The Genus Chlorosplenium (Helotiales, Leotiomycetes) from China with Notes on C. chlora Complex
Abstract
:1. Introduction
2. Materials and Methods
3. Results
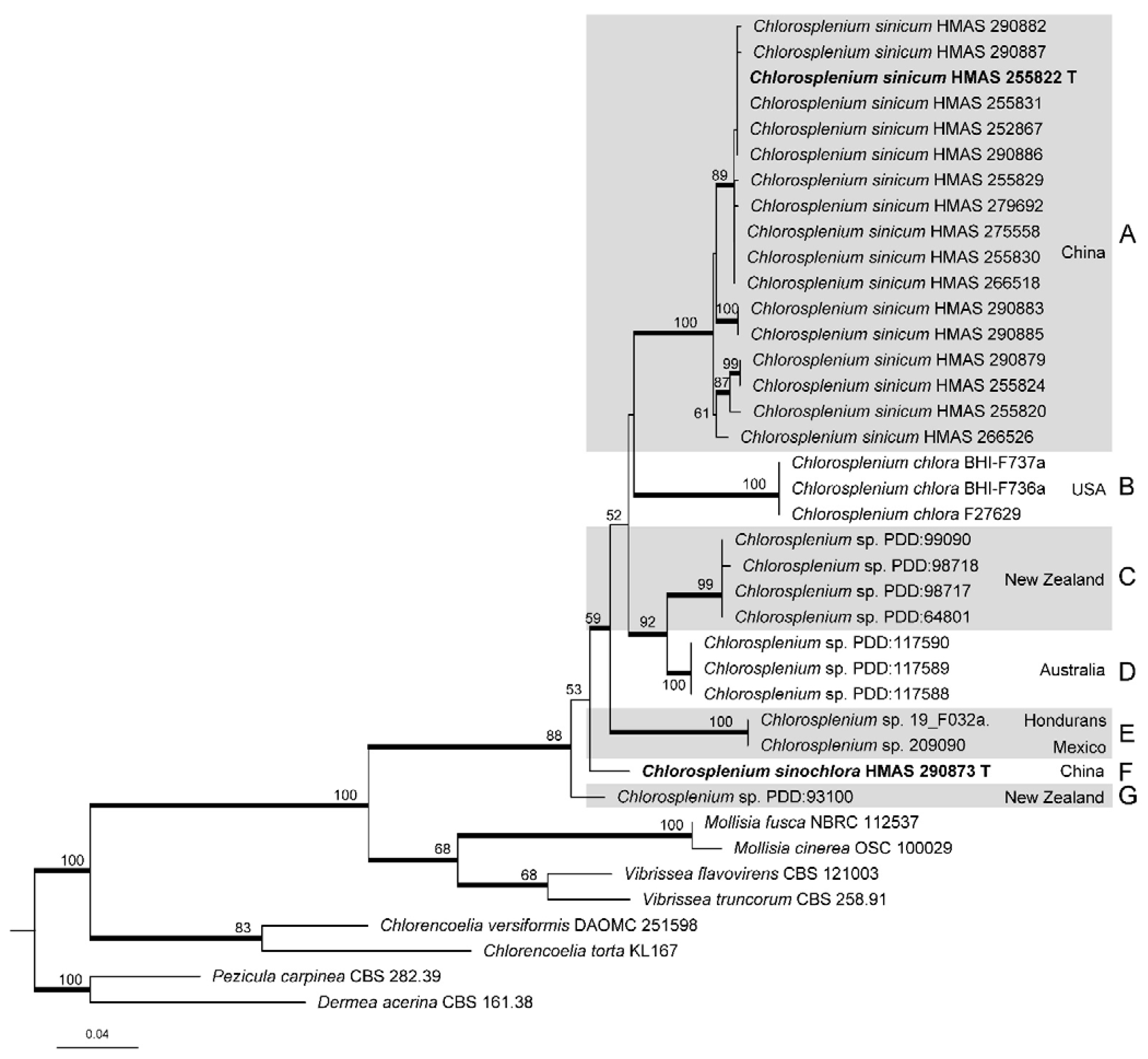
3.1. Phylogenetic Analyses
3.2. Taxonomy
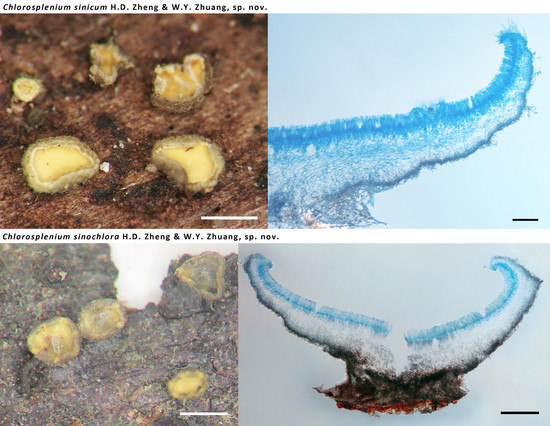
3.2.1. Chlorosplenium sinicum H.D. Zheng & W.Y. Zhuang, sp. nov.
- Fungal Names—FN570862
- Etymology—The specific epithet refers to the type locality of the species.
- Holotype—CHINA, Hunan, Yizhang, Mang Mt., on rotten wood, 28 Oct. 2015, Z.Q. Zeng et al. 10345 (HMAS 255822).
3.2.2. Chlorosplenium sinochlora H.D. Zheng & W.Y. Zhuang, sp. nov.
- Fungal Names—FN570863
- Etymology—The specific epithet refers to the origin of the species and its similarity with C. chlora.
- Holotype—CHINA, Hunan, Yizhang, Mang Mt., Jiangjunzhai, on rotten bark, 27 October 2015, Z.Q. Zeng et al. 10286 (HMAS 290873).
3.2.3. Excluded and Doubtful Species of Chlorosplenium
Chlorosplenium fusisporum Liou & Z.C. Chen
Chlorosplenium hyperici-maculati Svrček
3.2.4. Key to Accepted Species of Chlorosplenium
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dixon, J.R. Chlorosplenium and its segregates. I. Introduction and the genus Chlorosplenium. Mycotaxon 1974, 1, 65–104. [Google Scholar]
- Dixon, J.R. Chlorosplenium and its segregates II. The genera Chlorociboria and Chlorencoelia. Mycotaxon 1975, 1, 193–237. [Google Scholar]
- Johnston, P.R.; Quijada, L.; Smith, C.A.; Baral, H.O.; Hosoya, T.; Baschien, C.; Partel, K.; Zhuang, W.Y.; Haelewaters, D.; Park, D.; et al. A multigene phylogeny toward a new phylogenetic classification of Leotiomycetes. IMA Fungus 2019, 10, 1. [Google Scholar] [CrossRef] [PubMed]
- Kirk, P.M.; Cannon, P.F.; Minter, D.W.; Stalpers, J.A. Dictionary of the Fungi, 10th ed.; CABI: Wallingford, UK, 2008; p. 771. [Google Scholar]
- Ekanayaka, A.; Hyde, K.; Gentekaki, E.; McKenzie, E.; Zhao, Q.; Bulgakov, T.; Camporesi, E. Preliminary classification of Leotiomycetes. Mycosphere 2019, 10, 310–489. [Google Scholar] [CrossRef]
- Müller, E. Zur Pilzflora der Aletschwaldreservats (Kt. Wallis, Schweiz). Beiträge zur Kryptogamenflora der Schweiz 1977, 15, 1–126. [Google Scholar]
