Foliar Fungal Endophytes in a Tree Diversity Experiment Are Driven by the Identity but Not the Diversity of Tree Species
Abstract
:1. Introduction
2. Materials and Methods
2.1. Leaf Sampling
2.2. DNA Extraction and Sequencing
2.3. Sequence Processing
2.4. Statistical Analyses
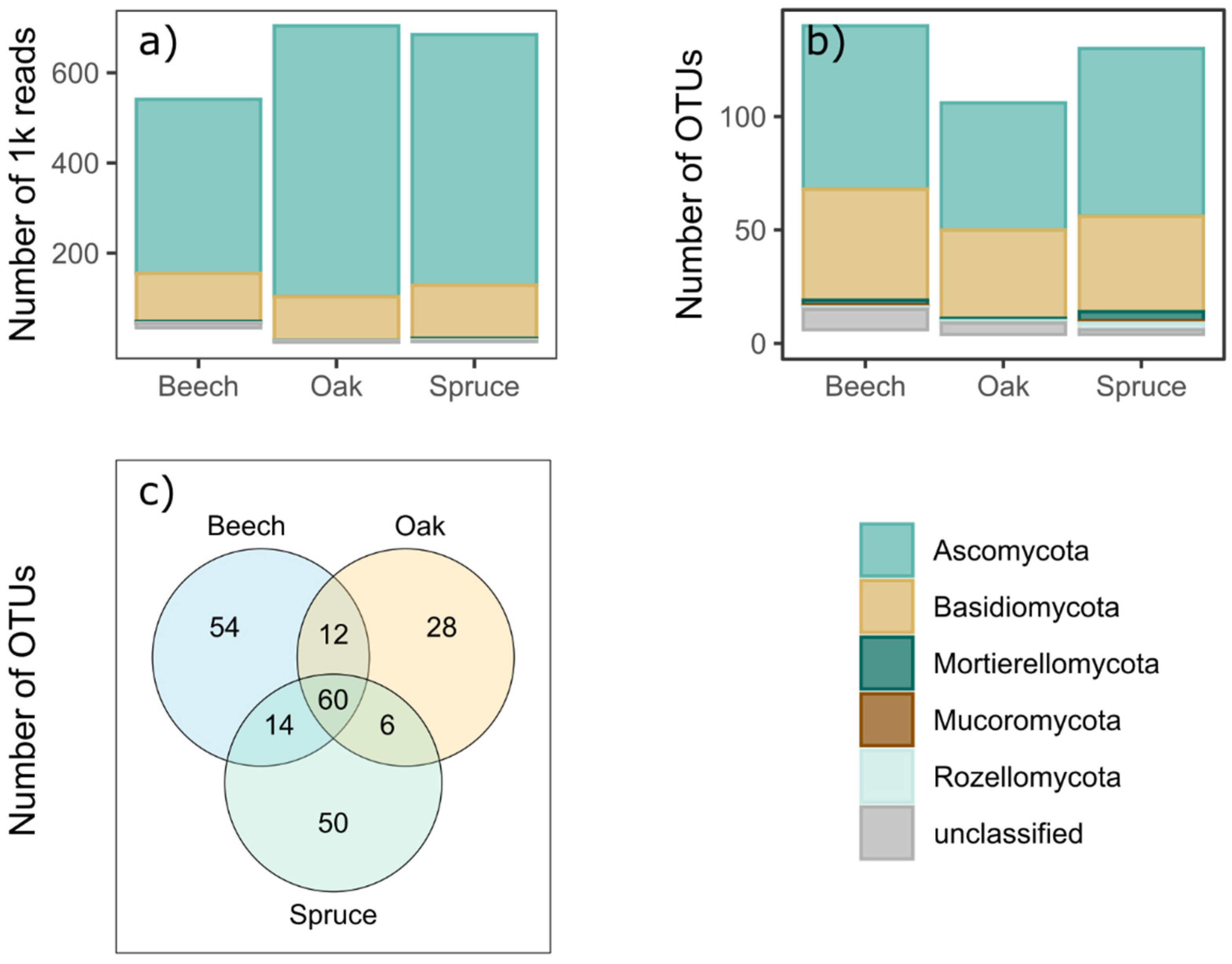
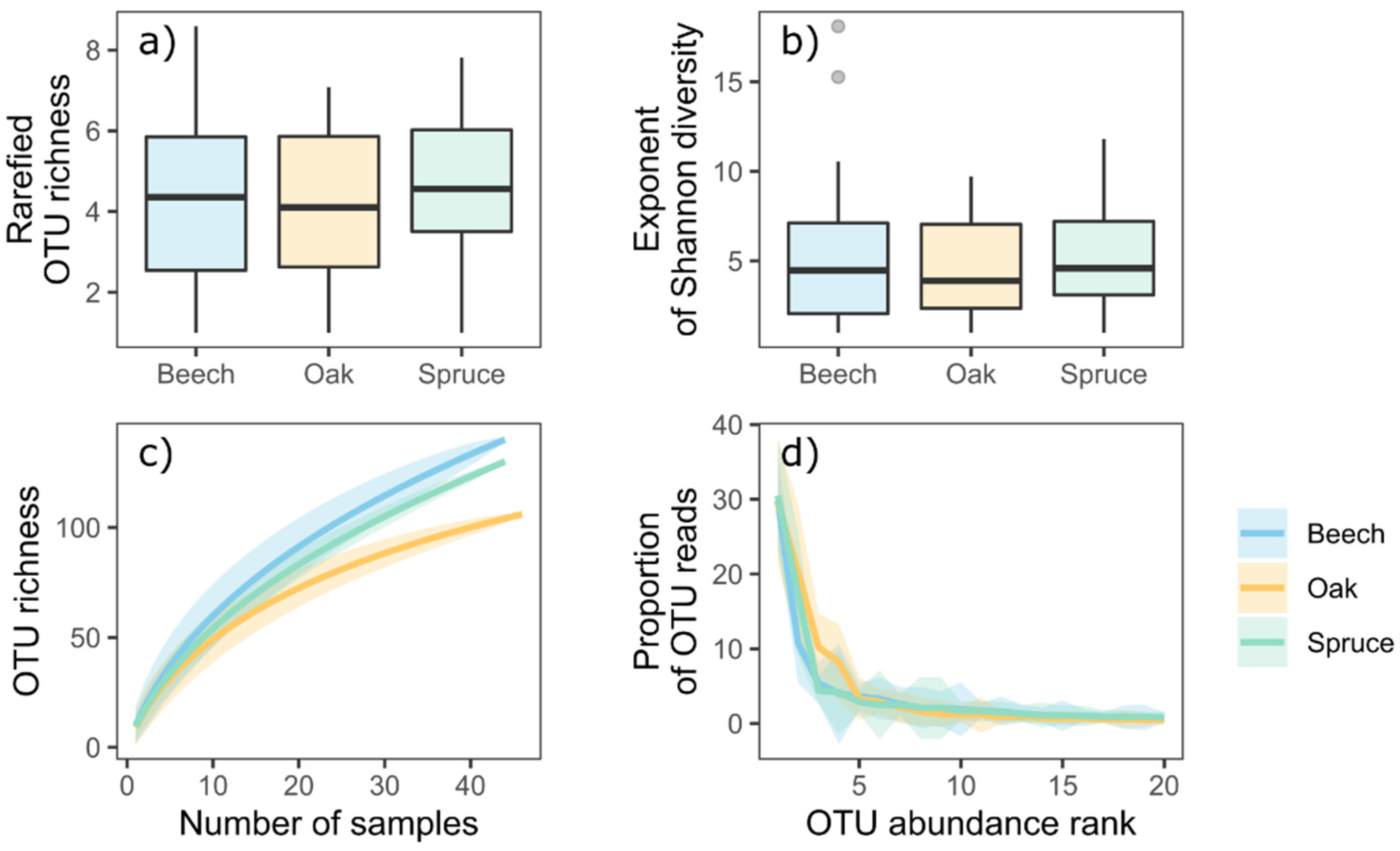
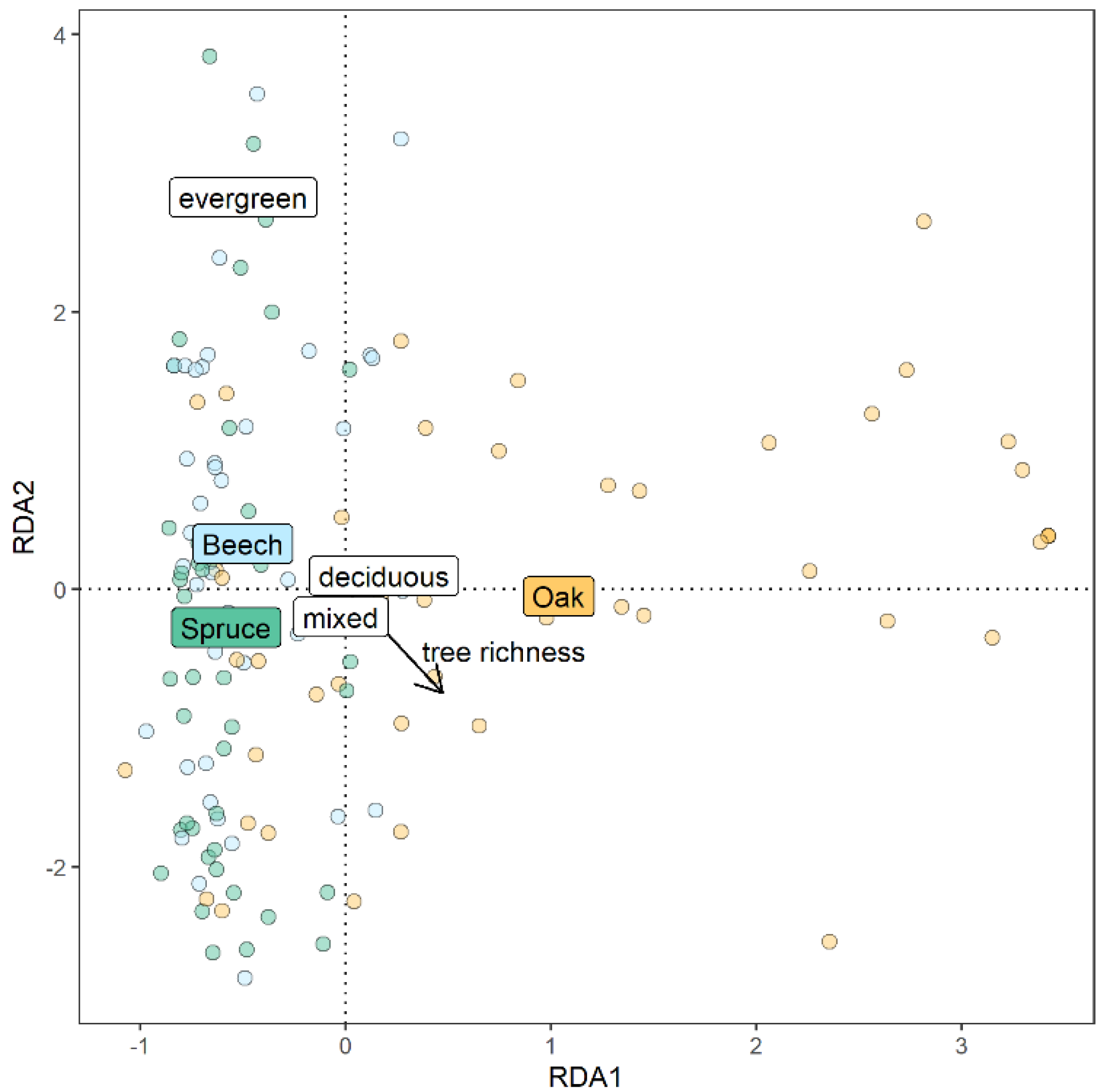
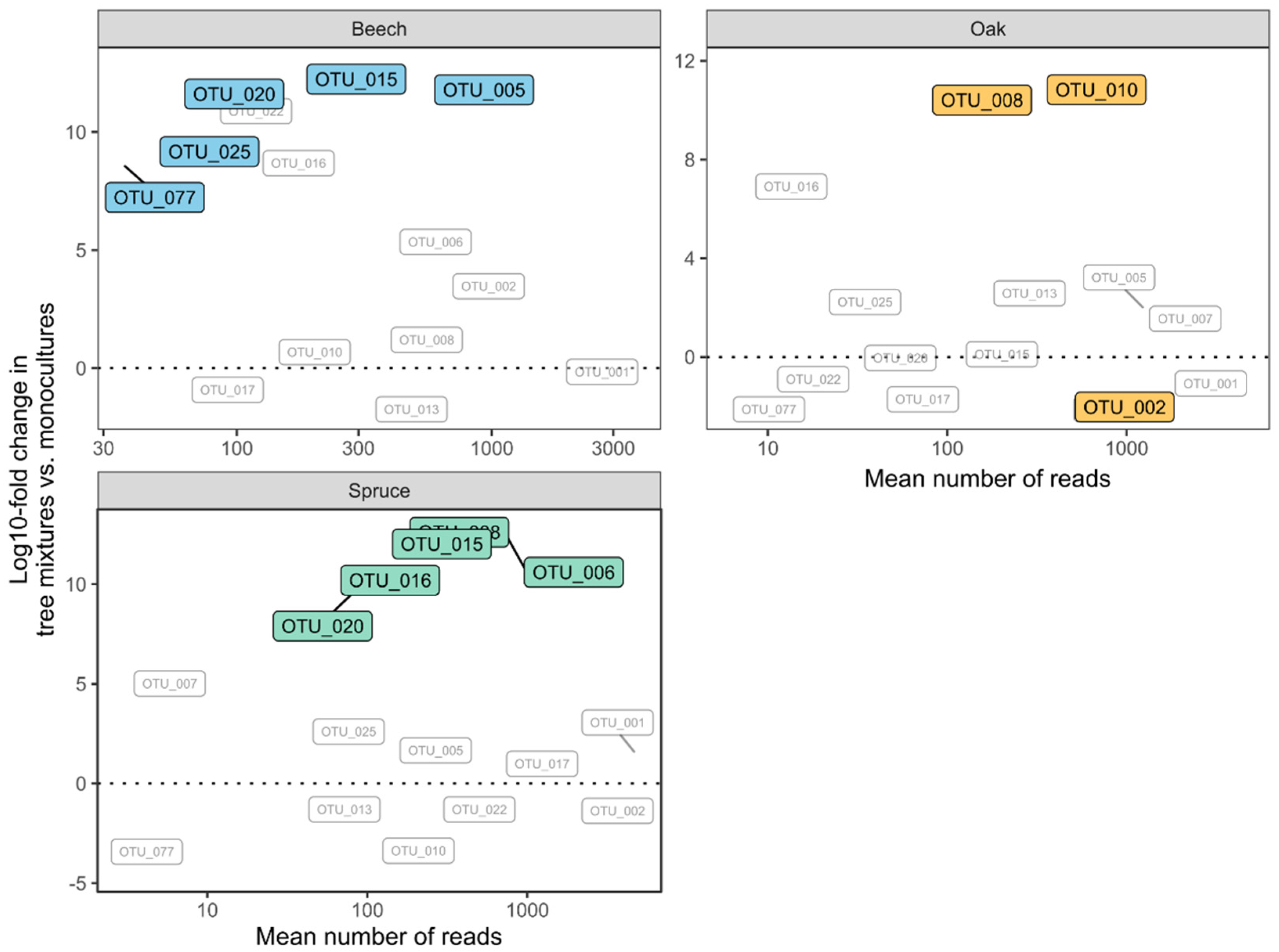
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Brockerhoff, E.G.; Barbaro, L.; Castagneyrol, B.; Forrester, D.I.; Gardiner, B.; González-Olabarria, J.R.; Lyver, P.O.; Meurisse, N.; Oxbrough, A.; Taki, H.; et al. Forest biodiversity, ecosystem functioning and the provision of ecosystem services. Biodivers. Conserv. 2017, 26, 3005–3035. [Google Scholar] [CrossRef] [Green Version]
- Wagg, C.; Schlaeppi, K.; Banerjee, S.; Kuramae, E.E.; van der Heijden, M.G.A. Fungal-bacterial diversity and microbiome complexity predict ecosystem functioning. Nat. Commun. 2019, 10, 4841. [Google Scholar] [CrossRef]
- Gehring, C.A.; Sthultz, C.M.; Flores-Rentería, L.; Whipple, A.V.; Whitham, T.G. Tree genetics defines fungal partner communities that may confer drought tolerance. Proc. Natl. Acad. Sci. USA 2017, 114, 11169–11174. [Google Scholar] [CrossRef] [Green Version]
