Making Blood from the Vessel: Extrinsic and Environmental Cues Guiding the Endothelial-to-Hematopoietic Transition
Abstract
:1. Introduction
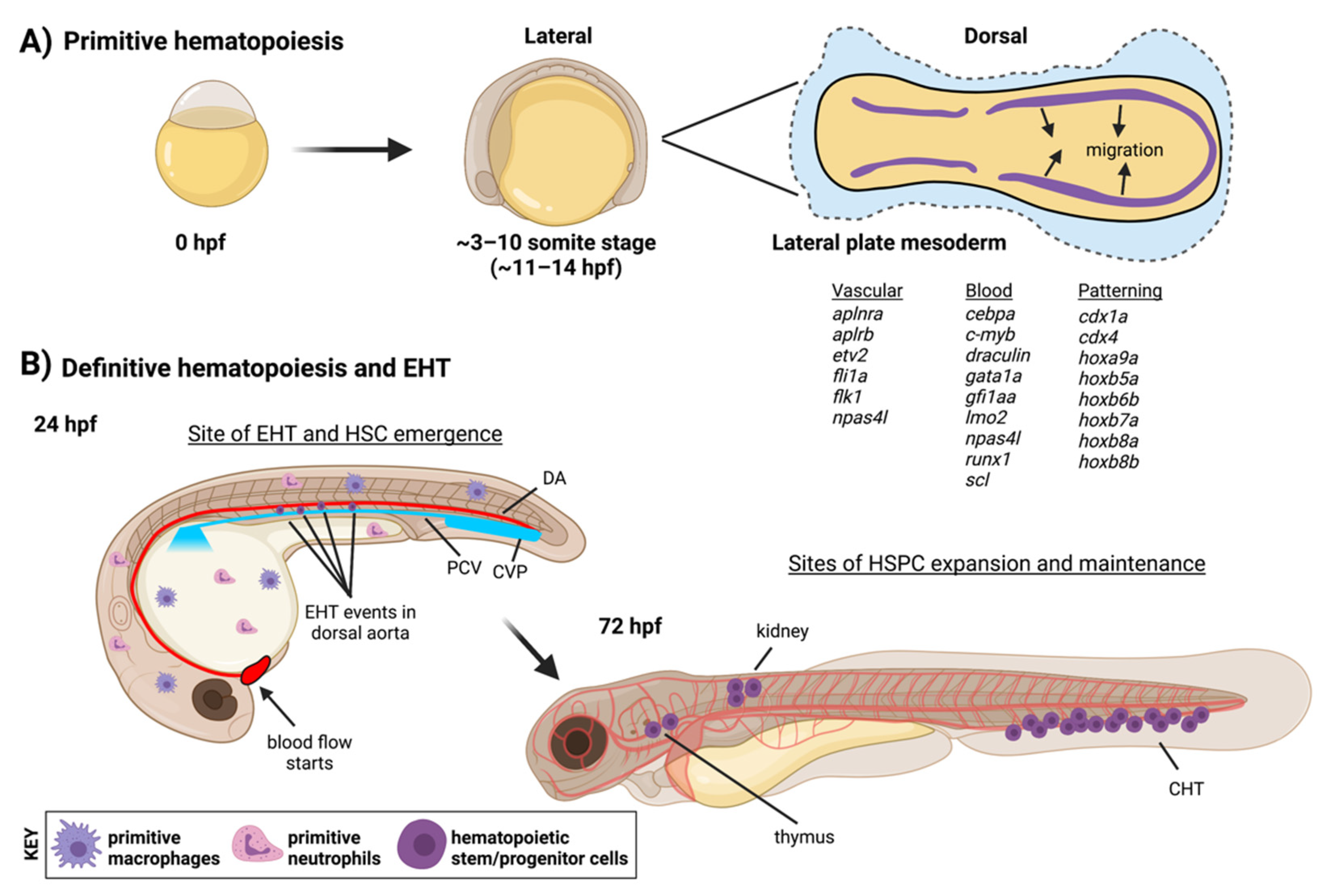
2. The Endothelial Origin of HSCs
2.1. Definitive Hematopoiesis in the Embryo
2.2. Importance, and Limits, of Intrinsic Factors
3. Extrinsic Cues: Mechanical Forces in the Environment
3.1. Influence of the Extracellular Matrix in EHT
3.2. Role of Blood Flow in Promoting EHT
3.3. Cell Contractility and Aortic Architecture during EHT
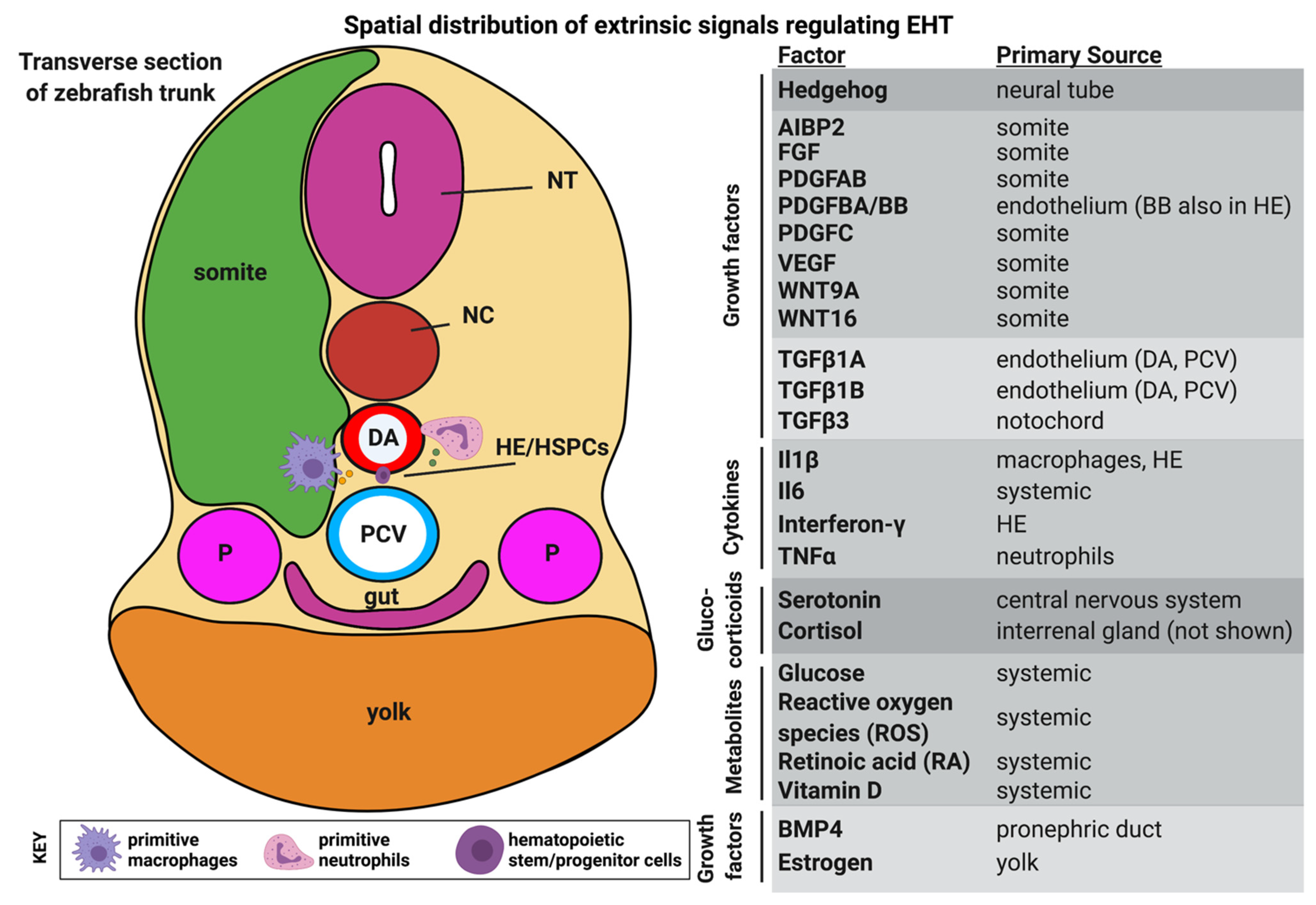
4. Extrinsic Cues: Metabolic, Hormonal and Inflammatory Signals
4.1. Glucose and Other Metabolites
4.2. Hormones
4.3. Sterile Inflammation
5. Extrinsic Cues: Finding New Relationships with ‘Omics Approaches
5.1. Bulk RNA-Sequencing: Hematopoietic Roles for GPCRs
5.2. Tomo-Seq, scRNA-Seq and Ligand/Receptor Predictions
5.3. Future Directions: Integrating Extrinsic Cues with Gene Regulatory Networks by Sequencing
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Clements, W.K.; Traver, D. Signalling Pathways That Control Vertebrate Haematopoietic Stem Cell Specification. Nat. Rev. Immunol. 2013, 13, 336–348. [Google Scholar] [CrossRef] [Green Version]
- Reischauer, S.; Stone, O.A.; Villasenor, A.; Chi, N.; Jin, S.-W.; Martin, M.; Lee, M.T.; Fukuda, N.; Marass, M.; Witty, A.; et al. Cloche Is a BHLH-PAS Transcription Factor That Drives Haemato-Vascular Specification. Nature 2016, 535, 294–298. [Google Scholar] [CrossRef]
- Gore, A.V.; Pillay, L.M.; Venero Galanternik, M.; Weinstein, B.M. The Zebrafish: A Fintastic Model for Hematopoietic Development and Disease. Wiley Interdiscip. Rev. Dev. Biol. 2018, 7, e312. [Google Scholar] [CrossRef]
- Swiers, G.; Rode, C.; Azzoni, E.; de Bruijn, M.F.T.R. A Short History of Hemogenic Endothelium. Blood Cells Mol. Dis. 2013, 51, 206–212. [Google Scholar] [CrossRef] [Green Version]
- Robert-Moreno, A.; Espinosa, L.; de la Pompa, J.L.; Bigas, A.; Simpson, P. RBPjkappa-Dependent Notch Function Regulates Gata2 and Is Essential for the Formation of Intra-Embryonic Hematopoietic Cells. Development 2005, 132, 1117–1126. [Google Scholar] [CrossRef] [Green Version]
- North, T.; Gu, T.L.; Stacy, T.; Wang, Q.; Howard, L.; Binder, M.; Marín-Padilla, M.; Speck, N.A. Cbfa2 Is Required for the Formation of Intra-Aortic Hematopoietic Clusters. Development 1999, 126, 2563–2575. [Google Scholar] [CrossRef]
- Swiers, G.; Baumann, C.; O’Rourke, J.; Giannoulatou, E.; Taylor, S.; Joshi, A.; Moignard, V.; Pina, C.; Bee, T.; Kokkaliaris, K.D.; et al. Early Dynamic Fate Changes in Haemogenic Endothelium Characterized at the Single-Cell Level. Nat. Commun. 2013, 4. [Google Scholar] [CrossRef] [Green Version]
- Ulloa, B.A.; Habbsa, S.S.; Potts, K.S.; Lewis, A.; McKinstry, M.; Payne, S.G.; Flores, J.C.; Nizhnik, A.; Feliz Norberto, M.; Mosimann, C.; et al. Definitive Hematopoietic Stem Cells Minimally Contribute to Embryonic Hematopoiesis. Cell Rep. 2021, 36, 109703. [Google Scholar] [CrossRef]
- Tian, Y.; Xu, J.; Feng, S.; He, S.; Zhao, S.; Zhu, L.; Jin, W.; Dai, Y.; Luo, L.; Qu, J.Y.; et al. The First Wave of T Lymphopoiesis in Zebrafish Arises from Aorta Endothelium Independent of Hematopoietic Stem Cells. J. Exp. Med. 2017, jem.20170488. [Google Scholar] [CrossRef] [Green Version]
- Zhan, Y.; Huang, Y.; Chen, J.; Cao, Z.; He, J.; Zhang, J.; Huang, H.; Ruan, H.; Luo, L.; Li, L. Caudal Dorsal Artery Generates Hematopoietic Stem and Progenitor Cells via the Endothelial-to-Hematopoietic Transition in Zebrafish. J. Genet. Genomics 2018, 45, 315–324. [Google Scholar] [CrossRef]
- Böiers, C.; Carrelha, J.; Lutteropp, M.; Luc, S.; Green, J.C.A.; Azzoni, E.; Woll, P.S.; Mead, A.J.; Hultquist, A.; Swiers, G.; et al. Lymphomyeloid Contribution of an Immune-Restricted Progenitor Emerging Prior to Definitive Hematopoietic Stem Cells. Cell Stem Cell 2013, 13, 535–548. [Google Scholar] [CrossRef] [Green Version]
- Beaudin, A.E.; Boyer, S.W.; Perez-Cunningham, J.; Hernandez, G.E.; Derderian, S.C.; Jujjavarapu, C.; Aaserude, E.; MacKenzie, T.; Forsberg, E.C. A Transient Developmental Hematopoietic Stem Cell Gives Rise to Innate-like B and T Cells. Cell Stem Cell 2016, 19, 768–783. [Google Scholar] [CrossRef] [Green Version]
- Bertrand, J.Y.; Kim, A.D.; Violette, E.P.; Stachura, D.L.; Cisson, J.L.; Traver, D. Definitive Hematopoiesis Initiates through a Committed Erythromyeloid Progenitor in the Zebrafish Embryo. Development 2007, 134, 4147–4156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frame, J.M.; Fegan, K.H.; Conway, S.J.; McGrath, K.E.; Palis, J. Definitive Hematopoiesis in the Yolk Sac Emerges from Wnt-Responsive Hemogenic Endothelium Independently of Circulation and Arterial Identity. Stem Cells 2016, 34, 431–444. [Google Scholar] [CrossRef] [Green Version]
- Plein, A.; Fantin, A.; Denti, L.; Pollard, J.W.; Ruhrberg, C. Erythro-Myeloid Progenitors Contribute Endothelial Cells to Blood Vessels. Nature 2018, 1. [Google Scholar] [CrossRef]
- Wattrus, S.J.; Zon, L.I. Stem Cell Safe Harbor: The Hematopoietic Stem Cell Niche in Zebrafish. Blood Adv. 2018, 2, 3063–3069. [Google Scholar] [CrossRef] [Green Version]
- Herbert, S.P.; Huisken, J.; Kim, T.N.; Feldman, M.E.; Houseman, B.T.; Wang, R.A.; Shokat, K.M.; Stainier, D.Y.R. Arterial-Venous Segregation by Selective Cell Sprouting: An Alternative Mode of Blood Vessel Formation. Science 2009, 326, 294–298. [Google Scholar] [CrossRef] [Green Version]
- Helker, C.S.M.; Schuermann, A.; Karpanen, T.; Zeuschner, D.; Belting, H.G.; Affolter, M.; Schulte-Merker, S.; Herzog, W. The Zebrafish Common Cardinal Veins Develop by a Novel Mechanism: Lumen Ensheathment. Development 2013, 140, 2776–2786. [Google Scholar] [CrossRef] [Green Version]
- Helker, C.S.M.; Schuermann, A.; Pollmann, C.; Chng, S.C.; Kiefer, F.; Reversade, B.; Herzog, W. The Hormonal Peptide Elabela Guides Angioblasts to the Midline during Vasculogenesis. Elife 2015, 4. [Google Scholar] [CrossRef]
- Gering, M.; Patient, R. Hedgehog Signaling Is Required for Adult Blood Stem Cell Formation in Zebrafish Embryos. Dev. Cell 2005, 8, 389–400. [Google Scholar] [CrossRef] [Green Version]