- Liou, S.-C.; Chen, Z.-C. Notes on Taiwan Discomycetes I. (Pezizales and Helotiales). Taiwania 1977, 22, 29–43. [Google Scholar]
- Svrček, M. New or less known Discomycetes. XXII. Česká Mykologie 1992, 46, 33–40. [Google Scholar]
- Zhuang, W.-Y.; Zheng, H.-D.; Zeng, Z.-Q. Species Catalogue of China, Volumen 3 Fungi, Cup-Fungi; Science Press: Beijing, China, 2018; p. 142. [Google Scholar]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Snisky, J.J., White, T.J., Eds.; Academic Press: London, UK, 1990; pp. 315–322. [Google Scholar]
- Moncalvo, J.-M.; Wang, H.-H.; Hseu, R.-S. Phylogenetic Relationships in Ganoderma Inferred from the Internal Transcribed Spacers and 25S Ribosomal DNA Sequences. Mycologia 1995, 87, 223–238. [Google Scholar] [CrossRef]
- Vilgalys, R.; Hester, M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 1990, 172, 4238–4246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stiller, J.W.; Hall, B.D. The origin of red algae: Implications for plastid evolution. Proc. Natl. Acad. Sci. USA 1997, 94, 4520–4525. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matheny, P.B.; Liu, Y.J.; Ammirati, J.F.; Hall, B.D. Using RPB1 sequences to improve phylogenetic inference among mushrooms (Inocybe, Agaricales). Am. J. Bot. 2002, 89, 688–698. [Google Scholar] [CrossRef] [PubMed]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Edgar, R.C. MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinform. 2004, 5, 113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef] [Green Version]
- Nylander, J.A.A. MrModeltest V2; Program distributed by the author; Evolutionary Biology Centre, Uppsala University: Uppsala, Sweden, 2004. [Google Scholar]
- Rambaut, A. FigTree, V1.4.4.; Institute of Evolutionary Biology, University of Edinburgh: Edinburgh, UK, 2018; Available online: http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 22 May 2019).
- Chlorosplenium chlora (Schwein). M.A. Curtis in GBIF Secretariat. GBIF Backbone Taxonomy. Checklist Dataset. Available online: https://doi.org/10.15468/39omei (accessed on 27 August 2021).