- Kivlin, S.N.; Emery, S.M.; Rudgers, J.A. Fungal symbionts alter plant responses to global change. Am. J. Bot. 2013, 100, 1445–1457. [Google Scholar] [CrossRef] [PubMed]
- Terhonen, E.; Blumenstein, K.; Kovalchuk, A.; Asiegbu, F.O. Forest Tree Microbiomes and Associated Fungal Endophytes: Functional Roles and Impact on Forest Health. Forests 2019, 10, 42. [Google Scholar] [CrossRef] [Green Version]
- Griffin, E.A.; Carson, W.P. Tropical tree endophytes: Cryptic drivers of forest diversity, species composition, and ecosystem function. In Endophytes of Forest Trees: Biology and Applications, 2nd ed.; Pirttila, A.M., Frank, A.C., Eds.; Springer: Dordrecht, The Netherlands, 2018; pp. 63–103. [Google Scholar]
- Griffin, E.A.; Wright, S.J.; Morin, P.J.; Carson, W.P. Pervasive interactions between foliar microbes and soil nutrients mediate leaf production and herbivore damage in a tropical forest. New Phytol. 2017, 216, 99–112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qie, J.; Jinwang, Q.; Hongna, M.; Honggang, S.; Chu, W. Foliar endophytic fungi: Diversity in species and functions in forest ecosystems. Symbiosis 2020, 80, 103–132. [Google Scholar] [CrossRef]
- Ferus, P.; Barta, M.; Konôpková, J. Endophytic fungus Beauveria bassiana can enhance drought tolerance in red oak seedlings. Trees 2019, 33, 1179–1186. [Google Scholar] [CrossRef]
- Bittleston, L.S.; Brockmann, F.; Wcislo, W.; van Bael, S.A. Endophytic fungi reduce leaf-cutting ant damage to seedlings. Biol. Lett. 2011, 7, 30–32. [Google Scholar] [CrossRef]
