The Structure and Functions of PRMT5 in Human Diseases
Abstract
:1. Introduction
2. Structure and General Function of PRMT5
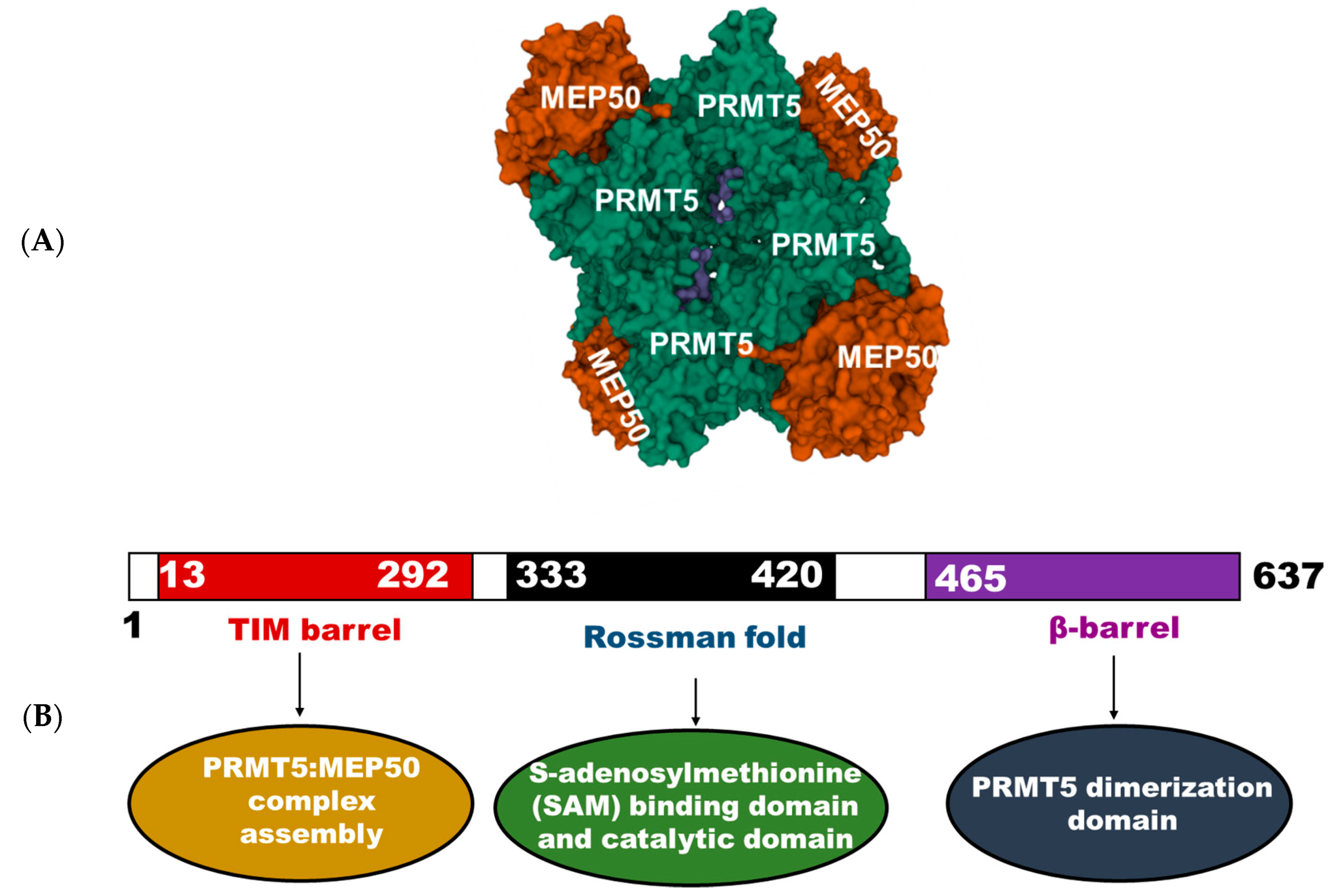
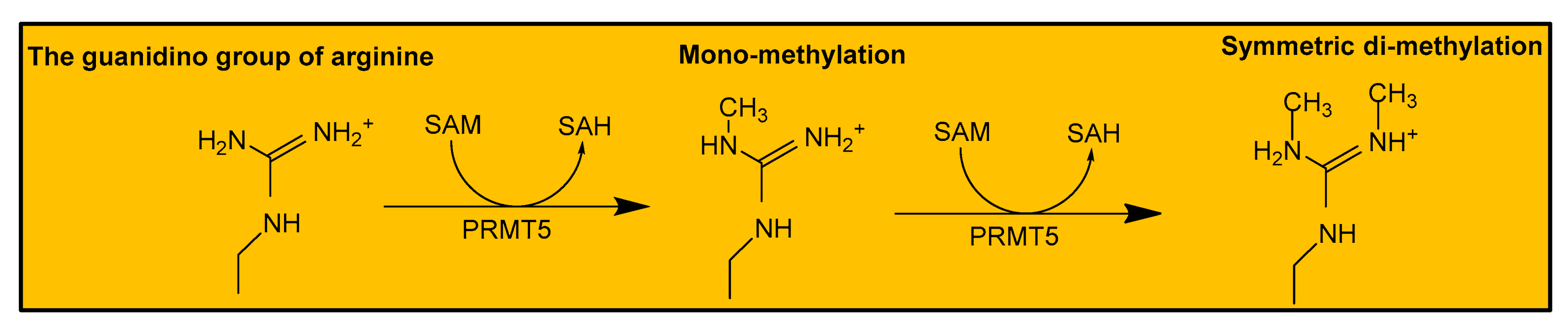
2.1. Human PRMT5 Structure and Mechanism of Action
2.2. Cellular Function of PRMT5
3. Regulation of PRMT5
3.1. Regulation by PTMs
3.2. Regulation by miRNA
3.3. Regulation by Interacting Proteins
4. Role of PRMT5 in Human Diseases
4.1. PRMT5 in Cancer
4.2. PRMT5 in Diabetes
4.3. PRMT5 in Cardiovascular Diseases
4.4. PRMT5 in Neurodegenerative Diseases
5. Targeting PRMT5 in Human Diseases
6. Perspective and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yang, Y.; Bedford, M.T. Protein arginine methyltransferases and cancer. Nat. Rev. Cancer 2012, 13, 37–50. [Google Scholar] [CrossRef]
- Morettin, A.; Baldwin, R.M.; Côté, J. Arginine methyltransferases as novel therapeutic targets for breast cancer. Mutagenesis 2015, 30, 177–189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bedford, M.T. Arginine methylation at a glance. J. Cell Sci. 2007, 120, 4243–4246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stopa, N.; Krebs, J.E.; Shechter, D. The PRMT5 arginine methyltransferase: Many roles in development, cancer and beyond. Cell. Mol. Life Sci. 2015, 72, 2041–2059. [Google Scholar] [CrossRef] [PubMed]
- Niewmierzycka, A.; Clarke, S. S-adenosylmethionine-dependent methylation in Saccharomyces cerevisiae. J. Biol. Chem. 1999, 274, 814–824. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Motolani, A.; Martin, M.; Sun, M.; Lu, T. Phosphorylation of the regulators, a complex facet of NF-κB signaling in cancer. Biomolecules 2020, 11, 15. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-H.; Cook, J.R.; Yang, Z.-H.; Mirochnitchenko, O.; Gunderson, S.I.; Felix, A.M.; Herth, N.; Hoffmann, R.; Pestka, S. PRMT7, a new protein arginine methyltransferase that synthesizes symmetric dimethylarginine. J. Biol. Chem. 2005, 280, 3656–3664. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zurita-Lopez, C.I.; Sandberg, T.; Kelly, R.; Clarke, S.G. Human protein arginine methyltransferase 7 (PRMT7) is a type III enzyme forming omega-NG-monomethylated arginine residues. J. Biol. Chem. 2012, 287, 7859–7870. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheung, N.; Chan, L.C.; Thompson, A.; Cleary, M.L.; So, C.W.E. Protein arginine-methyltransferase-dependent oncogenesis. Nat. Cell Biol. 2007, 9, 1208–1215. [Google Scholar] [CrossRef]
- Kim, J.H.; Yoo, B.C.; Yang, W.S.; Kim, E.; Hong, S.; Cho, J.Y. The role of protein arginine methyltransferases in inflammatory responses. Mediat. Inflamm. 2016, 2016, 4028353. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Le Romancer, M.; Treilleux, I.; Bouchekioua-Bouzaghou, K.; Sentis, S.; Corbo, L. Methylation, a key step for nongenomic estrogen signaling in breast tumors. Steroids 2010, 75, 560–564. [Google Scholar] [CrossRef]
- Mitchell, T.R.H.; Glenfield, K.; Jeyanthan, K.; Zhu, X.-D. Arginine methylation regulates telomere length and stability. Mol. Cell. Biol. 2009, 29, 4918–4934. [Google Scholar] [CrossRef] [Green Version]
- Blanc, R.S.; Richard, S. Arginine methylation: The coming of age. Mol. Cell 2017, 65, 8–24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, H.; Mundade, R.; Lange, K.C.; Lu, T. Protein arginine methylation of non-histone proteins and its role in diseases. Cell Cycle 2014, 13, 32–41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, D.; Yadav, N.; King, R.W.; Swanson, M.S.; Weinstein, E.J.; Bedford, M.T. Small molecule regulators of protein arginine methyltransferases. J. Biol. Chem. 2004, 279, 23892–23899. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Lianjun, Z.; Liu, C.; Han, F.; Chen, M.; Zhang, L.; Cui, X.; Qin, Y.; Bao, S.; Gao, F. Protein arginine methyltransferase 5 (Prmt5) is required for germ cell survival during mouse embryonic development. Biol. Reprod. 2015, 92, 104. [Google Scholar] [CrossRef]