- Clements, W.K.; Kim, A.D.; Ong, K.G.; Moore, J.C.; Lawson, N.D.; Traver, D. A Somitic Wnt16/Notch Pathway Specifies Haematopoietic Stem Cells. Nature 2011, 474, 220–225. [Google Scholar] [CrossRef] [Green Version]
- Kobayashi, I.; Kobayashi-Sun, J.; Kim, A.D.; Pouget, C.; Fujita, N.; Suda, T.; Traver, D. Jam1a-Jam2a Interactions Regulate Haematopoietic Stem Cell Fate through Notch Signalling. Nature 2014, 512, 319–323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Damm, E.W.; Clements, W.K. Pdgf Signalling Guides Neural Crest Contribution to the Haematopoietic Stem Cell Specification Niche. Nat. Cell Biol. 2017, 19, 457–467. [Google Scholar] [CrossRef] [Green Version]
- Bertrand, J.Y.; Chi, N.C.; Santoso, B.; Teng, S.; Stainier, D.Y.R.; Traver, D. Haematopoietic Stem Cells Derive Directly from Aortic Endothelium during Development. Nature 2010, 464, 108–111. [Google Scholar] [CrossRef] [Green Version]
- Kissa, K.; Herbomel, P. Blood Stem Cells Emerge from Aortic Endothelium by a Novel Type of Cell Transition. Nature 2010, 464, 112–115. [Google Scholar] [CrossRef]
- Boisset, J.C.; Van Cappellen, W.; Andrieu-Soler, C.; Galjart, N.; Dzierzak, E.; Robin, C. In Vivo Imaging of Haematopoietic Cells Emerging from the Mouse Aortic Endothelium. Nature 2010, 464, 116–120. [Google Scholar] [CrossRef] [PubMed]
- Ransom, D.G.; Haffter, P.; Odenthal, J.; Brownlie, A.; Vogelsang, E.; Kelsh, R.N.; Brand, M.; Van Eeden, F.J.M.; Furutani-Seiki, M.; Granato, M.; et al. Characterization of Zebrafish Mutants with Defects in Embryonic Hematopoiesis. Development 1996, 123, 311–319. [Google Scholar] [CrossRef]
- Weinstein, B.M.; Schier, A.F.; Abdelilah, S.; Malicki, J.; Solnica-Krezel, L.; Stemple, D.L.; Stainier, D.Y.; Zwartkruis, F.; Driever, W.; Fishman, M.C. Hematopoietic Mutations in the Zebrafish. Development 1996, 123, 303–309. [Google Scholar] [CrossRef]
- Genthe, J.R.; Clements, W.K. R-Spondin 1 Is Required for Specification of Hematopoietic Stem Cells through Wnt16 and Vegfa Signaling Pathways. Development 2017, 144, 590–600. [Google Scholar] [CrossRef] [Green Version]
- Wilkinson, R.N.; Pouget, C.; Gering, M.; Russell, A.J.; Davies, S.G.; Kimelman, D.; Patient, R. Hedgehog and Bmp Polarize Hematopoietic Stem Cell Emergence in the Zebrafish Dorsal Aorta. Dev. Cell 2009, 16, 909–916. [Google Scholar] [CrossRef] [Green Version]
- Ghersi, J.J.; Mahony, C.B.; Bertrand, J.Y. Bif1, a New BMP Signaling Inhibitor, Regulates Embryonic Hematopoiesis in the Zebrafish. Development 2019, 146. [Google Scholar] [CrossRef] [Green Version]
- Pouget, C.; Peterkin, T.; Simões, F.C.; Lee, Y.; Traver, D.; Patient, R. FGF Signalling Restricts Haematopoietic Stem Cell Specification via Modulation of the BMP Pathway. Nat. Commun. 2014, 5, 5588. [Google Scholar] [CrossRef] [Green Version]
- Lee, Y.; Manegold, J.E.; Kim, A.D.; Pouget, C.; Stachura, D.L.; Clements, W.K.; Traver, D. FGF Signalling Specifies Haematopoietic Stem Cells through Its Regulation of Somitic Notch Signalling. Nat. Commun. 2014, 5, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goessling, W.; North, T.E.; Loewer, S.; Lord, A.M.; Lee, S.; Stoick-Cooper, C.L.; Weidinger, G.; Puder, M.; Daley, G.Q.; Moon, R.T.; et al. Genetic Interaction of PGE2 and Wnt Signaling Regulates Developmental Specification of Stem Cells and Regeneration. Cell 2009, 136, 1136–1147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Monteiro, R.; Pinheiro, P.; Joseph, N.; Peterkin, T.; Koth, J.; Repapi, E.; Bonkhofer, F.; Kirmizitas, A.; Patient, R. Transforming Growth Factor β Drives Hemogenic Endothelium Programming and the Transition to Hematopoietic Stem Cells. Dev. Cell 2016, 38, 358–370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mahony, C.B.; Pasche, C.; Bertrand, J.Y. Oncostatin M and Kit-Ligand Control Hematopoietic Stem Cell Fate during Zebrafish Embryogenesis. Stem Cell Rep. 2018, 10, 1920–1934. [Google Scholar] [CrossRef] [PubMed]
- North, T.E.; De Bruijn, M.F.T.R.; Stacy, T.; Talebian, L.; Lind, E.; Robin, C.; Binder, M.; Dzierzak, E.; Speck, N.A. Runx1 Expression Marks Long-Term Repopulating Hematopoietic Stem Cells in the Midgestation Mouse Embryo. Immunity 2002, 16, 661–672. [Google Scholar] [CrossRef] [Green Version]
- Kalev-Zylinska, M.L.; Horsfield, J.A.; Flores, M.V.C.; Postlethwait, J.H.; Vitas, M.R.; Baas, A.M.; Crosier, P.S.; Crosier, K.E. Runx1 Is Required for Zebrafish Blood and Vessel Development and Expression of a Human RUNX-1-CBF2T1 Transgene Advances a Model for Studies of Leukemogenesis. Development 2002, 129, 2015–2030. [Google Scholar] [CrossRef]
- Wilson, N.K.; Foster, S.D.; Wang, X.; Knezevic, K.; Schütte, J.; Kaimakis, P.; Chilarska, P.M.; Kinston, S.; Ouwehand, W.H.; Dzierzak, E.; et al. Combinatorial Transcriptional Control in Blood Stem/Progenitor Cells: Genome-Wide Analysis of Ten Major Transcriptional Regulators. Cell Stem Cell 2010, 7, 532–544. [Google Scholar] [CrossRef] [Green Version]
- Dobrzycki, T.; Mahony, C.B.; Krecsmarik, M.; Koyunlar, C.; Rispoli, R.; Peulen-Zink, J.; Gussinklo, K.; Fedlaoui, B.; de Pater, E.; Patient, R.; et al. Deletion of a Conserved Gata2 Enhancer Impairs Haemogenic Endothelium Programming and Adult Zebrafish Haematopoiesis. Commun. Biol. 2020, 3. [Google Scholar] [CrossRef]
- Butko, E.; Distel, M.; Pouget, C.; Weijts, B.; Kobayashi, I.; Ng, K.; Mosimann, C.; Poulain, F.E.; McPherson, A.; Ni, C.-W.; et al. Gata2b Is a Restricted Early Regulator of Hemogenic Endothelium in the Zebrafish Embryo. Development 2015, 142, 1050–1061. [Google Scholar] [CrossRef] [Green Version]
- Cooney, J.D.; Hildick-Smith, G.J.; Shafizadeh, E.; McBride, P.F.; Carroll, K.J.; Anderson, H.; Shaw, G.C.; Tamplin, O.J.; Branco, D.S.; Dalton, A.J.; et al. Teleost Growth Factor Independence (Gfi) Genes Differentially Regulate Successive Waves of Hematopoiesis. Dev. Biol. 2013, 373, 431–441. [Google Scholar] [CrossRef] [Green Version]
- Bertrand, J.Y.; Cisson, J.L.; Stachura, D.L.; Traver, D. Notch Signaling Distinguishes 2 Waves of Definitive Hematopoiesis in the Zebrafish Embryo. Blood 2010, 115, 2777–2783. [Google Scholar] [CrossRef] [Green Version]