- Arendholz, W.-R.; Sharma, R. Some new or interesting Helotiales from the eastern Himalayas. Mycotaxon 1983, 17, 473–512. [Google Scholar]
- Park, P.-J.; Lee, J.-Y.; Otani, Y. Taxonomical Studies on Discomycetes in Korea (I). Korean J. Mycol. 1985, 13, 27–40. [Google Scholar]
- Saccardo, P.A. Sylloge Fungorum; Typis Seminarii: Patavii, Italy, 1889; Volume 8, p. 1143. [Google Scholar]
- Vila, J.; Llistosella, J.; Llimona, I.X. Contribuciό al coneixement dels fongs de l’estatge alpí dels pirineus de Catalunya. Il. Rev. Catalana Micol. 1998, 2, 93–113. [Google Scholar]




| Name | Voucher/Isolate | Country | GenBank Number | ||
|---|---|---|---|---|---|
| ITS | LSU | RPB1 | |||
| Chlorosplenium chlora (Schwein.) M.A. Curtis | BHI-F736 | USA | MG553993 | ||
| C. chlora | BHI-F737 | USA | MG553994 | ||
| C. chlora | F27629 | USA | MZ312166 | ||
| C. sinicum H.D. Zheng & W.Y. Zhuang | HMAS 279692 (8260) | China | MK425600 | ||
| C. sinicum | HMAS 266518 | China | MK425599 | ||
| C. sinicum | HMAS 252867 | China | MZ914636 | ||
| C. sinicum | HMAS 266526 | China | MZ914637 | ||
| C. sinicum | HMAS 275558 (9822) | China | MZ914635 | ||
| C. sinicum | HMAS 290882 (10543) | China | MZ914627 | ||
| C. sinicum | HMAS 290886 (12556) | China | MZ914630 | ||
| C. sinicum | HMAS 255830 (12559) | China | MZ914632 | ||
| C. sinicum | HMAS 255831 (12561) | China | MZ914634 | ||
| C. sinicum | HMAS 255822 T (10345) | China | MZ914624 | MZ920115 | MZ945729 |
| C. sinicum | HMAS 255829 (12558) | China | MZ914631 | MZ920119 | MZ945733 |
| C. sinicum | HMAS 290887 (12560) | China | MZ914633 | MZ920120 | MZ945734 |
| C. sinicum | HMAS 255820 (10249) | China | MZ914623 | MZ920114 | MZ945728 |
| C. sinicum | HMAS 255824 (10398) | China | MZ914625 | MZ920116 | MZ945730 |
| C. sinicum | HMAS 290879 (10433) | China | MZ914626 | MZ920117 | MZ945731 |
| C. sinicum | HMAS 290885 (11441) | China | MZ914628 | MZ920118 | MZ945732 |
| C. sinicum | HMAS 290883 (YN16-34) | China | MZ914629 | MZ920121 | MZ945735 |
| C. sinochlora H.D. Zheng & W.Y. Zhuang | HMAS 290873 T (10286) | China | MZ914622 | MZ920122 | MZ945736 |
| Chlorosplenium sp. | HONDURAS19-F032 | Honduras | MT571528 | ||
