Expression of Kisspeptin 1 in the Brain of the Adult Sea Lamprey Petromyzon marinus
Abstract
1. Introduction
2. Materials and Methods
2.1. Alignment of the Kiss1 and Kiss2 Sea Lamprey Precursor Sequences with Kiss Precursor Sequences from other Chordates and Phylogenetic Analyses
2.2. Animals
2.3. Cloning and Sequencing of the Sea Lamprey Kiss1 and Kiss2 cDNAs
2.4. In Situ Hybridization on Tissue Sections
2.5. Imaging and Figure Preparation
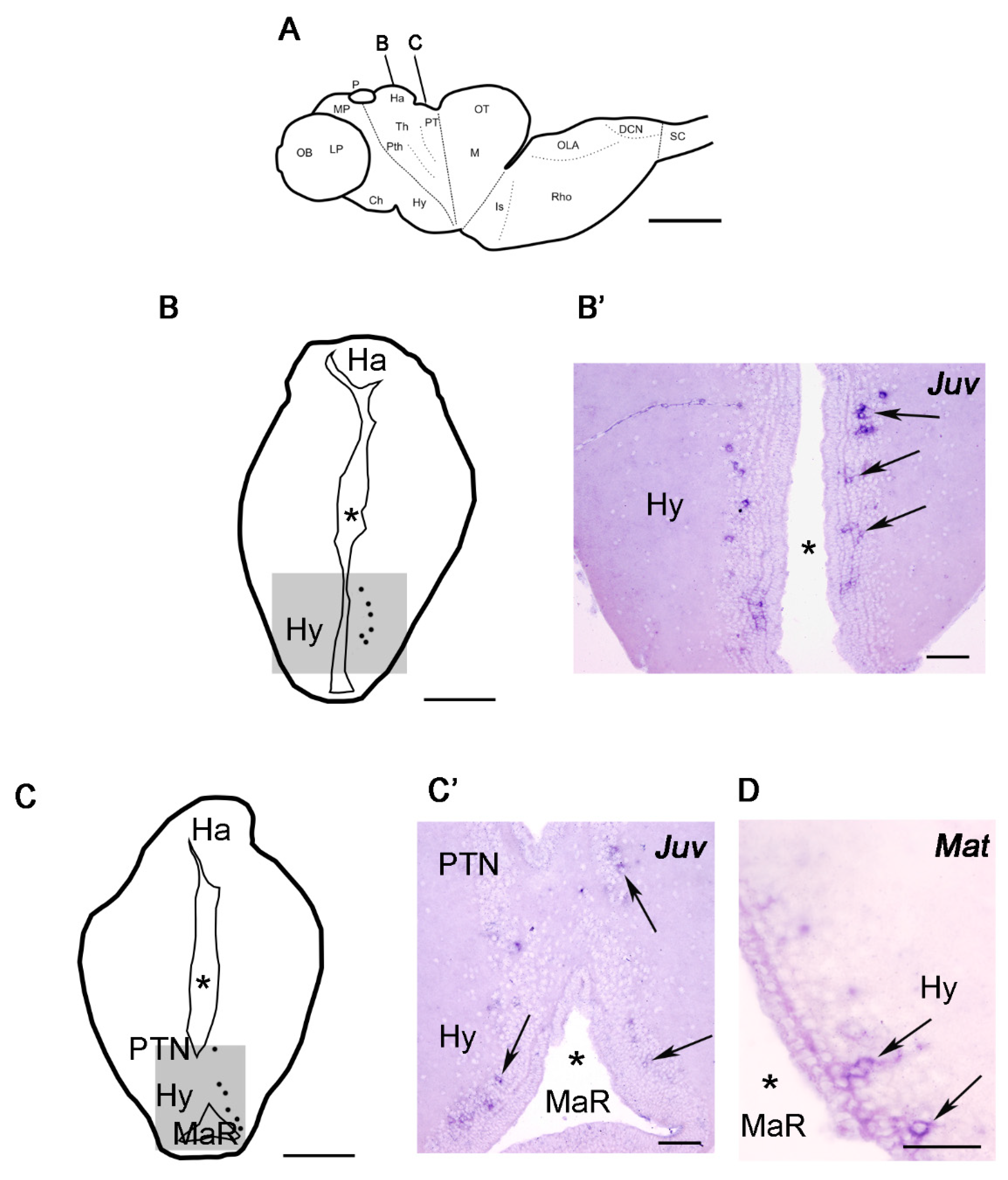
3. Results
3.1. Sequence Analyses
3.2. Expression of the Kiss1 and Kiss2 Transcripts in the Adult Sea Lamprey Brain
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Osugi, T.; Son, Y.L.; Ubuka, T.; Satake, H.; Tsutsui, K. RFamide peptides in agnathans and basal chordates. Gen. Comp. Endocrinol. 2016, 227, 94–100. [Google Scholar] [CrossRef] [PubMed]
- Ohtaki, T.; Shintani, Y.; Honda, S.; Matsumoto, H.; Hori, A.; Kanehashi, K.; Terao, Y.; Kumano, S.; Takatsu, Y.; Masuda, Y.; et al. Metastasis suppressor gene KiSS-1 encodes peptide ligand of a G-protein-coupled receptor. Nat. Cell Biol. 2001, 411, 613–617. [Google Scholar] [CrossRef] [PubMed]
- Pasquier, J.; Lafont, A.-G.; Rousseau, K.; Quérat, B.; Chemineau, P.; Dufour, S. Looking for the bird Kiss: Evolutionary scenario in sauropsids. BMC Evol. Biol. 2014, 14, 30. [Google Scholar] [CrossRef]
- Lee, Y.R.; Tsunekawa, K.; Moon, M.J.; Um, H.N.; Hwang, J.-I.; Osugi, T.; Otaki, N.; Sunakawa, Y.; Kim, K.; Vaudry, H.; et al. Molecular Evolution of Multiple Forms of Kisspeptins and GPR54 Receptors in Vertebrates. Endocrinology 2009, 150, 2837–2846. [Google Scholar] [CrossRef]
- Felip, A.; Zanuy, S.; Pineda, R.; Pinilla, L.; Carrillo, M.; Tena-Sempere, M.; Gómez, A. Evidence for two distinct KiSS genes in non-placental vertebrates that encode kisspeptins with different gonadotropin-releasing activities in fish and mammals. Mol. Cell. Endocrinol. 2009, 312, 61–71. [Google Scholar] [CrossRef]
- Roa, J.; Castellano, J.M.; Navarro, V.; Handelsman, D.; Pinilla, L.; Tena-Sempere, M. Kisspeptins and the control of gonadotropin secretion in male and female rodents. Peptides 2009, 30, 57–66. [Google Scholar] [CrossRef]
- Sower, S.A. Landmark discoveries in elucidating the origins of the hypothalamic-pituitary system from the perspective of a basal vertebrate, sea lamprey. Gen. Comp. Endocrinol. 2018, 264, 3–15. [Google Scholar] [CrossRef]
- Pasquier, J.; Lafont, A.-G.; Leprince, J.; Vaudry, H.; Rousseau, K.; Dufour, S. First evidence for a direct inhibitory effect of kisspeptins on LH expression in the eel, Anguilla anguilla. Gen. Comp. Endocrinol. 2011, 173, 216–225. [Google Scholar] [CrossRef]