- Kim, A.D.; Melick, C.H.; Clements, W.K.; Stachura, D.L.; Distel, M.; Panáková, D.; MacRae, C.; Mork, L.A.; Crump, J.G.; Traver, D. Discrete Notch Signaling Requirements in the Specification of Hematopoietic Stem Cells. EMBO J. 2014, 33, 2363–2373. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lizama, C.O.; Hawkins, J.S.; Schmitt, C.E.; Bos, F.L.; Zape, J.P.; Cautivo, K.M.; Borges Pinto, H.; Rhyner, A.M.; Yu, H.; Donohoe, M.E.; et al. Repression of Arterial Genes in Hemogenic Endothelium Is Sufficient for Haematopoietic Fate Acquisition. Nat. Commun. 2015, 6, 7739. [Google Scholar] [CrossRef] [Green Version]
- Sood, R.; English, M.A.; Belele, C.L.; Jin, H.; Bishop, K.; Haskins, R.; McKinney, M.C.; Chahal, J.; Weinstein, B.M.; Wen, Z.; et al. Development of Multilineage Adult Hematopoiesis in the Zebrafish with a Runx1 Truncation Mutation. Blood 2010, 115, 2806–2809. [Google Scholar] [CrossRef] [PubMed]
- Pelster, B.; Burggren, W.W. Disruption of Hemoglobin Oxygen Transport Does Not Impact Oxygen-Dependent Physiological Processes in Developing Embryos of Zebra Fish (Danio Rerio). Circ. Res. 1996, 79, 358–362. [Google Scholar] [CrossRef] [PubMed]
- Bresciani, E.; Carrington, B.; Yu, K.; Kim, E.M.K.; Zhen, T.; Guzman, V.S.; Broadbridge, E.; Bishop, K.; Kirby, M.; Harper, U.; et al. Redundant Mechanisms Driven Independently by RUNX1 and GATA2 for Hematopoietic Development. Blood Adv. 2021. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Jia, X.; Yuan, H.; Ma, K.; Chen, Y.; Jin, Y.; Deng, M.; Pan, W.; Chen, S.; Chen, Z.; et al. DNA Methyltransferase 1 Functions through C/Ebpa to Maintain Hematopoietic Stem and Progenitor Cells in Zebrafish. J. Hematol. Oncol. 2015, 8, 15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gore, A.V.; Athans, B.; Iben, J.R.; Johnson, K.; Russanova, V.; Castranova, D.; Pham, V.N.; Butler, M.G.; Williams-Simons, L.; Nichols, J.T.; et al. Epigenetic Regulation of Hematopoiesis by DNA Methylation. Elife 2016, 5. [Google Scholar] [CrossRef]
- Burns, C.E.; Galloway, J.L.; Smith, A.C.H.; Keefe, M.D.; Cashman, T.J.; Paik, E.J.; Mayhall, E.A.; Amsterdam, A.H.; Zon, L.I. A Genetic Screen in Zebrafish Defines a Hierarchical Network of Pathways Required for Hematopoietic Stem Cell Emergence. Blood 2009, 113, 5776–5782. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, H.T.; Kathrein, K.L.; Barton, A.; Gitlin, Z.; Huang, Y.H.; Ward, T.P.; Hofmann, O.; Dibiase, A.; Song, A.; Tyekucheva, S.; et al. A Network of Epigenetic Regulators Guides Developmental Haematopoiesis in Vivo. Nat. Cell Biol. 2013, 15, 1516–1525. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, Y.; Wang, W.; Ma, D.; Liang, G.; Kang, Z.; Xue, Y.; Zhang, Y.; Wang, L.; Heng, J.; Zhang, Y.; et al. Smarca5-Mediated Epigenetic Programming Facilitates Fetal HSPC Development in Vertebrates. Blood 2021, 137, 190–202. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Mazor, T.; Huang, H.; Huang, H.T.; Kathrein, K.L.; Woo, A.J.; Chouinard, C.R.; Labadorf, A.; Akie, T.E.; Moran, T.B.; et al. Direct Recruitment of Polycomb Repressive Complex 1 to Chromatin by Core Binding Transcription Factors. Mol. Cell 2012, 45, 330–343. [Google Scholar] [CrossRef] [Green Version]
- Zhong, Y.; Ye, Q.; Chen, C.; Wang, M.; Wang, H. Ezh2 Promotes Clock Function and Hematopoiesis Independent of Histone Methyltransferase Activity in Zebrafish. Nucleic Acids Res. 2018, 46, 3382–3399. [Google Scholar] [CrossRef]
- Soto, R.A.; Najia, M.A.T.; Hachimi, M.; Frame, J.M.; Yette, G.A.; Lummertz da Rocha, E.; Stankunas, K.; Daley, G.Q.; North, T.E. Sequential Regulation of Hemogenic Fate and Hematopoietic Stem and Progenitor Cell Formation from Arterial Endothelium by Ezh1/2. Stem Cell Rep. 2021. [Google Scholar] [CrossRef]
- Batta, K.; Florkowska, M.; Kouskoff, V.; Lacaud, G. Direct Reprogramming of Murine Fibroblasts to Hematopoietic Progenitor Cells. Cell Rep. 2014, 9, 1871–1884. [Google Scholar] [CrossRef]
- Sturgeon, C.M.; Ditadi, A.; Awong, G.; Kennedy, M.; Keller, G. Wnt Signaling Controls the Specification of Definitive and Primitive Hematopoiesis from Human Pluripotent Stem Cells. Nat. Biotechnol. 2014, 32, 554–561. [Google Scholar] [CrossRef] [Green Version]
- Ditadi, A.; Sturgeon, C.M.; Tober, J.; Awong, G.; Kennedy, M.; Yzaguirre, A.D.; Azzola, L.; Ng, E.S.; Stanley, E.G.; French, D.L.; et al. Human Definitive Haemogenic Endothelium and Arterial Vascular Endothelium Represent Distinct Lineages. Nat. Cell Biol. 2015, 17, 580–591. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lis, R.; Karrasch, C.C.; Poulos, M.G.; Kunar, B.; Redmond, D.; Duran, J.G.B.; Badwe, C.R.; Schachterle, W.; Ginsberg, M.; Xiang, J.; et al. Conversion of Adult Endothelium to Immunocompetent Haematopoietic Stem Cells. Nature 2017, 545, 439–445. [Google Scholar] [CrossRef] [PubMed]
- Cahan, P.; Li, H.; Morris, S.A.; Lummertz Da Rocha, E.; Daley, G.Q.; Collins, J.J. CellNet: Network Biology Applied to Stem Cell Engineering. Cell 2014, 158, 903–915. [Google Scholar] [CrossRef] [Green Version]
- Doulatov, S.; Vo, L.T.; Chou, S.S.; Kim, P.G.; Arora, N.; Li, H.; Hadland, B.K.; Bernstein, I.D.; Collins, J.J.; Zon, L.I.; et al. Induction of Multipotential Hematopoietic Progenitors from Human Pluripotent Stem Cells via Respecification of Lineage-Restricted Precursors. Cell Stem Cell 2013, 13, 459–470. [Google Scholar] [CrossRef] [Green Version]
- Sugimura, R.; Jha, D.K.; Han, A.; Soria-Valles, C.; Da Rocha, E.L.; Lu, Y.F.; Goettel, J.A.; Serrao, E.; Rowe, R.G.; Malleshaiah, M.; et al. Haematopoietic Stem and Progenitor Cells from Human Pluripotent Stem Cells. Nature 2017, 545, 432–438. [Google Scholar] [CrossRef] [Green Version]
- Uenishi, G.I.; Jung, H.S.; Kumar, A.; Park, M.A.; Hadland, B.K.; McLeod, E.; Raymond, M.; Moskvin, O.; Zimmerman, C.E.; Theisen, D.J.; et al. NOTCH Signaling Specifies Arterial-Type Definitive Hemogenic Endothelium from Human Pluripotent Stem Cells. Nat. Commun. 2018, 9, 1828. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jung, H.S.; Uenishi, G.; Park, M.A.; Liu, P.; Suknuntha, K.; Raymond, M.; Choi, Y.J.; Thomson, J.A.; Ong, I.M.; Slukvin, I.I. SOX17 Integrates HOXA and Arterial Programs in Hemogenic Endothelium to Drive Definitive Lympho-Myeloid Hematopoiesis. Cell Rep. 2021, 34. [Google Scholar] [CrossRef] [PubMed]
