Recent Advances in the Photoautotrophic Metabolism of Cyanobacteria: Biotechnological Implications
Abstract
1. Introduction
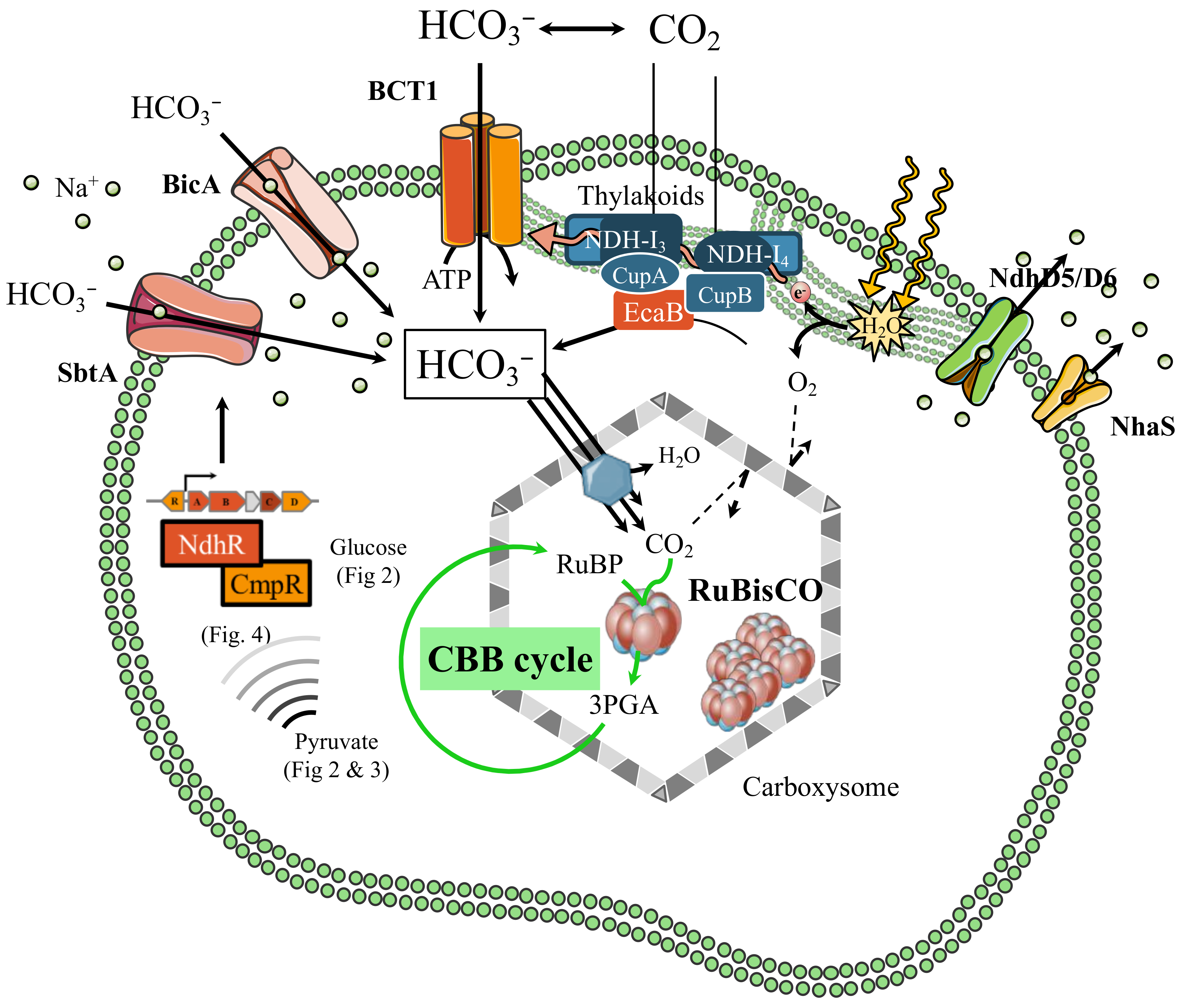
2. Cyanobacteria Perform Oxygenic Photosynthesis to Power up Their Photoautotrophic Metabolism
3. RubisCO: The Key Enzyme Player in CO2 Fixation