- Prabhu, L.; Wei, H.; Chen, L.; Demir, Ö.; Sandusky, G.; Sun, E.; Wang, J.; Mo, J.; Zeng, L.; Fishel, M.; et al. Adapting AlphaLISA high throughput screen to discover a novel small-molecule inhibitor targeting protein arginine methyltransferase 5 in pancreatic and colorectal cancers. Oncotarget 2017, 8, 39963–39977. [Google Scholar] [CrossRef] [Green Version]
- Pollack, B.P.; Kotenko, S.V.; He, W.; Izotova, L.S.; Barnoski, B.L.; Pestka, S. The human homologue of the yeast proteins Skb1 and Hsl7p interacts with Jak kinases and contains protein methyltransferase activity. J. Biol. Chem. 1999, 274, 31531–31542. [Google Scholar] [CrossRef] [Green Version]
- Antonysamy, S.; Bonday, Z.; Campbell, R.M.; Doyle, B.; Druzina, Z.; Gheyi, T.; Han, B.; Jungheim, L.N.; Qian, Y.; Rauch, C.; et al. Crystal structure of the human PRMT5: MEP50 complex. Proc. Natl. Acad. Sci. USA 2012, 109, 17960–17965. [Google Scholar] [CrossRef] [Green Version]
- Sun, L.; Wang, M.; Lv, Z.; Yang, N.; Liu, Y.; Bao, S.; Gong, W.; Xu, R.-M. Structural insights into protein arginine symmetric dimethylation by PRMT5. Proc. Natl. Acad. Sci. USA 2011, 108, 20538–20543. [Google Scholar] [CrossRef] [Green Version]
- Eddershaw, A.R.; Stubbs, C.J.; Edwardes, L.V.; Underwood, E.; Hamm, G.R.; Davey, P.R.J.; Clarkson, P.N.; Syson, K. Characterization of the kinetic mechanism of human protein arginine methyltransferase 5. Biochemistry 2020, 59, 4775–4786. [Google Scholar] [CrossRef]
- Musiani, D.; Bok, J.; Massignani, E.; Wu, L.; Tabaglio, T.; Ippolito, M.R.; Cuomo, A.; Ozbek, U.; Zorgati, H.; Ghoshdastider, U.; et al. Proteomics profiling of arginine methylation defines PRMT5 substrate specificity. Sci. Signal. 2019, 12, eaat8388. [Google Scholar] [CrossRef]
- Wang, M.; Xu, R.-M.; Thompson, P.R. Substrate specificity, processivity, and kinetic mechanism of protein arginine methyltransferase 5. Biochemistry 2013, 52, 5430–5440. [Google Scholar] [CrossRef]
- Koh, C.M.; Bezzi, M.; Guccione, E. The where and the how of PRMT5. Curr. Mol. Biol. Rep. 2015, 1, 19–28. [Google Scholar] [CrossRef]
- Tee, W.-W.; Pardo, M.; Theunissen, T.; Yu, L.; Choudhary, J.; Hajkova, P.; Surani, M.A. Prmt5 is essential for early mouse development and acts in the cytoplasm to maintain ES cell pluripotency. Genes Dev. 2010, 24, 2772–2777. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berrens, R.V.; Reik, W. Prmt5: A guardian of the germline protects future generations. EMBO J. 2015, 34, 689–690. [Google Scholar] [CrossRef] [PubMed]
- Litzler, L.; Zahn, A.; Meli, A.; Hébert, S.; Patenaude, A.-M.; Methot, S.P.; Sprumont, A.; Bois, T.; Kitamura, D.; Costantino, S.; et al. PRMT5 is essential for B cell development and germinal center dynamics. Nat. Commun. 2019, 10, 22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tanaka, H.; Fujita, N.; Tsuruo, T. 3-phosphoinositide-dependent protein kinase-1-mediated IκB kinase β (IKKB) phosphorylation activates NF-κB signaling. J. Biol. Chem. 2005, 280, 40965–40973. [Google Scholar] [CrossRef] [Green Version]
- Scaglione, A.; Patzig, J.; Liang, J.; Frawley, R.; Bok, J.; Mela, A.; Yattah, C.; Zhang, J.; Teo, S.X.; Zhou, T.; et al. PRMT5-mediated regulation of developmental myelination. Nat. Commun. 2018, 9, 2840. [Google Scholar] [CrossRef] [PubMed]
- Tan, D.Q.; Li, Y.; Yang, C.; Li, J.; Tan, S.H.; Chin, D.W.; Nakamura-Ishizu, A.; Yang, H.; Suda, T. Abstract 971: PRMT5 modulates splicing for genome integrity and preserves proteostasis of hematopoietic stem cells. Tumor Biol. 2019, 26, 2316–2328. [Google Scholar] [CrossRef]