- Hartley, S.E.; Gange, A.C. Impacts of plant symbiotic fungi on insect herbivores: Mutualism in a multitrophic context. Annu. Rev. Entomol. 2009, 54, 323–342. [Google Scholar] [CrossRef] [PubMed]
- Arnold, A.E.; Mejía, L.C.; Kyllo, D.; Rojas, E.I.; Maynard, Z.; Robbins, N.; Herre, E.A. Fungal endophytes limit pathogen damage in a tropical tree. Proc. Natl. Acad. Sci. USA 2003, 100, 15649–15654. [Google Scholar] [CrossRef] [Green Version]
- Bamisile, B.S.; Dash, C.K.; Akutse, K.S.; Keppanan, R.; Wang, L. Fungal Endophytes: Beyond Herbivore Management. Front. Microbiol. 2018, 9, 544. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guerreiro, M.A.; Brachmann, A.; Begerow, D.; Peršoh, D. Transient leaf endophytes are the most active fungi in 1-year-old beech leaf litter. Fungal Divers. 2018, 89, 237–251. [Google Scholar] [CrossRef] [Green Version]
- Laforest-Lapointe, I.; Paquette, A.; Messier, C.; Kembel, S.W. Leaf bacterial diversity mediates plant diversity and ecosystem function relationships. Nature 2017, 546, 145–147. [Google Scholar] [CrossRef] [PubMed]
- Mori, A.S.; Lertzman, K.P.; Gustafsson, L. Biodiversity and ecosystem services in forest ecosystems: A research agenda for applied forest ecology. J. Appl. Ecol. 2017, 54, 12–27. [Google Scholar] [CrossRef]