- Mestas, J.; Hughes, C.C.W. Of Mice and Not Men: Differences between Mouse and Human Immunology. J. Immunol. 2004, 172, 2731–2738. [Google Scholar] [CrossRef] [Green Version]
- Stachura, D.L.; Reyes, J.R.; Bartunek, P.; Paw, B.H.; Zon, L.I.; Traver, D. Zebrafish Kidney Stromal Cell Lines Support Multilineage Hematopoiesis. Blood 2009, 114, 279–289. [Google Scholar] [CrossRef] [Green Version]
- Stachura, D.L.; Svoboda, O.; Lau, R.P.; Balla, K.M.; Zon, L.I.; Bartunek, P.; Traver, D. Clonal Analysis of Hematopoietic Progenitor Cells in the Zebrafish. Blood 2011, 118, 1274–1282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Svoboda, O.; Stachura, D.L.; MacHonova, O.; Zon, L.I.; Traver, D.; Bartunek, P. Ex Vivo Tools for the Clonal Analysis of Zebrafish Hematopoiesis. Nat. Protoc. 2016, 11, 1007–1020. [Google Scholar] [CrossRef] [Green Version]
- North, T.E.; Goessling, W.; Walkley, C.R.; Lengerke, C.; Kopani, K.R.; Lord, A.M.; Weber, G.J.; Bowman, T.V.; Jang, I.H.; Grosser, T.; et al. Prostaglandin E2 Regulates Vertebrate Haematopoietic Stem Cell Homeostasis. Nature 2007, 447, 1007–1011. [Google Scholar] [CrossRef]
- Cutler, C.; Multani, P.; Robbins, D.; Kim, H.T.; Le, T.; Hoggatt, J.; Pelus, L.M.; Desponts, C.; Chen, Y.-B.; Rezner, B.; et al. Prostaglandin-Modulated Umbilical Cord Blood Hematopoietic Stem Cell Transplantation. Blood 2013, 122, 3074–3081. [Google Scholar] [CrossRef]
- Kechagia, J.Z.; Ivaska, J.; Roca-Cusachs, P. Integrins as Biomechanical Sensors of the Microenvironment. Nat. Rev. Mol. Cell Biol. 2019, 20, 457–473. [Google Scholar] [CrossRef]
- Moro, A.; Driscoll, T.P.; Boraas, L.C.; Armero, W.; Kasper, D.M.; Baeyens, N.; Jouy, C.; Mallikarjun, V.; Swift, J.; Ahn, S.J.; et al. MicroRNA-Dependent Regulation of Biomechanical Genes Establishes Tissue Stiffness Homeostasis. Nat. Cell Biol. 2019, 1. [Google Scholar] [CrossRef]
- Ni, F.; Yu, W.-M.; Wang, X.; Fay, M.E.; Young, K.M.; Qiu, Y.; Lam, W.A.; Sulchek, T.A.; Cheng, T.; Scadden, D.T.; et al. Ptpn21 Controls Hematopoietic Stem Cell Homeostasis and Biomechanics. Cell Stem Cell 2019, 24, 608–620.e6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Theodore, L.N.; Hagedorn, E.J.; Cortes, M.; Natsuhara, K.; Liu, S.Y.; Perlin, J.R.; Yang, S.; Daily, M.L.; Zon, L.I.; North, T.E. Distinct Roles for Matrix Metalloproteinases 2 and 9 in Embryonic Hematopoietic Stem Cell Emergence, Migration, and Niche Colonization. Stem Cell Rep. 2017, 8, 1226–1241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iida, A.; Sakaguchi, K.; Sato, K.; Sakurai, H.; Nishimura, D.; Iwaki, A.; Takeuchi, M.; Kobayashi, M.; Misaki, K.; Yonemura, S.; et al. Metalloprotease-Dependent Onset of Blood Circulation in Zebrafish. Curr. Biol. 2010, 20, 1110–1116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Winograd-Katz, S.E.; Fässler, R.; Geiger, B.; Legate, K.R. The Integrin Adhesome: From Genes and Proteins to Human Disease. Nat. Rev. Mol. Cell Biol. 2014, 15, 273–288. [Google Scholar] [CrossRef] [PubMed]
- Rho, S.-S.; Kobayashi, I.; Oguri-Nakamura, E.; Ando, K.; Fujiwara, M.; Kamimura, N.; Hirata, H.; Iida, A.; Iwai, Y.; Mochizuki, N.; et al. Rap1b Promotes Notch-Signal-Mediated Hematopoietic Stem Cell Development by Enhancing Integrin-Mediated Cell Adhesion. Dev. Cell 2019. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Xue, W.; Li, M.; Dong, M.; Wang, J.; Wang, X.; Li, X.; Chen, K.; Zhang, W.; Wu, S.; et al. VCAM-1+ Macrophages Guide the Homing of HSPCs to a Vascular Niche. Nature 2018, 564, 119–124. [Google Scholar] [CrossRef] [PubMed]
- Adamo, L.; Naveiras, O.; Wenzel, P.L.; McKinney-Freeman, S.; Mack, P.J.; Gracia-Sancho, J.; Suchy-Dicey, A.; Yoshimoto, M.; Lensch, M.W.; Yoder, M.C.; et al. Biomechanical Forces Promote Embryonic Haematopoiesis. Nature 2009, 459, 1131–1135. [Google Scholar] [CrossRef] [Green Version]
- North, T.E.; Goessling, W.; Peeters, M.; Li, P.; Ceol, C.; Lord, A.M.; Weber, G.J.; Harris, J.; Cutting, C.C.; Huang, P.; et al. Hematopoietic Stem Cell Development Is Dependent on Blood Flow. Cell 2009, 137, 736–748. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Diaz, M.F.; Li, N.; Lee, H.J.; Adamo, L.; Evans, S.M.; Willey, H.E.; Arora, N.; Torisawa, Y.; Vickers, D.A.; Morris, S.A.; et al. Biomechanical Forces Promote Blood Development through Prostaglandin E 2 and the CAMP–PKA Signaling Axis. J. Exp. Med. 2015, 212, 665–680. [Google Scholar] [CrossRef] [PubMed]
- Kim, P.G.; Nakano, H.; Das, P.P.; Chen, M.J.; Rowe, R.G.; Chou, S.S.; Ross, S.J.; Sakamoto, K.M.; Zon, L.I.; Schlaeger, T.M.; et al. Flow-Induced Protein Kinase A–CREB Pathway Acts via BMP Signaling to Promote HSC Emergence. J. Exp. Med. 2015, 212, 633–648. [Google Scholar] [CrossRef] [PubMed]
- Jing, L.; Tamplin, O.J.; Chen, M.J.; Deng, Q.; Patterson, S.; Kim, P.G.; Durand, E.M.; McNeil, A.; Green, J.M.; Matsuura, S.; et al. Adenosine Signaling Promotes Hematopoietic Stem and Progenitor Cell Emergence. J. Exp. Med. 2015, 212, 649–663. [Google Scholar] [CrossRef] [PubMed]
- Anton, H.; Harlepp, S.; Ramspacher, C.; Wu, D.; Monduc, F.; Bhat, S.; Liebling, M.; Paoletti, C.; Charvin, G.; Freund, J.B.; et al. Pulse Propagation by a Capacitive Mechanism Drives Embryonic Blood Flow. Development 2013, 140, 4426–4434. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goetz, J.G.; Steed, E.; Ferreira, R.R.; Roth, S.; Ramspacher, C.; Boselli, F.; Charvin, G.; Liebling, M.; Wyart, C.; Schwab, Y.; et al. Endothelial Cilia Mediate Low Flow Sensing during Zebrafish Vascular Development. Cell Rep. 2014, 6, 799–808. [Google Scholar] [CrossRef]
- Sugden, W.W.; Meissner, R.; Aegerter-Wilmsen, T.; Tsaryk, R.; Leonard, E.V.; Bussmann, J.; Hamm, M.J.; Herzog, W.; Jin, Y.; Jakobsson, L.; et al. Endoglin Controls Blood Vessel Diameter through Endothelial Cell Shape Changes in Response to Haemodynamic Cues. Nat. Cell Biol. 2017. [Google Scholar] [CrossRef] [Green Version]
- Liu, Z.; Tu, H.; Kang, Y.; Xue, Y.; Ma, D.; Zhao, C.; Li, H.; Wang, L.; Liu, F. Primary Cilia Regulate Hematopoietic Stem and Progenitor Cell Specification through Notch Signaling in Zebrafish. Nat. Commun. 2019, 10, 1839. [Google Scholar] [CrossRef]