- Wei, H.; Wang, B.; Miyagi, M.; She, Y.; Gopalan, B.; Huang, D.-B.; Ghosh, G.; Stark, G.R.; Lu, T. PRMT5 dimethylates R30 of the p65 subunit to activate NF-κB. Proc. Natl. Acad. Sci. USA 2013, 110, 13516–13521. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hartley, A.-V.; Lu, T. Modulating the modulators: Regulation of protein arginine methyltransferases by post-translational modifications. Drug Discov. Today 2020, 25, 1735–1743. [Google Scholar] [CrossRef]
- Motolani, A.A.; Sun, M.; Martin, M.; Sun, S.; Lu, T. Discovery of small molecule inhibitors for histone methyltransferases in cancer. In Translational Research in Cancer; BoD—Books on Demand: Norderstedt, Germany, 2021. [Google Scholar] [CrossRef]
- Nie, M.; Wang, Y.; Guo, C.; Li, X.; Wang, Y.; Deng, Y.; Yao, B.; Gui, T.; Ma, C.; Liu, M.; et al. CARM1-mediated methylation of protein arginine methyltransferase 5 represses human γ-globin gene expression in erythroleukemia cells. J. Biol. Chem. 2018, 293, 17454–17463. [Google Scholar] [CrossRef] [Green Version]
- Espejo, A.B.; Gao, G.; Black, K.; Gayatri, S.; Veland, N.; Kim, J.; Chen, T.; Sudol, M.; Walker, C.; Bedford, M.T. PRMT5 C-terminal phosphorylation modulates a 14-3-3/PDZ interaction switch. J. Biol. Chem. 2017, 292, 2255–2265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hartley, A.-V.; Wang, B.; Jiang, G.; Wei, H.; Sun, M.; Prabhu, L.; Martin, M.; Safa, A.; Sun, S.; Liu, Y.; et al. Regulation of a PRMT5/NF-κB axis by phosphorylation of PRMT5 at serine 15 in colorectal cancer. Int. J. Mol. Sci. 2020, 21, 3684. [Google Scholar] [CrossRef]
- Lattouf, H.; Poulard, C.; le Romancer, M. PRMT5 prognostic value in cancer. Oncotarget 2019, 10, 3151–3153. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.-T.; Zeng, L.-F.; He, Q.-Y.; Tao, W.A.; Zha, Z.-G.; Hu, C.-D. The E3 ubiquitin ligase CHIP mediates ubiquitination and proteasomal degradation of PRMT5. Biochim. Biophys. Acta BBA Bioenerg. 2016, 1863, 335–346. [Google Scholar] [CrossRef]
- Jin, J.; Martin, M.; Hartley, A.-V.; Lu, T. PRMTs and miRNAs: Functional cooperation in cancer and beyond. Cell Cycle 2019, 18, 1676–1686. [Google Scholar] [CrossRef] [PubMed]
- Pal, S.; Baiocchi, R.A.; Byrd, J.C.; Grever, M.R.; Jacob, S.T.; Sif, S. Low levels of miR-92b/96 induce PRMT5 translation and H3R8/H4R3 methylation in mantle cell lymphoma. EMBO J. 2007, 26, 3558–3569. [Google Scholar] [CrossRef]
- Wang, L.; Pal, S.; Sif, S. Protein arginine methyltransferase 5 suppresses the transcription of the RB family of tumor suppressors in leukemia and lymphoma cells. Mol. Cell. Biol. 2008, 28, 6262–6277. [Google Scholar] [CrossRef] [Green Version]
- Lacroix, M.; El Messaoudi, S.; Rodier, G.; le Cam, A.; Sardet, C.; Fabbrizio, E. The histone-binding protein COPR5 is required for nuclear functions of the protein arginine methyltransferase PRMT5. EMBO Rep. 2008, 9, 452–458. [Google Scholar] [CrossRef] [Green Version]
- Pal, S.; Vishwanath, S.N.; Erdjument-Bromage, H.; Tempst, P.; Sif, S. Human SWI/SNF-associated PRMT5 methylates histone H3 arginine 8 and negatively regulates expression of ST7 and NM23 tumor suppressor genes. Mol. Cell. Biol. 2004, 24, 9630–9645. [Google Scholar] [CrossRef] [Green Version]
- Le Guezennec, X.; Vermeulen, M.; Brinkman, A.B.; Hoeijmakers, W.A.M.; Cohen, A.; Lasonder, E.; Stunnenberg, H.G. MBD2/NuRD and MBD3/NuRD, two distinct complexes with different biochemical and functional properties. Mol. Cell. Biol. 2006, 26, 843–851. [Google Scholar] [CrossRef] [Green Version]
- Matkar, S.; Thiel, A.; Hua, X. Menin: A scaffold protein that controls gene expression and cell signaling. Trends Biochem. Sci. 2013, 38, 394–402. [Google Scholar] [CrossRef] [Green Version]