| Chlorosplenium sp. | 209090 | Mexico | MG976227 | ||
| Chlorosplenium sp. | PDD:117588 | Australia | MW191764 | ||
| Chlorosplenium sp. | PDD:117589 | Australia | MW191759 | ||
| Chlorosplenium sp. | PDD:117590 | Australia | MW191760 | ||
| Chlorosplenium sp. | PDD:64801 | New Zealand | MW191757 | ||
| Chlorosplenium sp. | PDD:98717 | New Zealand | MW191758 | ||
| Chlorosplenium sp. | PDD:98718 | New Zealand | MW191763 | ||
| Chlorosplenium sp. | PDD:99090 | New Zealand | MW191761 | ||
| Chlorosplenium sp. | PDD:93100 | New Zealand | MW191762 | ||
| Chlorencoelia torta (Schwein.) J.R. Dixon | KL167 | China | LT158424 | ||
| C. versiformis (Pers.) J.R. Dixon | DAOMC 251598 | Canada | MH457140 | ||
| Dermea acerina (Peck) Rehm | CBS 161.38/AFTOL-ID 941 | Canada | MH855942 | MH867440 | DQ471164 |
| Mollisia cinerea (Batsch) P. Karst. | OSC 100029/AFTOL-ID 76 | Unknown | DQ491498 | DQ470942 | DQ471122 |
| M. fusca (Pers.) P. Karst. | TNS:F17463/NBRC 112537 | Japan | LC425049 | ||
| Pezicula carpinea (Pers.) Tul. ex Fuckel | CBS 282.39/AFTOL-ID 938 | Canada | KR859272 | DQ470967 | DQ842032 |
| Vibrissea flavovirens (Pers.) Korf & J.R. Dixon | CBS 121003 | Germany | MT026430 | ||
| V. truncorum (Alb. & Schwein.) Fr. | CBS 258.91/AFTOL-ID 1322 | Canada | MT026377 | FJ176874 | FJ238438 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zheng, H.-D.; Zhuang, W.-Y. The Genus Chlorosplenium (Helotiales, Leotiomycetes) from China with Notes on C. chlora Complex. Life 2021, 11, 1167. https://doi.org/10.3390/life11111167
Zheng H-D, Zhuang W-Y. The Genus Chlorosplenium (Helotiales, Leotiomycetes) from China with Notes on C. chlora Complex. Life. 2021; 11(11):1167. https://doi.org/10.3390/life11111167
Chicago/Turabian StyleZheng, Huan-Di, and Wen-Ying Zhuang. 2021. "The Genus Chlorosplenium (Helotiales, Leotiomycetes) from China with Notes on C. chlora Complex" Life 11, no. 11: 1167. https://doi.org/10.3390/life11111167
APA StyleZheng, H.-D., & Zhuang, W.-Y. (2021). The Genus Chlorosplenium (Helotiales, Leotiomycetes) from China with Notes on C. chlora Complex. Life, 11(11), 1167. https://doi.org/10.3390/life11111167