- Wang, L.; Zhang, P.; Wei, Y.; Gao, Y.; Patient, R.; Liu, F. A Blood Flow—Dependent Klf2a -NO Signaling Cascade Is Required for Stabilization of Hematopoietic Stem Cell Programming in Zebrafish Embryos. Blood 2011, 118, 4102–4111. [Google Scholar] [CrossRef] [Green Version]
- Novodvorsky, P.; Watson, O.; Gray, C.; Wilkinson, R.N.; Reeve, S.; Smythe, C.; Beniston, R.; Plant, K.; Maguire, R.; Rothman, A.M.K.; et al. Klf2ash317mutant Zebrafish Do Not Recapitulate Morpholino-Induced Vascular and Haematopoietic Phenotypes. PLoS ONE 2015, 10. [Google Scholar] [CrossRef] [Green Version]
- Rossi, A.; Kontarakis, Z.; Gerri, C.; Nolte, H.; Hölper, S.; Krüger, M.; Stainier, D.Y.R. Genetic Compensation Induced by Deleterious Mutations but Not Gene Knockdowns. Nature 2015, 524, 230–233. [Google Scholar] [CrossRef]
- Dekker, R.J.; Van Soest, S.; Fontijn, R.D.; Salamanca, S.; De Groot, P.G.; VanBavel, E.; Pannekoek, H.; Horrevoets, A.J.G. Prolonged Fluid Shear Stress Induces a Distinct Set of Endothelial Cell Genes, Most Specifically Lung Krüppel-like Factor (KLF2). Blood 2002, 100, 1689–1698. [Google Scholar] [CrossRef] [PubMed]
- Goddard, L.M.; Duchemin, A.L.; Ramalingan, H.; Wu, B.; Chen, M.; Bamezai, S.; Yang, J.; Li, L.; Morley, M.P.; Wang, T.; et al. Hemodynamic Forces Sculpt Developing Heart Valves through a KLF2-WNT9B Paracrine Signaling Axis. Dev. Cell 2017, 43, 274–289.e5. [Google Scholar] [CrossRef] [PubMed]
- Steed, E.; Faggianelli, N.; Roth, S.; Ramspacher, C.; Concordet, J.P.; Vermot, J. Klf2a Couples Mechanotransduction and Zebrafish Valve Morphogenesis through Fibronectin Synthesis. Nat. Commun. 2016, 7, 11646. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grainger, S.; Richter, J.; Palazón, R.E.; Pouget, C.; Lonquich, B.; Wirth, S.; Grassme, K.S.; Herzog, W.; Swift, M.R.; Weinstein, B.M.; et al. Wnt9a Is Required for the Aortic Amplification of Nascent Hematopoietic Stem Cells. Cell Rep. 2016, 17, 1595–1606. [Google Scholar] [CrossRef] [Green Version]
- Grainger, S.; Nguyen, N.; Richter, J.; Setayesh, J.; Lonquich, B.; Oon, C.H.; Wozniak, J.M.; Barahona, R.; Kamei, C.N.; Houston, J.; et al. EGFR Is Required for Wnt9a–Fzd9b Signalling Specificity in Haematopoietic Stem Cells. Nat. Cell Biol. 2019, 21, 721–730. [Google Scholar] [CrossRef] [PubMed]
- Dupont, S.; Morsut, L.; Aragona, M.; Enzo, E.; Giulitti, S.; Cordenonsi, M.; Zanconato, F.; Le Digabel, J.; Forcato, M.; Bicciato, S.; et al. Role of YAP/TAZ in Mechanotransduction. Nature 2011, 474, 179–184. [Google Scholar] [CrossRef]
- Wang, L.; Luo, J.-Y.; Li, B.; Tian, X.Y.; Chen, L.-J.; Huang, Y.; Liu, J.; Deng, D.; Lau, C.W.; Wan, S.; et al. Integrin-YAP/TAZ-JNK Cascade Mediates Atheroprotective Effect of Unidirectional Shear Flow. Nature 2016, 540, 579–582. [Google Scholar] [CrossRef]
- Nakajima, H.; Yamamoto, K.; Agarwala, S.; Terai, K.; Fukui, H.; Fukuhara, S.; Ando, K.; Miyazaki, T.; Yokota, Y.; Schmelzer, E.; et al. Flow-Dependent Endothelial YAP Regulation Contributes to Vessel Maintenance. Dev. Cell 2017, 40, 523–536.e6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jansson, L.; Larsson, J. Normal Hematopoietic Stem Cell Function in Mice with Enforced Expression of the Hippo Signaling Effector YAP1. PLoS ONE 2012, 7, e32013. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Donato, E.; Biagioni, F.; Bisso, A.; Caganova, M.; Amati, B.; Campaner, S. YAP and TAZ Are Dispensable for Physiological and Malignant Haematopoiesis. Leukemia 2018, 32, 2037–2040. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goode, D.K.; Obier, N.; Vijayabaskar, M.S.; Lie-A-Ling, M.; Lilly, A.J.; Hannah, R.; Lichtinger, M.; Batta, K.; Florkowska, M.; Patel, R.; et al. Dynamic Gene Regulatory Networks Drive Hematopoietic Specification and Differentiation. Dev. Cell 2016, 36, 572–587. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lundin, V.; Sugden, W.W.; Theodore, L.N.; Sousa, P.M.; Han, A.; Chou, S.; Wrighton, P.J.; Cox, A.G.; Ingber, D.E.; Goessling, W.; et al. YAP Regulates Hematopoietic Stem Cell Formation in Response to the Biomechanical Forces of Blood Flow. Dev. Cell 2020, 52. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Trahan, G.D.; Howell, E.D.; Speck, N.A.; Jones, K.L.; Gillen, A.E.; Riemondy, K.; Hesselberth, J.; Bryder, D.; Ernst, P. Enhancing Hematopoiesis from Murine Embryonic Stem Cells through MLL1-Induced Activation of a Rac/Rho/Integrin Signaling Axis. Stem Cell Rep. 2020, 14, 285–299. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yue, R.; Li, H.; Liu, H.; Li, Y.; Wei, B.; Gao, G.; Jin, Y.; Liu, T.; Wei, L.; Du, J.; et al. Thrombin Receptor Regulates Hematopoiesis and Endothelial-to-Hematopoietic Transition. Dev. Cell 2012, 22, 1092–1100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lancino, M.; Majello, S.; Herbert, S.; De Chaumont, F.; Tinevez, J.Y.; Olivo-Marin, J.C.; Herbomel, P.; Schmidt, A. Anisotropic Organization of Circumferential Actomyosin Characterizes Hematopoietic Stem Cells Emergence in the Zebrafish. Elife 2018, 7, 1–36. [Google Scholar] [CrossRef] [PubMed]
- Campinho, P.; Lamperti, P.; Boselli, F.; Vilfan, A.; Vermot, J. Blood Flow Limits Endothelial Cell Extrusion in the Zebrafish Dorsal Aorta. Cell Rep. 2020, 31. [Google Scholar] [CrossRef] [PubMed]
- Poullet, N.; Golushko, I.; Lorman, V.; Travnickova, J.; Bureau, C.; Chalin, D.; Rochal, S.; Parmeggiani, A.; Kissa, K. Mechanical Instabilities of Aorta Drive Blood Stem Cell Production: A Live Study. Cell. Mol. Life Sci. 2020, 77, 3453–3464. [Google Scholar] [CrossRef] [Green Version]
- Chalin, D.; Bureau, C.; Parmeggiani, A.; Rochal, S.; Kissa, K.; Golushko, I. Modeling and Live Imaging of Mechanical Instabilities in the Zebrafish Aorta during Hematopoiesis. Sci. Rep. 2021, 11. [Google Scholar] [CrossRef]
- Potente, M.; Carmeliet, P. The Link between Angiogenesis and Endothelial Metabolism. Annu. Rev. Physiology. 2017, 10, 43–66. [Google Scholar] [CrossRef]
- Harris, J.M.; Esain, V.; Frechette, G.M.; Harris, L.J.; Cox, A.G.; Cortes, M.; Garnaas, M.K.; Carroll, K.J.; Cutting, C.C.; Khan, T.; et al. Glucose Metabolism Impacts the Spatiotemporal Onset and Magnitude of HSC Induction in Vivo. Blood 2013, 121, 2483–2493. [Google Scholar] [CrossRef] [Green Version]