- Griffin, E.A.; Harrison, J.G.; McCormick, M.K.; Burghardt, K.T.; Parker, J.D. Tree Diversity Reduces Fungal Endophyte Richness and Diversity in a Large-Scale Temperate Forest Experiment. Diversity 2019, 11, 234. [Google Scholar] [CrossRef] [Green Version]
- Saadani, M.; Hönig, L.; Bien, S.; Koehler, M.; Rutten, G.; Wubet, T.; Braun, U.; Bruelheide, H. Local Tree Diversity Suppresses Foliar Fungal Infestation and Decreases Morphological But Not Molecular Richness in a Young Subtropical Forest. J. Fungi 2021, 7, 173. [Google Scholar] [CrossRef] [PubMed]
- Hantsch, L.; Bien, S.; Radatz, S.; Braun, U.; Auge, H.; Bruelheide, H. Tree diversity and the role of non-host neighbour tree species in reducing fungal pathogen infestation. J. Ecol. 2014, 102, 1673–1687. [Google Scholar] [CrossRef]
- Peršoh, D.; Melcher, M.; Flessa, F.; Rambold, G. First fungal community analyses of endophytic ascomycetes associated with Viscum album ssp. austriacum and its host Pinus sylvestris. Fungal Biol. 2010, 114, 585–596. [Google Scholar] [CrossRef] [PubMed]
- Schoch, C.L.; Seifert, K.A.; Huhndorf, S.; Robert, V.; Spouge, J.L.; Levesque, C.A.; Chen, W. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc. Natl. Acad. Sci. USA 2012, 109, 6241–6246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stielow, J.B.; Lévesque, C.A.; Seifert, K.A.; Meyer, W.; Iriny, L.; Smits, D.; Renfurm, R.; Verkley, G.J.M.; Groenewald, M.; Chaduli, D.; et