- Capella-Gutierrez, S.; Silla-Martinez, J.M.; Gabaldon, T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef]
- Lemoine, F.; Correia, D.; Lefort, V.; Doppelt-Azeroual, O.; Mareuil, F.; Cohen-Boulakia, S.; Gascuel, O. NGPhylogeny.fr: New generation phylogenetic services for non-specialists. Nucleic Acids Res. 2019, 47, W260–W265. [Google Scholar] [CrossRef]
- Frickey, T.; Lupas, A. CLANS: A Java application for visualizing protein families based on pairwise similarity. Bioinformatics 2004, 20, 3702–3704. [Google Scholar] [CrossRef]
- Yun, S.; Furlong, M.; Sim, M.; Cho, M.; Park, S.; Cho, E.B.; Reyes-Alcaraz, A.; Hwang, J.-I.; Kim, J.; Seong, J.Y. Prevertebrate Local Gene Duplication Facilitated Expansion of the Neuropeptide GPCR Superfamily. Mol. Biol. Evol. 2015, 32, 2803–2817. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sobrido-Cameán, D.; Guerra, L.A.Y.; Lamanna, F.; Conde-Fernández, C.; Kaessmann, H.; Elphick, M.R.; Anadón, R.; Rodicio, M.C.; Barreiro-Iglesias, A. Galanin in an Agnathan: Precursor Identification and Localisation of Expression in the Brain of the Sea Lamprey Petromyzon marinus. Front. Neuroanat. 2019, 13, 83. [Google Scholar] [CrossRef]
- Sobrido-Cameán, D.; Yáñez-Guerra, L.A.; Robledo, D.; López-Varela, E.; Rodicio, M.C.; Elphick, M.R.; Anadón, R.; Barreiro-Iglesias, A. Cholecystokinin in the central nervous system of the sea lamprey Petromyzon marinus: Precursor identification and neuroanatomical relationships with other neuronal signalling systems. Anat. Embryol. 2020, 225, 249–284. [Google Scholar] [CrossRef]
- Sobrido-Cameán, D.; Yáñez-Guerra, L.A.; Deber, A.; Freire-Delgado, M.; Cacheiro-Vázquez, R.; Rodicio, M.C.; Tostivint, H.; Anadón, R.; Barreiro-Iglesias, A. Differential expression of somatostatin genes in the central nervous system of the sea lamprey. Anat. Embryol. 2021, 226, 1031–1052. [Google Scholar] [CrossRef]
- Foster, S.R.; Hauser, A.S.; Vedel, L.; Strachan, R.T.; Huang, X.-P.; Gavin, A.C.; Shah, S.D.; Nayak, A.P.; Haugaard-Kedström, L.; Penn, R.B.; et al. Discovery of Human Signaling Systems: Pairing Peptides to G Protein-Coupled Receptors. Cell 2019, 179, 895–908.e21. [Google Scholar] [CrossRef]
- Kanda, S.; Akazome, Y.; Matsunaga, T.; Yamamoto, N.; Yamada, S.; Tsukamura, H.; Maeda, K.-I.; Oka, Y. Identification of KiSS-1 Product Kisspeptin and Steroid-Sensitive Sexually Dimorphic Kisspeptin Neurons in Medaka (Oryzias latipes). Endocrinology 2008, 149, 2467–2476. [Google Scholar] [CrossRef]