- Gerri, C.; Marass, M.; Rossi, A.; Stainier, D.Y.R. Hif-1a and Hif-2a Regulate Hemogenic Endothelium and Hematopoietic Stem Cell Formation in Zebrafish. Blood 2018, 131, 963–973. [Google Scholar] [CrossRef] [Green Version]
- Imanirad, P.; Solaimani Kartalaei, P.; Crisan, M.; Vink, C.; Yamada-Inagawa, T.; de Pater, E.; Kurek, D.; Kaimakis, P.; van der Linden, R.; Speck, N.; et al. HIF1α Is a Regulator of Hematopoietic Progenitor and Stem Cell Development in Hypoxic Sites of the Mouse Embryo. Stem Cell Res. 2014, 12, 24–35. [Google Scholar] [CrossRef] [Green Version]
- Gu, Q.; Yang, X.; Lv, J.; Zhang, J.; Xia, B.; Kim, J.D.; Wang, R.; Xiong, F.; Meng, S.; Clements, T.P.; et al. AIBP-Mediated Cholesterol Efflux Instructs Hematopoietic Stem and Progenitor Cell Fate. Science 2019, 363, 1085–1088. [Google Scholar] [CrossRef]
- Zhang, C.; Chen, Y.; Sun, B.; Wang, L.; Yang, Y.; Ma, D.; Lv, J.; Heng, J.; Ding, Y.; Xue, Y.; et al. M6A Modulates Haematopoietic Stem and Progenitor Cell Specification. Nature 2017, 549, 273–276. [Google Scholar] [CrossRef] [PubMed]
- Bielczyk-Maczyńska, E.; Lam Hung, L.; Ferreira, L.; Fleischmann, T.; Weis, F.; Fernández-Pevida, A.; Harvey, S.A.; Wali, N.; Warren, A.J.; Barroso, I.; et al. The Ribosome Biogenesis Protein Nol9 Is Essential for Definitive Hematopoiesis and Pancreas Morphogenesis in Zebrafish. PLoS Genet. 2015, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kasper, D.M.; Moro, A.; Ristori, E.; Narayanan, A.; Hill-Teran, G.; Fleming, E.; Moreno-Mateos, M.; Vejnar, C.E.; Zhang, J.; Lee, D.; et al. MicroRNAs Establish Uniform Traits during the Architecture of Vertebrate Embryos. Dev. Cell 2017, 40, 552–565.e5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kasper, D.M.; Hintzen, J.; Wu, Y.; Ghersi, J.J.; Mandl, H.K.; Salinas, K.E.; Armero, W.; He, Z.; Sheng, Y.; Xie, Y.; et al. The N-Glycome Regulates the Endothelial-Tohematopoietic Transition. Science. 2020, 370. [Google Scholar] [CrossRef] [PubMed]
- Carroll, K.J.; Esain, V.; Garnaas, M.K.; Cortes, M.; Dovey, M.C.; Nissim, S.; Frechette, G.M.; Liu, S.Y.; Kwan, W.; Cutting, C.C.; et al. Estrogen Defines the Dorsal-Ventral Limit of VEGF Regulation to Specify the Location of the Hemogenic Endothelial Niche. Dev. Cell 2014, 29, 437–453. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cortes, M.; Liu, S.Y.; Kwan, W.; Alexa, K.; Goessling, W.; North, T.E. Accumulation of the Vitamin D Precursor Cholecalciferol Antagonizes Hedgehog Signaling to Impair Hemogenic Endothelium Formation. Stem Cell Reports 2015, 5, 471–479. [Google Scholar] [CrossRef] [Green Version]
- Cortes, M.; Chen, M.J.; Stachura, D.L.; Liu, S.Y.; Kwan, W.; Wright, F.; Vo, L.T.; Theodore, L.N.; Esain, V.; Frost, I.M.; et al. Developmental Vitamin D Availability Impacts Hematopoietic Stem Cell Production. Cell Rep. 2016, 17, 458–468. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Xue, Y.; Cao, C.; Huang, J.; Hong, Q.; Hai, T.; Jia, Q.; Wang, X.; Qin, G.; Yao, J.; et al. Thyroid Hormone Regulates Hematopoiesis via the TR-KLF9 Axis. Blood 2017, 130, 2161–2170. [Google Scholar] [CrossRef] [Green Version]
- Esain, V.; Kwan, W.; Carroll, K.J.; Cortes, M.; Liu, S.Y.; Frechette, G.M.; Sheward, L.M.V.; Nissim, S.; Goessling, W.; North, T.E. Cannabinoid Receptor-2 Regulates Embryonic Hematopoietic Stem Cell Development via Prostaglandin E2 and P-Selectin Activity. Stem Cells 2015, 33, 2596–2612. [Google Scholar] [CrossRef] [Green Version]
- Kwan, W.; Cortes, M.; Frost, I.; Esain, V.; Theodore, L.N.; Liu, S.Y.; Budrow, N.; Goessling, W.; North, T.E. The Central Nervous System Regulates Embryonic HSPC Production via Stress-Responsive Glucocorticoid Receptor Signaling. Cell Stem Cell 2016, 19, 370–382. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rhinn, M.; Dollé, P. Retinoic Acid Signalling during Development. Development 2012, 139, 843–858. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chanda, B.; Ditadi, A.; Iscove, N.N.; Keller, G. XRetinoic Acid Signaling Is Essential for Embryonic Hematopoietic Stem Cell Development. Cell 2013, 155, 215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Jong, J.L.O.; Davidson, A.J.; Wang, Y.; Palis, J.; Opara, P.; Pugach, E.; Daley, G.Q.; Zon, L.I. Interaction of Retinoic Acid and Scl Controls Primitive Blood Development. Blood 2010, 116, 201–209. [Google Scholar] [CrossRef] [Green Version]
- Ma, A.C.H.; Chung, M.I.S.; Liang, R.; Leung, A.Y.H. A DEAB-Sensitive Aldehyde Dehydrogenase Regulates Hematopoietic Stem and Progenitor Cells Development during Primitive Hematopoiesis in Zebrafish Embryos. Leukemia 2010, 24, 2090–2099. [Google Scholar] [CrossRef] [PubMed]
- Pillay, L.M.; Mackowetzky, K.J.; Widen, S.A.; Waskiewicz, A.J. Somite-Derived Retinoic Acid Regulates Zebrafish Hematopoietic Stem Cell Formation. PLoS ONE 2016, 11. [Google Scholar] [CrossRef] [PubMed]
- Philip Creamer, J.; Dege, C.; Ren, Q.; Ho, J.T.K.; Valentine, M.C.; Druley, T.E.; Sturgeon, C.M. Human Definitive Hematopoietic Specification from Pluripotent Stem Cells Is Regulated by Mesodermal Expression of CDX4. Blood 2017, 129, 2988–2992. [Google Scholar] [CrossRef]
- Davidson, A.J.; Ernst, P.; Wang, Y.; Dekens, M.P.S.; Kingsley, P.D.; Palis, J.; Korsmeyer, S.J.; Daley, G.Q.; Zon, L.I. Cdx4 Mutants Fail to Specify Blood Progenitors and Can Be Rescued by Multiple Hox Genes. Nature 2003, 425, 300–306. [Google Scholar] [CrossRef] [PubMed]
- Davidson, A.J.; Zon, L.I. The Caudal-Related Homeobox Genes Cdx1a and Cdx4 Act Redundantly to Regulate Hox Gene Expression and the Formation of Putative Hematopoietic Stem Cells during Zebrafish Embryogenesis. Dev. Biol. 2006, 292, 506–518. [Google Scholar] [CrossRef] [Green Version]
- Espín-Palazón, R.; Stachura, D.L.; Campbell, C.A.; García-Moreno, D.; Del Cid, N.; Kim, A.D.; Candel, S.; Meseguer, J.; Mulero, V.; Traver, D. Proinflammatory Signaling Regulates Hematopoietic Stem Cell Emergence. Cell 2014, 159, 1070–1085. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Esain, V.; Teng, L.; Xu, J.; Kwan, W.; Frost, I.M.; Yzaguirre, A.D.; Cai, X.; Cortes, M.; Maijenburg, M.W.; et al. Inflammatory Signaling Regulates Embryonic Hematopoietic Stem and Progenitor Cell Production. Genes Dev. 2014, 28, 2597–2612. [Google Scholar] [CrossRef] [Green Version]