al. One fungus, which genes? Development and assessment of universal primers for potential secondary fungal DNA barcodes. Persoonia 2015, 35, 242–263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gossner, M.M.; Beenken, L.; Arend, K.; Begerow, D.; Peršoh, D. Insect herbivory facilitates the establishment of an invasive plant pathogen. ISME Commun. 2021, 1, 1–8. [Google Scholar] [CrossRef]
- Gardes, M.; Bruns, T.D. ITS primers with enhanced specificity for basidiomycetes—application to the identification of mycorrhizae and rusts. Mol. Ecol. 1993, 2, 113–118. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA Genes for phylogenetics. In PCR—Protocols and Applications—A Laboratory Manual; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press, Inc.: San Diego, CA, USA, 1990; ISBN 0-12-372180-6. [Google Scholar]
- Röhl, O.; Peršoh, D.; Mittelbach, M.; Elbrecht, V.; Brachmann, A.; Nuy, J.; Boenigk, J.; Leese, F.; Begerow, D. Distinct sensitivity of fungal freshwater guilds to water quality. Mycol. Prog. 2017, 16, 155–169. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, W.; Fu, L.; Niu, B.; Wu, S.; Wooley, J. Ultrafast clustering algorithms for metagenomic sequence analysis. Brief. Bioinform. 2012, 13, 656–668. [Google Scholar] [CrossRef]