- Ancelin, K.; Lange, U.C.; Hajkova, P.; Schneider, R.J.; Bannister, A.; Kouzarides, T.; Surani, A. Blimp1 associates with Prmt5 and directs histone arginine methylation in mouse germ cells. Nat. Cell Biol. 2006, 8, 623–630. [Google Scholar] [CrossRef] [PubMed]
- Tabata, T.; Kokura, K.; Dijke, P.T.; Ishii, S. Ski co-repressor complexes maintain the basal repressed state of the TGF-β target gene, SMAD7, via HDAC3 and PRMT5. Genes Cells 2008, 14, 17–28. [Google Scholar] [CrossRef] [PubMed]
- Owens, J.L.; Beketova, E.; Liu, S.; Tinsley, S.L.; Asberry, A.M.; Deng, X.; Huang, J.; Li, C.; Wan, J.; Hu, C.-D. PRMT5 cooperates with pICln to function as a master epigenetic activator of DNA double-strand break repair genes. iScience 2020, 23, 100750. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Friesen, W.J.; Paushkin, S.; Wyce, A.; Massenet, S.; Pesiridis, G.S.; van Duyne, G.; Rappsilber, J.; Mann, M.; Dreyfuss, G. The methylosome, a 20S complex containing JBP1 and pICln, produces dimethylarginine-modified Sm proteins. Mol. Cell. Biol. 2001, 21, 8289–8300. [Google Scholar] [CrossRef] [Green Version]
- Guderian, G.; Peter, C.; Wiesner, J.; Sickmann, A.; Schulze-Osthoff, K.; Fischer, U.; Grimmler, M. RioK1, a new interactor of protein arginine methyltransferase 5 (PRMT5), competes with pICln for binding and modulates PRMT5 complex composition and substrate specificity. J. Biol. Chem. 2011, 286, 1976–1986. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, H.; Ronai, Z.A. PRMT5 function and targeting in cancer. Cell Stress 2020, 4, 199–215. [Google Scholar] [CrossRef]
- Chung, J.; Karkhanis, V.; Baiocchi, R.A.; Sif, S. Protein arginine methyltransferase 5 (PRMT5) promotes survival of lymphoma cells via activation of WNT/β-catenin and AKT/GSK3β proliferative signaling. J. Biol. Chem. 2019, 294, 7692–7710. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Chitnis, N.; Nakagawa, H.; Kita, Y.; Natsugoe, S.; Yang, Y.; Li, Z.; Wasik, M.; Klein-Szanto, A.J.P.; Rustgi, A.K.; et al. PRMT5 is required for lymphomagenesis triggered by multiple oncogenic drivers. Cancer Discov. 2015, 5, 288–303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gullà, A.; Hideshima, T.; Bianchi, G.; Fulciniti, M.; Samur, M.K.; Qi, J.; Tai, Y.-T.; Harada, T.; Morelli, E.; Amodio, N.; et al. Protein arginine methyltransferase 5 has prognostic relevance and is a druggable target in multiple myeloma. Leukemia 2018, 32, 996–1002. [Google Scholar] [CrossRef] [PubMed]
- Shailesh, H.; Zakaria, Z.Z.; Baiocchi, R.; Sif, S. Protein arginine methyltransferase 5 (PRMT5) dysregulation in cancer. Oncotarget 2018, 9, 36705–36718. [Google Scholar] [CrossRef] [Green Version]
- Shailesh, H.; Siveen, K.S.; Sif, S. Protein arginine methyltransferase 5 (PRMT5) activates WNT/β-catenin signalling in breast cancer cells via epigenetic silencing of DKK1 and DKK3. J. Cell. Mol. Med. 2021, 25, 1583–1600. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Zhu, Y.; Zhou, Z.; Xu, J.; Jin, S.; Xu, K.; Zhang, H.; Sun, Q.; Wang, J.; Xu, J. PRMT5 promotes cell proliferation by inhibiting BTG2 expression via the ERK signaling pathway in hepatocellular carcinoma. Cancer Med. 2018, 7, 869–882. [Google Scholar] [CrossRef]
- Sheng, X.; Wang, Z. Protein arginine methyltransferase 5 regulates multiple signaling pathways to promote lung cancer cell proliferation. BMC Cancer 2016, 16, 567. [Google Scholar] [CrossRef] [Green Version]
- Jing, P.; Zhao, N.; Ye, M.; Zhang, Y.; Zhang, Z.; Sun, J.; Wang, Z.; Zhang, J.; Gu, Z. Protein arginine methyltransferase 5 promotes lung cancer metastasis via the epigenetic regulation of miR-99 family/FGFR3 signaling. Cancer Lett. 2018, 427, 38–48. [Google Scholar] [CrossRef]