- Sawamiphak, S.; Kontarakis, Z.; Stainier, D.Y.R. Interferon Gamma Signaling Positively Regulates Hematopoietic Stem Cell Emergence. Dev. Cell 2014, 31, 640–653. [Google Scholar] [CrossRef] [Green Version]
- He, Q.; Zhang, C.; Wang, L.; Zhang, P.; Ma, D.; Lv, J.; Liu, F. Inflammatory Signaling Regulates Hematopoietic Stem and Progenitor Cell Emergence in Vertebrates. Blood 2015, 125, 1098–1106. [Google Scholar] [CrossRef] [Green Version]
- Lim, S.E.; Esain, V.; Kwan, W.; Theodore, L.N.; Cortes, M.; Frost, I.M.; Liu, S.Y.; North, T.E. HIF1α-Induced PDGFRβ Signaling Promotes Developmental HSC Production via IL-6 Activation. Exp. Hematol. 2017, 46, 83–95.e6. [Google Scholar] [CrossRef] [Green Version]
- Frame, J.M.; Kubaczka, C.; Long, T.L.; Esain, V.; Soto, R.A.; Hachimi, M.; Jing, R.; Shwartz, A.; Goessling, W.; Daley, G.Q.; et al. Metabolic Regulation of Inflammasome Activity Controls Embryonic Hematopoietic Stem and Progenitor Cell Production. Dev. Cell 2020. [Google Scholar] [CrossRef]
- Tyrkalska, S.D.; Pérez-Oliva, A.B.; Rodríguez-Ruiz, L.; Martínez-Morcillo, F.J.; Alcaraz-Pérez, F.; Martínez-Navarro, F.J.; Lachaud, C.; Ahmed, N.; Schroeder, T.; Pardo-Sánchez, I.; et al. Inflammasome Regulates Hematopoiesis through Cleavage of the Master Erythroid Transcription Factor GATA1. Immunity 2019, 51, 50–63.e5. [Google Scholar] [CrossRef] [PubMed]
- Lefkopoulos, S.; Polyzou, A.; Derecka, M.; Bergo, V.; Clapes, T.; Cauchy, P.; Jerez-Longres, C.; Onishi-Seebacher, M.; Yin, N.; Martagon-Calderón, N.A.; et al. Repetitive Elements Trigger RIG-I-like Receptor Signaling That Regulates the Emergence of Hematopoietic Stem and Progenitor Cells. Immunity 2020, 53, 934–951.e9. [Google Scholar] [CrossRef] [PubMed]
- Weinreb, J.T.; Ghazale, N.; Pradhan, K.; Gupta, V.; Potts, K.S.; Tricomi, B.; Daniels, N.J.; Padgett, R.A.; De Oliveira, S.; Verma, A.; et al. Excessive R-Loops Trigger an Inflammatory Cascade Leading to Increased HSPC Production. Dev. Cell 2021, 56, 627–640.e5. [Google Scholar] [CrossRef]
- Solaimani Kartalaei, P.; Yamada-Inagawa, T.; Vink, C.S.; de Pater, E.; van der Linden, R.; Marks-Bluth, J.; van der Sloot, A.; van den Hout, M.; Yokomizo, T.; van Schaick-Solernó, M.L.; et al. Whole-Transcriptome Analysis of Endothelial to Hematopoietic Stem Cell Transition Reveals a Requirement for Gpr56 in HSC Generation. J. Exp. Med. 2015, 212, 93–106. [Google Scholar] [CrossRef] [Green Version]
- Maglitto, A.; Mariani, S.A.; de Pater, E.; Rodriguez-Seoane, C.; Vink, C.S.; Piao, X.; Lukke, M.L.; Dzierzak, E. Unexpected Redundancy of Gpr56 and Gpr97 during Hematopoietic Cell Development and Differentiation. Blood Adv. 2021, 5, 829–842.e5. [Google Scholar] [CrossRef]
- Zhang, P.; He, Q.; Chen, D.; Liu, W.; Wang, L.; Zhang, C.; Ma, D.; Li, W.; Liu, B.; Liu, F. G Protein-Coupled Receptor 183 Facilitates Endothelial-to-Hematopoietic Transition via Notch1 Inhibition. Cell Res. 2015, 25, 1093–1107. [Google Scholar] [CrossRef] [Green Version]
- Kwon, H.B.; Mackie, D.I.; Bonnavion, R.; Mercier, A.L.; Helker, C.S.M.; Son, T.; Guenter, S.; Serafin, D.S.; Kim, K.W.; Offermanns, S.; et al. The Orphan G-Protein Coupled Receptor 182 Is a Negative Regulator of Definitive Hematopoiesis through Leukotriene B4 Signaling. ACS Pharmacol. Transl. Sci. 2020, 3, 676–689. [Google Scholar] [CrossRef]
- Xue, Y.; Liu, D.; Cui, G.; Ding, Y.; Ai, D.; Gao, S.; Zhang, Y.; Suo, S.; Wang, X.; Lv, P.; et al. A 3D Atlas of Hematopoietic Stem and Progenitor Cell Expansion by Multi-Dimensional RNA-Seq Analysis. Cell Rep. 2019, 27, 1567–1578.e5. [Google Scholar] [CrossRef] [Green Version]
- Yvernogeau, L.; Klaus, A.; Maas, J.; Morin-Poulard, I.; Weijts, B.; Schulte-Merker, S.; Berezikov, E.; Junker, J.P.; Robin, C. Multispecies RNA Tomography Reveals Regulators of Hematopoietic Stem Cell Birth in the Embryonic Aorta. Blood 2020, 136, 831–844. [Google Scholar] [CrossRef] [PubMed]
- Baron, C.S.; Kester, L.; Klaus, A.; Boisset, J.C.; Thambyrajah, R.; Yvernogeau, L.; Kouskoff, V.; Lacaud, G.; Van Oudenaarden, A.; Robin, C. Single-Cell Transcriptomics Reveal the Dynamic of Haematopoietic Stem Cell Production in the Aorta. Nat. Commun. 2018, 9, 2517. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.; He, J.; Bai, Z.; Li, Z.; Gong, Y.; Liu, C.; Ni, Y.; Du, J.; Ma, C.; Bian, L.; et al. Tracing the First Hematopoietic Stem Cell Generation in Human Embryo by Single-Cell RNA Sequencing. Cell Res. 2019, 29, 881–894. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, M.J.; Lummertz da Rocha, E.; Cahan, P.; Kubaczka, C.; Hunter, P.; Sousa, P.; Mullin, N.K.; Fujiwara, Y.; Nguyen, M.; Tan, Y.; et al. Transcriptome Dynamics of Hematopoietic Stem Cell Formation Revealed Using a Combinatorial Runx1 and Ly6a Reporter System. Stem Cell Rep. 2020. [Google Scholar] [CrossRef]
- Weinreb, C.; Rodriguez-Fraticelli, A.; Camargo, F.D.; Klein, A.M. Lineage Tracing on Transcriptional Landscapes Links State to Fate during Differentiation. Science 2020, 367. [Google Scholar] [CrossRef] [PubMed]
- Lummertz da Rocha, E.; Rowe, R.G.; Lundin, V.; Malleshaiah, M.; Jha, D.K.; Rambo, C.R.; Li, H.; North, T.E.; Collins, J.J.; Daley, G.Q. Reconstruction of Complex Single-Cell Trajectories Using CellRouter. Nat. Commun. 2018, 9, 892. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herman, J.S.; Sagar; Grün, D. FateID Infers Cell Fate Bias in Multipotent Progenitors from Single-Cell RNA-Seq Data. Nat. Methods 2018, 15, 379–386. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.; Cahan, P. SingleCellNet: A Computational Tool to Classify Single Cell RNA-Seq Data across Platforms and Across Species. Cell Syst. 2019, 9, 207–213.e2. [Google Scholar] [CrossRef]
- Macaulay, I.C.; Svensson, V.; Labalette, C.; Ferreira, L.; Hamey, F.; Voet, T.; Teichmann, S.A.; Cvejic, A. Single-Cell RNA-Sequencing Reveals a Continuous Spectrum of Differentiation in Hematopoietic Cells. Cell Rep. 2016, 14, 966–977. [Google Scholar] [CrossRef] [Green Version]