- Kõljalg, U.; Nilsson, R.H.; Abarenkov, K.; Tedersoo, L.; Taylor, A.F.S.; Bahram, M.; Bates, S.T.; Bruns, T.D.; Bengtsson-Palme, J.; Callaghan, T.M.; et al. Towards a unified paradigm for sequence-based identification of fungi. Mol. Ecol. 2013, 22, 5271–5277. [Google Scholar] [CrossRef] [Green Version]
- Legendre, P.; Gallagher, E.D. Ecologically meaningful transformations for ordination of species data. Oecologia 2001, 129, 271–280. [Google Scholar] [CrossRef]
- Legendre, P.; Oksanen, J.; ter Braak, C.J.F. Testing the significance of canonical axes in redundancy analysis. Methods Ecol. Evol. 2011, 2, 269–277. [Google Scholar] [CrossRef]
- Sørensen, T. A method of establishing groups of equal amplitude in plant sociology based on similarity of species and its application to analyses of the vegetation on Danish commons. K. Dan. Vidensk. Selsk. Biol. Skr. 1948, 5, 1–34. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing; 2020. Version 4.0.5. Available online: https://www.R-project.org(accessed on 7 October 2021).
- Kindt, R.; Coe, R. Tree Diversity Analysis. A Manual and Software for Common Statistical Methods for Ecological and Biodiversity Studies; World Agroforestry Centre (ICRAF): Nairobi, Kenya, 2005. [Google Scholar]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Springer-Verlag New York: New York, NY, USA, 2009; ISBN 1282509918. [Google Scholar]
- Jari Oksanen, F.; Blanchet, G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Vegan: Community Ecology Package; version 2.5-7, 2019.
