021; 11(11):1172. https://doi.org/10.3390/life11111172
Chicago/Turabian StyleBall, Richard Y., Ryan Cardenas, Mark S. Winterbone, Marcelino Y. Hanna, Chris Parker, Rachel Hurst, Daniel S. Brewer, Lauren D’Sa, Rob Mills, Colin S. Cooper, and et al. 2021. "The Urine Biomarker PUR-4 Is Positively Associated with the Amount of Gleason 4 in Human Prostate Cancers" Life 11, no. 11: 1172. https://doi.org/10.3390/life11111172
APA StyleBall, R. Y., Cardenas, R., Winterbone, M. S., Hanna, M. Y., Parker, C., Hurst, R., Brewer, D. S., D’Sa, L., Mills, R., Cooper, C. S., & Clark, J. (2021). The Urine Biomarker PUR-4 Is Positively Associated with the Amount of Gleason 4 in Human Prostate Cancers. Life, 11(11), 1172. https://doi.org/10.3390/life11111172