- Hartley, A.-V.; Wang, B.; Mundade, R.; Jiang, G.; Sun, M.; Wei, H.; Sun, S.; Liu, Y.; Lu, T. PRMT5-mediated methylation of YBX1 regulates NF-κB activity in colorectal cancer. Sci. Rep. 2020, 10, 15934. [Google Scholar] [CrossRef]
- Bao, X.; Zhao, S.; Liu, T.; Liu, Y.; Liu, Y.; Yang, X. Overexpression of PRMT5 promotes tumor cell growth and is associated with poor disease prognosis in epithelial ovarian cancer. J. Histochem. Cytochem. 2013, 61, 206–217. [Google Scholar] [CrossRef] [Green Version]
- Lattouf, H.; Kassem, L.; Jacquemetton, J.; Choucair, A.; Poulard, C.; Tredan, O.; Corbo, L.; Diab-Assaf, M.; Hussein, N.; Treilleux, I.; et al. LKB1 regulates PRMT5 activity in breast cancer. Int. J. Cancer 2019, 144, 595–606. [Google Scholar] [CrossRef] [Green Version]
- Shimizu, D.; Kanda, M.; Sugimoto, H.; Shibata, M.; Tanaka, H.; Takami, H.; Iwata, N.; Hayashi, M.; Tanaka, C.; Kobayashi, D.; et al. The protein arginine methyltransferase 5 promotes malignant phenotype of hepatocellular carcinoma cells and is associated with adverse patient outcomes after curative hepatectomy. Int. J. Oncol. 2017, 50, 381–386. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pak, M.G.; Lee, H.W.; Roh, M.S. High nuclear expression of protein arginine methyltransferase-5 is a potentially useful marker to estimate submucosal invasion in endoscopically resected early colorectal carcinoma. Pathol. Int. 2015, 65, 541–548. [Google Scholar] [CrossRef]
- Chatterjee, S.; Khunti, K.; Davies, M.J. Type 2 diabetes. Lancet 2017, 389, 2239–2251. [Google Scholar] [CrossRef]
- Galicia-Garcia, U.; Benito-Vicente, A.; Jebari, S.; Larrea-Sebal, A.; Siddiqi, H.; Uribe, K.B.; Ostolaza, H.; Martín, C. Pathophysiology of type 2 diabetes mellitus. Int. J. Mol. Sci. 2020, 21, 6275. [Google Scholar] [CrossRef]
- Jia, Z.; Yue, F.; Chen, X.; Narayanan, N.; Qiu, J.; Syed, S.A.; Imbalzano, A.N.; Deng, M.; Yu, P.; Hu, C.; et al. Protein arginine methyltransferase PRMT5 regulates fatty acid metabolism and lipid droplet biogenesis in white adipose tissues. Adv. Sci. 2020, 7, 2002602. [Google Scholar] [CrossRef]
- Leblanc, S.E.; Konda, S.; Wu, Q.; Hu, Y.-J.; Oslowski, C.M.; Sif, S.; Imbalzano, A.N. Protein arginine methyltransferase 5 (Prmt5) promotes gene expression of peroxisome proliferator-activated receptor γ2 (PPARγ2) and its target genes during adipogenesis. Mol. Endocrinol. 2012, 26, 583–597. [Google Scholar] [CrossRef] [PubMed]
- Muhammad, A.B.; Xing, B.; Liu, C.; Naji, A.; Ma, X.; Simmons, R.; Hua, X. Menin and PRMT5 suppress GLP1 receptor transcript and PKA-mediated phosphorylation of FOXO1 and CREB. Am. J. Physiol. Metab. 2017, 313, E148–E166. [Google Scholar] [CrossRef] [PubMed]
- Tsai, W.-W.; Niessen, S.; Goebel, N.; Yates, J.R.; Guccione, E.; Montminy, M. PRMT5 modulates the metabolic response to fasting signals. Proc. Natl. Acad. Sci. USA 2013, 110, 8870–8875. [Google Scholar] [CrossRef] [Green Version]
- Ma, J.; He, X.; Cao, Y.; O’Dwyer, K.; Szigety, K.M.; Wu, Y.; Gurung, B.; Feng, Z.; Katona, B.; Hua, X.; et al. Islet-specific Prmt5 excision leads to reduced insulin expression and glucose intolerance in mice. J. Endocrinol. 2020, 244, 41–52. [Google Scholar] [CrossRef]
- Nabel, E.G. Cardiovascular disease. N. Engl. J. Med. 2003, 349, 60–72. [Google Scholar] [CrossRef]
- Cai, S.; Liu, R.; Wang, P.; Li, J.; Xie, T.; Wang, M.; Cao, Y.; Li, Z.; Liu, P. PRMT5 prevents cardiomyocyte hypertrophy via symmetric dimethylating HoxA9 and repressing HoxA9 expression. Front. Pharmacol. 2020, 11, 2140. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Yi, B.; Sun, J. Inhibition of cardiomyocyte hypertrophy by protein arginine methyltransferase 5. J. Biol. Chem. 2014, 289, 24325–24335. [Google Scholar] [CrossRef] [Green Version]