- Kitahashi, T.; Ogawa, S.; Parhar, I.S. Cloning and Expression of kiss2 in the Zebrafish and Medaka. Endocrinology 2009, 150, 821–831. [Google Scholar] [CrossRef]
- Mitani, Y.; Kanda, S.; Akazome, Y.; Zempo, B.; Oka, Y. Hypothalamic Kiss1 but Not Kiss2 Neurons Are Involved in Estrogen Feedback in Medaka (Oryzias latipes). Endocrinology 2010, 151, 1751–1759. [Google Scholar] [CrossRef]
- Servili, A.; Le Page, Y.; Leprince, J.; Caraty, A.; Escobar, S.; Parhar, I.S.; Seong, J.Y.; Vaudry, H.; Kah, O. Organization of Two Independent Kisspeptin Systems Derived from Evolutionary-Ancient Kiss Genes in the Brain of Zebrafish. Endocrinology 2011, 152, 1527–1540. [Google Scholar] [CrossRef]
- Zmora, N.; Stubblefield, J.; Zulperi, Z.; Biran, J.; Levavi-Sivan, B.; Muñoz-Cueto, J.A.; Zohar, Y. Differential and Gonad Stage-Dependent Roles of Kisspeptin1 and Kisspeptin2 in Reproduction in the Modern Teleosts, Morone Species1. Biol. Reprod. 2012, 86, 177. [Google Scholar] [CrossRef]
- Escobar, S.; Felip, A.; Gueguen, M.-M.; Zanuy, S.; Carrillo, M.; Kah, O.; Servili, A. Expression of kisspeptins in the brain and pituitary of the european sea bass (Dicentrarchus labrax). J. Comp. Neurol. 2013, 521, 933–948. [Google Scholar] [CrossRef]
- Escobar, S.; Servili, A.; Espigares, F.; Gueguen, M.-M.; Brocal, I.; Felip, A.; Gómez, A.; Carrillo, M.; Zanuy, S.; Kah, O. Expression of Kisspeptins and Kiss Receptors Suggests a Large Range of Functions for Kisspeptin Systems in the Brain of the European Sea Bass. PLoS ONE 2013, 8, e70177. [Google Scholar] [CrossRef]
- Zhang, R.; Nie, H.; Duan, S.; Yan, P.; Izaz, A.; Wang, R.; Zhou, Y.; Wu, X. Cloning, characterisation and expression profile of kisspeptin1 and the kisspeptin1 receptor in the hypothalamic–pituitary–ovarian axis of Chinese alligator Alligator sinensis during the reproductive cycle. Reprod. Fertil. Dev. 2020, 32, 792–804. [Google Scholar] [CrossRef]
- Clarkson, J.; Herbison, A.E. Postnatal Development of Kisspeptin Neurons in Mouse Hypothalamus; Sexual Dimorphism and Projections to Gonadotropin-Releasing Hormone Neurons. Endocrinology 2006, 147, 5817–5825. [Google Scholar] [CrossRef]
- Van Gulick, E.R.; Marquis, T.J.; Sower, S.A. Co-localization of three gonadotropin-releasing hormone transcripts in larval, parasitic, and adult sea lamprey brains. Gen. Comp. Endocrinol. 2018, 264, 84–93. [Google Scholar] [CrossRef]
- Song, Y.; Duan, X.; Chen, J.; Huang, W.; Zhu, Z.; Hu, W. The Distribution of Kisspeptin (Kiss)1- and Kiss2-Positive Neurones and Their Connections with Gonadotrophin-Releasing Hormone-3 Neurones in the Zebrafish Brain. J. Neuroendocr. 2015, 27, 198–211. [Google Scholar] [CrossRef]