- Tang, Q.; Iyer, S.; Lobbardi, R.; Moore, J.C.; Chen, H.; Lareau, C.; Hebert, C.; Shaw, M.L.; Neftel, C.; Suva, M.L.; et al. Dissecting Hematopoietic and Renal Cell Heterogeneity in Adult Zebrafish at Single-Cell Resolution Using RNA Sequencing. J. Exp. Med. 2017, 214, 2875–2887. [Google Scholar] [CrossRef]
- Kobayashi, I.; Kondo, M.; Yamamori, S.; Kobayashi-Sun, J.; Taniguchi, M.; Kanemaru, K.; Katakura, F.; Traver, D. Enrichment of Hematopoietic Stem/Progenitor Cells in the Zebrafish Kidney. Sci. Rep. 2019, 9. [Google Scholar] [CrossRef] [Green Version]
- Xia, J.; Kang, Z.; Xue, Y.; Ding, Y.; Gao, S.; Zhang, Y.; Lv, P.; Wang, X.; Ma, D.; Wang, L.; et al. A Single-Cell Resolution Developmental Atlas of Hematopoietic Stem and Progenitor Cell Expansion in Zebrafish. Proc. Natl. Acad. Sci. USA 2021, 118, e2015748118. [Google Scholar] [CrossRef]
- Gao, P.; Chen, C.; Howell, E.D.; Li, Y.; Tober, J.; Uzun, Y.; He, B.; Gao, L.; Zhu, Q.; Siekmann, A.F.; et al. Transcriptional Regulatory Network Controlling the Ontogeny of Hematopoietic Stem Cells. Genes Dev. 2020. [Google Scholar] [CrossRef]
- Bonkhofer, F.; Rispoli, R.; Pinheiro, P.; Krecsmarik, M.; Schneider-Swales, J.; Tsang, I.H.C.; de Bruijn, M.; Monteiro, R.; Peterkin, T.; Patient, R. Blood Stem Cell-Forming Haemogenic Endothelium in Zebrafish Derives from Arterial Endothelium. Nat. Commun. 2019, 10, 3577. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Efremova, M.; Vento-Tormo, M.; Teichmann, S.A.; Vento-Tormo, R. CellPhoneDB: Inferring Cell–Cell Communication from Combined Expression of Multi-Subunit Ligand–Receptor Complexes. Nat. Protoc. 2020, 15, 1484–1506. [Google Scholar] [CrossRef]
- Browaeys, R.; Saelens, W.; Saeys, Y. NicheNet: Modeling Intercellular Communication by Linking Ligands to Target Genes. Nat. Methods 2020, 17, 159–162. [Google Scholar] [CrossRef]
- Zhu, Q.; Gao, P.; Tober, J.; Bennett, L.; Chen, C.; Uzun, Y.; Li, Y.; Howell, E.D.; Mumau, M.; Yu, W.; et al. Developmental Trajectory of Prehematopoietic Stem Cell Formation from Endothelium. Blood 2020, 136, 845–856. [Google Scholar] [CrossRef]
- Oatley, M.; Bölükbası, Ö.V.; Svensson, V.; Shvartsman, M.; Ganter, K.; Zirngibl, K.; Pavlovich, P.V.; Milchevskaya, V.; Foteva, V.; Natarajan, K.N.; et al. Single-Cell Transcriptomics Identifies CD44 as a Marker and Regulator of Endothelial to Haematopoietic Transition. Nat. Commun. 2020, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, Y.; Hirschi, K.K. Regulation of Hemogenic Endothelial Cell Development and Function. Annu. Rev. Physiol. 2021, 10, 17–37. [Google Scholar] [CrossRef]
- Chen, Q.; Liu, Y.; Jeong, H.-W.; Stehling, M.; Dinh, V.V.; Zhou, B.; Adams, R.H. Apelin+ Endothelial Niche Cells Control Hematopoiesis and Mediate Vascular Regeneration after Myeloablative Injury. Cell Stem Cell 2019. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tamplin, O.J.; Durand, E.M.; Carr, L.A.; Childs, S.J.; Hagedorn, E.J.; Li, P.; Yzaguirre, A.D.; Speck, N.A.; Zon, L.I. Hematopoietic Stem Cell Arrival Triggers Dynamic Remodeling of the Perivascular Niche. Cell 2015, 160, 241–252. [Google Scholar] [CrossRef] [Green Version]
- Xue, Y.; Lv, J.; Zhang, C.; Wang, L.; Ma, D.; Liu, F. The Vascular Niche Regulates Hematopoietic Stem and Progenitor Cell Lodgment and Expansion via Klf6a-Ccl25b. Dev. Cell 2017, 42, 349–362.e4. [Google Scholar] [CrossRef]

| Last Author, Year | Species | Type of Sequencing | Sorted Population(s) | Accession Number(s) | Ref. |
|---|---|---|---|---|---|
| Zhang et al., 2015 | zebrafish | bulk RNAseq | flk1:mCherry+ (ECs), flk1:mCherry+/ runx1en:GFP+ (HE); and runx1en:GFP+(HSPCs) | N/A | [144] |
| Kartalaei et al., 2015 | mouse | bulk RNAseq | E10.5 AGM ECs(CD31+, cKit−,Ly6aGFP−), HE (CD31+, cKit−,Ly6aGFP+), HSCs(CD31+, cKit+,Ly6aGF+) and HPs( CD31+, cKit+,Ly6aGF+) | GSE63316 | [142] |
| Bonkhofer et al., 2019 | zebrafish | bulk RNAseq and ATACseq | TgBAC(runx1P2:Citrine);Tg(kdrl: mCherry) to sort for HE, non-HE arterial ECs and venous ECs, including runx1 morphants | GSE132259, GSE132258 | [160] |
| Baron et al., 2018 | mouse | scRNA-seq | CD31+, cKit+ cells from E10 and E11 aorta after intra-aortic antibody staining, together with other aortic subfractions by surface markers | GSE112642 | [148] |
| Zeng et al., 2019 | human | scRNA-seq | Dissected AGM from ~30-day old human embryo (depleted of red blood cells) | GSE135202 | [149] |
| Yvernogeau et al., 2020 | zebrafish, mouse, chicken, human | Tomo-seq | zebrafish Tg(kdrl:mCherry/cd41:EGFP) at 28- and 40-hpf for HE/HSPC identification through trunk; E10.5 and E11.5 mouse trunk, E3 chicken embryo trunk, 35-day-old human embryo trunk | N/A | [147] |
| Chen et al., 2020 | mouse | scRNA-seq | Lin−, cKit+ cells with both Runx1-mKO2 and Ly6a-GFP transgenic reporters (HSCs) | GSE145638 | [150] |
| Zhu et al., 2020 | mouse | scRNA-seq | Purified EC, HE and intra-aortic cluster cells with surface markers and Runx1-GFP expression | GSE137117 | [163] |
| Oatley et al., 2020 | mouse | scRNA-seq | VE-cadherin+ cells from E10 AGM | E-MTAB-6987 | [164] |
| Kasper et al., 2020 | zebrafish | scRNA-seq | Dissected trunks from 27hpf wildtype and miR-223 mutants, sorted on Tg(kdrl:mCherry) ECs (also has miR-223 GFP+ population for clustering) | GSE135246 | [118] |
| Soto et al., 2021 | zebrafish | scRNA-seq | Tg(kdrl:EGFP) ECs at 30hpf from ezh1 wildtype, heterozygous and homozygous mutants | GSE173972 | [56] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sugden, W.W.; North, T.E. Making Blood from the Vessel: Extrinsic and Environmental Cues Guiding the Endothelial-to-Hematopoietic Transition. Life 2021, 11, 1027. https://doi.org/10.3390/life11101027
Sugden WW, North TE. Making Blood from the Vessel: Extrinsic and Environmental Cues Guiding the Endothelial-to-Hematopoietic Transition. Life. 2021; 11(10):1027. https://doi.org/10.3390/life11101027
Chicago/Turabian StyleSugden, Wade W., and Trista E. North. 2021. "Making Blood from the Vessel: Extrinsic and Environmental Cues Guiding the Endothelial-to-Hematopoietic Transition" Life 11, no. 10: 1027. https://doi.org/10.3390/life11101027
APA StyleSugden, W. W., & North, T. E. (2021). Making Blood from the Vessel: Extrinsic and Environmental Cues Guiding the Endothelial-to-Hematopoietic Transition. Life, 11(10), 1027. https://doi.org/10.3390/life11101027