- Tan, B.; Liu, Q.; Yang, L.; Yang, Y.; Liu, D.; Liu, L.; Meng, F. Low expression of PRMT5 in peripheral blood may serve as a potential independent risk factor in assessments of the risk of stable CAD and AMI. BMC Cardiovasc. Disord. 2019, 19, 31. [Google Scholar] [CrossRef] [PubMed]
- Harris, D.P.; Bandyopadhyay, S.; Maxwell, T.J.; Willard, B.; DiCorleto, P.E. Tumor necrosis factor (TNF)-α induction of CXCL10 in endothelial cells requires protein arginine methyltransferase 5 (PRMT5)-mediated nuclear factor (NF)-κB p65 methylation. J. Biol. Chem. 2014, 289, 15328–15339. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harris, D.P.; Chandrasekharan, U.M.; Bandyopadhyay, S.; Willard, B.; DiCorleto, P.E. PRMT5-mediated methylation of NF-κB p65 at Arg174 is required for endothelial CXCL11 gene induction in response to TNF-α and IFN-γ costimulation. PLoS ONE 2016, 11, e0148905. [Google Scholar] [CrossRef] [Green Version]
- Han, X.; Li, R.; Zhang, W.; Yang, X.; Wheeler, C.G.; Friedman, G.K.; Province, P.; Ding, Q.; You, Z.; Fathallah-Shaykh, H.M.; et al. Expression of PRMT5 correlates with malignant grade in gliomas and plays a pivotal role in tumor growth in vitro. J. Neuro-Oncol. 2014, 118, 61–72. [Google Scholar] [CrossRef] [Green Version]
- Haass, C.; Selkoe, D.J. Soluble protein oligomers in neurodegeneration: Lessons from the Alzheimer’s amyloid β-peptide. Nat. Rev. Mol. Cell Biol. 2007, 8, 101–112. [Google Scholar] [CrossRef]
- Quan, X.; Yue, W.; Luo, Y.; Cao, J.; Wang, H.; Wang, Y.; Lu, Z. The protein arginine methyltransferase PRMT5 regulates Aβ-induced toxicity in human cells and Caenorhabditis elegans models of Alzheimer’s disease. J. Neurochem. 2015, 134, 969–977. [Google Scholar] [CrossRef] [PubMed]
- Lukiw, W.J. Bacteroides fragilis lipopolysaccharide and inflammatory signaling in Alzheimer’s disease. Front. Microbiol. 2016, 7, 1544. [Google Scholar] [CrossRef] [Green Version]
- Zhan, X.; Stamova, B.; Sharp, F.R. Lipopolysaccharide associates with amyloid plaques, neurons and oligodendrocytes in Alzheimer’s disease brain: A review. Front. Aging Neurosci. 2018, 10, 42. [Google Scholar] [CrossRef] [Green Version]
- Ross, C.A.; Aylward, E.H.; Wild, E.; Langbehn, D.; Long, J.; Warner, J.H.; Scahill, R.; Leavitt, B.R.; Stout, J.; Paulsen, J.; et al. Huntington disease: Natural history, biomarkers and prospects for therapeutics. Nat. Rev. Neurol. 2014, 10, 204–216. [Google Scholar] [CrossRef] [Green Version]
- Ratovitski, T.; Arbez, N.; Stewart, J.C.; Chighladze, E.; Ross, C.A. PRMT5- mediated symmetric arginine dimethylation is attenuated by mutant huntingtin and is impaired in Huntington’s disease (HD). Cell Cycle 2015, 14, 1716–1729. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, F.; Rui, L. PRMT5 in gene regulation and hematologic malignancies. Genes Dis. 2019, 6, 247–257. [Google Scholar] [CrossRef] [PubMed]
- Demetriadou, C.; Pavlou, D.; Mpekris, F.; Achilleos, C.; Stylianopoulos, T.; Zaravinos, A.; Papageorgis, P.; Kirmizis, A. NAA40 contributes to colorectal cancer growth by controlling PRMT5 expression. Cell Death Dis. 2019, 10, 236. [Google Scholar] [CrossRef]
- Watts, J.M.; Bradley, T.J.; Thomassen, A.; Brunner, A.M.; Minden, M.D.; Papadantonakis, M.N.; Abedin, S.; Baines, A.J.; Barbash, O.; Gorman, S.; et al. A phase I/II study to investigate the safety and clinical activity of the protein arginine methyltransferase 5 inhibitor GSK3326595 in subjects with myelodysplastic syndrome and acute myeloid leukemia. Blood 2019, 134, 2656. [Google Scholar] [CrossRef]
- Hamard, P.-J.; Santiago, G.E.; Liu, F.; Karl, D.L.; Martinez, C.; Man, N.; Mookhtiar, A.K.; Duffort, S.; Greenblatt, S.; Verdun, R.E.; et al. PRMT5 regulates DNA repair by controlling the alternative splicing of histone-modifying enzymes. Cell Rep. 2018, 24, 2643–2657. [Google Scholar] [CrossRef] [Green Version]