- Pombal, M.A.; El Manira, A.; Grillner, S. Afferents of the lamprey striatum with special reference to the dopaminergic system: A combined tracing and immunohistochemical study. J. Comp. Neurol. 1997, 386, 71–91. [Google Scholar] [CrossRef]
- Pérez-Fernández, J.; Stephenson-Jones, M.; Suryanarayana, S.M.; Robertson, B.; Grillner, S. Evolutionarily conserved organization of the dopaminergic system in lamprey: SNc/VTA afferent and efferent connectivity and D2 receptor expression. J. Comp. Neurol. 2014, 522, 3775–3794. [Google Scholar] [CrossRef]
- von Twickel, A.; Kowatschew, D.; Saltürk, M.; Schauer, M.; Robertson, B.; Korsching, S.; Walkowiak, W.; Grillner, S.; Pérez-Fernández, J. Individual Dopaminergic Neurons of Lamprey SNc/VTA Project to Both the Striatum and Optic Tectum but Restrict Co-release of Glutamate to Striatum Only. Curr. Biol. 2019, 29, 677–685. [Google Scholar] [CrossRef]
- Ryczko, D.; Grätsch, S.; Alpert, M.H.; Cone, J.J.; Kasemir, J.; Ruthe, A.; Beauséjour, P.-A.; Auclair, F.; Roitman, M.F.; Alford, S.; et al. Descending Dopaminergic Inputs to Reticulospinal Neurons Promote Locomotor Movements. J. Neurosci. 2020, 40, 8478–8490. [Google Scholar] [CrossRef]
- Barreiro-Iglesias, A.; Villar-Cerviño, V.; Anadón, R.; Rodicio, M.C. Descending brain-spinal cord projections in a primitive vertebrate, the lamprey: Cerebrospinal fluid-contacting and dopaminergic neurons. J. Comp. Neurol. 2008, 511, 711–723. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sobrido-Cameán, D.; Yáñez-Guerra, L.A.; Deber, A.; Rodicio, M.C.; Barreiro-Iglesias, A. Expression of Kisspeptin 1 in the Brain of the Adult Sea Lamprey Petromyzon marinus. Life 2021, 11, 1174. https://doi.org/10.3390/life11111174
Sobrido-Cameán D, Yáñez-Guerra LA, Deber A, Rodicio MC, Barreiro-Iglesias A. Expression of Kisspeptin 1 in the Brain of the Adult Sea Lamprey Petromyzon marinus. Life. 2021; 11(11):1174. https://doi.org/10.3390/life11111174
Chicago/Turabian StyleSobrido-Cameán, Daniel, Luis Alfonso Yáñez-Guerra, Alexandre Deber, María Celina Rodicio, and Antón Barreiro-Iglesias. 2021. "Expression of Kisspeptin 1 in the Brain of the Adult Sea Lamprey Petromyzon marinus" Life 11, no. 11: 1174. https://doi.org/10.3390/life11111174
APA StyleSobrido-Cameán, D., Yáñez-Guerra, L. A., Deber, A., Rodicio, M. C., & Barreiro-Iglesias, A. (2021). Expression of Kisspeptin 1 in the Brain of the Adult Sea Lamprey Petromyzon marinus. Life, 11(11), 1174. https://doi.org/10.3390/life11111174