- Bahnmann, B.; Mašínová, T.; Halvorsen, R.; Davey, M.L.; Sedlák, P.; Tomšovský, M.; Baldrian, P. Effects of oak, beech and spruce on the distribution and community structure of fungi in litter and soils across a temperate forest. Soil Biol. Biochem. 2018, 119, 162–173. [Google Scholar] [CrossRef]
- Elizabeth Arnold, A.; Lutzoni, F. Diversity and host range of foliar fungal endophytes: Are tropical leaves biodiversity hotspots? Ecology 2007, 88, 541–549. [Google Scholar] [CrossRef]
- Suryanarayanan, T.S.; Devarajan, P.T.; Girivasan, K.P.; Govindarajulu, M.B.; Kumaresan, V.; Murali, T.S.; Rajamani, T.; Thirunavukkarasu, N.; Venkatesan, G. The Host Range of Multi-Host Endophytic Fungi. Curr. Sci. 2018, 115, 1963. [Google Scholar] [CrossRef]
- The Fungal Community: Its Organization and Role in the Ecosystem: Foliar Endophyte Communities and Leaf Traits in Tropical Trees, 4th ed.; Dighton, J.; White, J.F. (Eds.) CRC Press: Boca Raton, FL, USA; Taylor & Francis: Boca Raton, FL, USA, 2017. [Google Scholar]
- Rajala, T.; Velmala, S.M.; Tuomivirta, T.; Haapanen, M.; Müller, M.; Pennanen, T. Endophyte communities vary in the needles of Norway spruce clones. Fungal Biol. 2013, 117, 182–190. [Google Scholar] [CrossRef]
- Albrectsen, B.R.; Siddique, A.B.; Decker, V.H.G.; Unterseher, M.; Robinson, K.M. Both plant genotype and herbivory shape aspen endophyte communities. Oecologia 2018, 187, 535–545. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nguyen, D.; Boberg, J.; Ihrmark, K.; Stenström, E.; Stenlid, J. Do foliar fungal communities of Norway spruce shift along a tree species diversity gradient in mature European forests? Fungal Ecol. 2016, 23, 97–108. [Google Scholar] [CrossRef] [Green Version]
- Harrison, J.G.; Griffin, E.A. The diversity and distribution of endophytes across biomes, plant phylogeny and host tissues: How far have we come and where do we go from here? Environ. Microbiol. 2020, 22, 2107–2123. [Google Scholar] [CrossRef] [Green Version]
- Vincent, J.B.; Weiblen, G.D.; May, G. Host associations and beta diversity of fungal endophyte communities in New Guinea rainforest trees. Mol. Ecol. 2016, 25, 825–841. [Google Scholar] [CrossRef]
- Oliva, J.; Ridley, M.; Redondo, M.A.; Caballol, M. Competitive exclusion amongst endophytes determines shoot blight severity on pine. Funct. Ecol. 2020, 35, 239–254. [Google Scholar] [CrossRef]
- Newcombe, G. Endophytes in Forest Management: Four Challenges. In Endophytes of Forest Trees: Biology and Applications; Pirttilä, A.M., Frank, A.C., Eds.; Springer: Dordrecht, The Netherlands, 2011; pp. 251–262. ISBN 978-94-007-1599-8. [Google Scholar]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kambach, S.; Sadlowski, C.; Peršoh, D.; Guerreiro, M.A.; Auge, H.; Röhl, O.; Bruelheide, H. Foliar Fungal Endophytes in a Tree Diversity Experiment Are Driven by the Identity but Not the Diversity of Tree Species. Life 2021, 11, 1081. https://doi.org/10.3390/life11101081
Kambach S, Sadlowski C, Peršoh D, Guerreiro MA, Auge H, Röhl O, Bruelheide H. Foliar Fungal Endophytes in a Tree Diversity Experiment Are Driven by the Identity but Not the Diversity of Tree Species. Life. 2021; 11(10):1081. https://doi.org/10.3390/life11101081
Chicago/Turabian StyleKambach, Stephan, Christopher Sadlowski, Derek Peršoh, Marco Alexandre Guerreiro, Harald Auge, Oliver Röhl, and Helge Bruelheide. 2021. "Foliar Fungal Endophytes in a Tree Diversity Experiment Are Driven by the Identity but Not the Diversity of Tree Species" Life 11, no. 10: 1081. https://doi.org/10.3390/life11101081
APA StyleKambach, S., Sadlowski, C., Peršoh, D., Guerreiro, M. A., Auge, H., Röhl, O., & Bruelheide, H. (2021). Foliar Fungal Endophytes in a Tree Diversity Experiment Are Driven by the Identity but Not the Diversity of Tree Species. Life, 11(10), 1081. https://doi.org/10.3390/life11101081