- Millar, H.J.; Brehmer, D.; Verhulst, T.; Haddish-Berhane, N.; Greway, T.; Gaffney, D.; Packman, K. In vivo efficacy and pharmacodynamic modulation of JNJ-64619178, a selective PRMT5 inhibitor, in human lung and hematologic preclinical models. Cancer Res. 2019, 79, 950. [Google Scholar]
- Ahnert, J.R.; Perez, C.A.; Wong, K.M.; Maitland, M.L.; Tsai, F.; Berlin, J.; Liao, K.H.; Wang, I.-M.; Markovtsova, L.; Jacobs, I.A.; et al. PF-06939999, a potent and selective PRMT5 inhibitor, in patients with advanced or metastatic solid tumors: A phase 1 dose escalation study. J. Clin. Oncol. 2021, 39, 3019. [Google Scholar] [CrossRef]
- Pastore, F.; Bhagwat, N.; Pastore, A.; Radzisheuskaya, A.; Karzai, A.; Krishnan, A.; Li, B.; Bowman, R.L.; Xiao, W.; Viny, A.D.; et al. PRMT5 inhibition modulates E2F1 methylation and gene-regulatory networks leading to therapeutic efficacy in JAK2V617F-mutant MPN. Cancer Discov. 2020, 10, 1742–1757. [Google Scholar] [CrossRef]
- Bonday, Z.Q.; Cortez, G.S.; Grogan, M.J.; Antonysamy, S.; Weichert, K.; Bocchinfuso, W.P.; Li, F.; Kennedy, S.; Li, B.; Mader, M.M.; et al. LLY-283, a potent and selective inhibitor of arginine methyltransferase 5, PRMT5, with antitumor activity. ACS Med. Chem. Lett. 2018, 9, 612–617. [Google Scholar] [CrossRef] [PubMed]
- Banasavadi-Siddegowda, Y.K.; Welker, A.; An, M.; Yang, X.; Zhou, W.; Shi, G.; Imitola, J.; Li, C.; Hsu, S.; Wang, J.; et al. PRMT5 as a druggable target for glioblastoma therapy. Neuro-Oncology 2017, 20, 753–763. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.R.; Kulyk, S.; Lawson, J.D.; Engstrom, L.D.; Aranda, R.; Briere, D.M.; Gunn, R.; Moya, K.; Rahbaek, L.; Waters, L.; et al. Abstract LB003: Fragment based discovery of MRTX9768, a synthetic lethal-based inhibitor designed to bind the PRMT5-MTA complex and selectively target MTAP/CDKN2A-deleted tumors. Cancer Chem. 2021, 81, LB003. [Google Scholar] [CrossRef]
- Carter, J.; Ito, K.; Thodima, V.; Bhagwat, N.; Rager, J.; Burr, N.S.; Kaufman, J.; Ruggeri, B.; Scherle, P.; Vaddi, K. PRMT5 inhibition downregulates MYB and NOTCH1 signaling, key molecular drivers of adenoid cystic carcinoma. Cancer Res. 2021, 81 (Suppl. S13), 1138. [Google Scholar] [CrossRef]
| Member | Type | Methylation Pattern | References |
|---|---|---|---|
| PRMT1 | I | Monomethylation and asymmetric dimethylation | [3] |
| PRMT2 | I | Monomethylation and asymmetric dimethylation | [3] |
| PRMT3 | I | Monomethylation and asymmetric dimethylation | [3] |
| PRMT4 | I | Monomethylation and asymmetric dimethylation | [3] |
| PRMT5 | II | Monomethylation and symmetric dimethylation | [3] |
| PRMT6 | I | Monomethylation and asymmetric dimethylation | [3] |
| PRMT7 | II and III | Monomethylation OR symmetric dimethylation | [3] |
| PRMT8 | I | Monomethylation and asymmetric dimethylation | [3] |
| PRMT9/FBXO11/PRMT10 | II | Monomethylation and symmetric dimethylation | [3] |
| Regulation | Regulators | Mechanism | Effect | Reference |
|---|---|---|---|---|
| PTMs | Coactivator-associated arginine methyltransferase 1 (CARM1) | Methylation of R505 | Promotes PRMT5 homodimerization | [34] |
| Protein kinase B/Akt | Phosphorylation of T634 | Aids interaction with 14-3-3-proteins | [35] | |
| Janus kinase 2 (JAK2) | Phosphorylation of Y304 and Y307 | Increases substrate binding | ||
| Protein Kinase C iota (PKCι) | Phosphorylation of S15 | Promotes NF-κB activation | [36] | |
| Liver kinase B1 (LKB1) | Phosphorylation of T139 and T144 | Increases methyltransferase activity and interaction with co-factors | [37] | |
| miRNAs | miR-19a, miR-25, miR-32 miR-92b, and miR-96 | Binds to 3′UTR of PRMT5 mRNA | Reduces PRMT5 levels | [40] |
| Protein Interactions | Coordinator Of PRMT5 (COPR5) | Serves as an adaptor for PRMT5 recruitment to chromatin | Causes PRMT5 preferential methylation of H4R3 | [42] |
| Human SWItch/Sucrose Non-Fermentable (hSWI/SNF) | Recruits PRMT5 to H3R8 and H4R3 at ST7 and NM23 promoters | Reduces expression of ST7 and NM23 | [43] | |
| Methyl-CpG-binding domain protein 2/nucleosome remodeling and deacetylase (MBD2/NuRD) | Recruits PRMT5 to CpG islands of p14ARF and p16INK4a | Reduces expression of p14ARF and p16INK4a | [44] | |
| Menin | Recruits PRMT5 to Gas1 promoter | Reduces Gas1 gene expression and enhances Sonic Hedgehog signaling | [45] | |
| B-Lymphocyte induced maturation protein-1 (Blimp1) | Recruits PRMT5 to H2AR3 and H4R3 | Repression of genes in cell cycle, cell signaling, metabolism and transcription | [46] | |
| pICln | Recruits PRMT5 to H4R3 and spliceosomal proteins | Repression of genes in DNA double-stranded break; Assembly of pre-mRNA splicing machinery | [48,49] | |
| RIO kinase 1 (RioK1) | Recruits PRMT5 to nucleolin for methylation | Ribosomal synthesis and maturation | [50] |
| Company | Compound Name | Mode of Inhibition | Clinical Trial Stage | Trial Duration | Country | Types of Indications | References |
|---|---|---|---|---|---|---|---|
| Epizyme (sponsored by GSK) | EPZ015938/GSK3326595 (Pemrametostat) | Direct | Phase I/II | Phase I: 08/30/2016–04/29/2025 Phase II (03/21/2021–12/31/2022) | USA | Phase I: Solid tumors and non-Hodgkin’s lymphoma (with drug: pembrolizumab) Phase II: Early-stage breast cancer | (NCT02783300/NCT04676516) [87] |
| Johnson & Johnson | JNJ-64619178 (Onametostat) | Direct | Phase I | 07/13/2018–12/30/2022 | USA | Solid tumor, adult; non-Hodgkin lymphoma; myelodysplastic syndromes | NCT03573310 [89] |
| Pfizer | PF-06939999 | Direct | Phase I | 03/14/2019–04/14/2026 | USA | Advanced or metastatic non-small cell lung cancer, head and neck squamous cell carcinoma, esophageal cancer, endometrial cancer, cervical cancer, bladder cancer (monotherapy, in combination with docetaxel) | NCT03854227 [90] |
| Prelude | PRT811 | Direct | Phase I | 11/06/2019–10/2022 | USA | Advanced solid tumors, CNS lymphoma, and recurrent high-grade gliomas | NCT04089449 |
| Prelude | PRT543 | Direct | Phase I | 02/11/2019–08/11/2022 | USA | Relapsed/refractory advanced solid tumors; relapsed/refractory diffuse large B-cell lymphoma; relapsed/refractory myelodysplasia; relapsed/refractory myelofibrosis; adenoid cystic carcinoma; relapsed/refractory mantle cell lymphoma; relapsed/refractory acute myeloid leukemia; refractory chronic myelomonocytic leukemia | NCT03886831 [95] |
| EQon Pharmaceuticals | PR5-LL-CM01 | Direct | Preclinical | USA | Pancreatic cancer/colorectal cancer (breast cancer) | [17] | |
| GSK(license with Epizyme) | GSK3186000A | Direct | Preclinical | USA | Leukemia | [89] | |
| Eli Lilly | LLY-283 | Direct | Preclinical | USA | Skin cancer | [91] | |
| Merck | CMP-5 | Direct | Preclinical | USA | Glioblastoma | [93] | |
| Miratis | MRTX9768 | Targets PRMT5-MTA complex | Preclinical | USA | MTAPdel cancer cells | [94] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Motolani, A.; Martin, M.; Sun, M.; Lu, T. The Structure and Functions of PRMT5 in Human Diseases. Life 2021, 11, 1074. https://doi.org/10.3390/life11101074
Motolani A, Martin M, Sun M, Lu T. The Structure and Functions of PRMT5 in Human Diseases. Life. 2021; 11(10):1074. https://doi.org/10.3390/life11101074
Chicago/Turabian StyleMotolani, Aishat, Matthew Martin, Mengyao Sun, and Tao Lu. 2021. "The Structure and Functions of PRMT5 in Human Diseases" Life 11, no. 10: 1074. https://doi.org/10.3390/life11101074
APA StyleMotolani, A., Martin, M., Sun, M., & Lu, T. (2021). The Structure and Functions of PRMT5 in Human Diseases. Life, 11(10), 1074. https://doi.org/10.3390/life11101074