4. Carboxysome A Complex Intracellular Compartment for CO2 Fixation
4.1. Carboxysomes Stochiometry
4.2. Carboxysome Positioning Systems
5. Phosphoenol-Pyruvate-Carboxylase: Another Important Player in CO2 Fixation
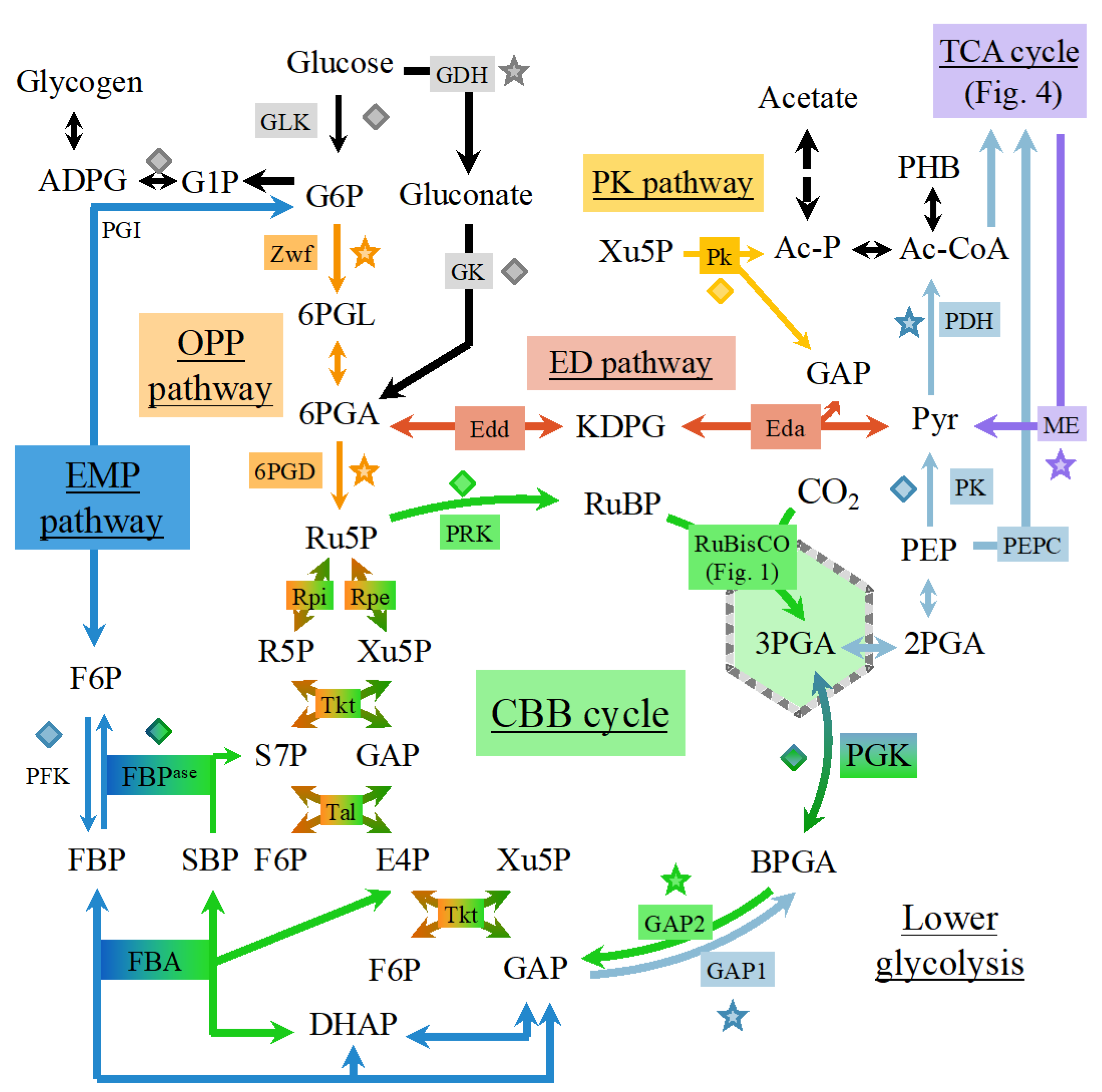
6. Plasticity of the Cyanobacterial Carbon Metabolism
6.1. Occurrence of Multiple Glycolytic Pathways
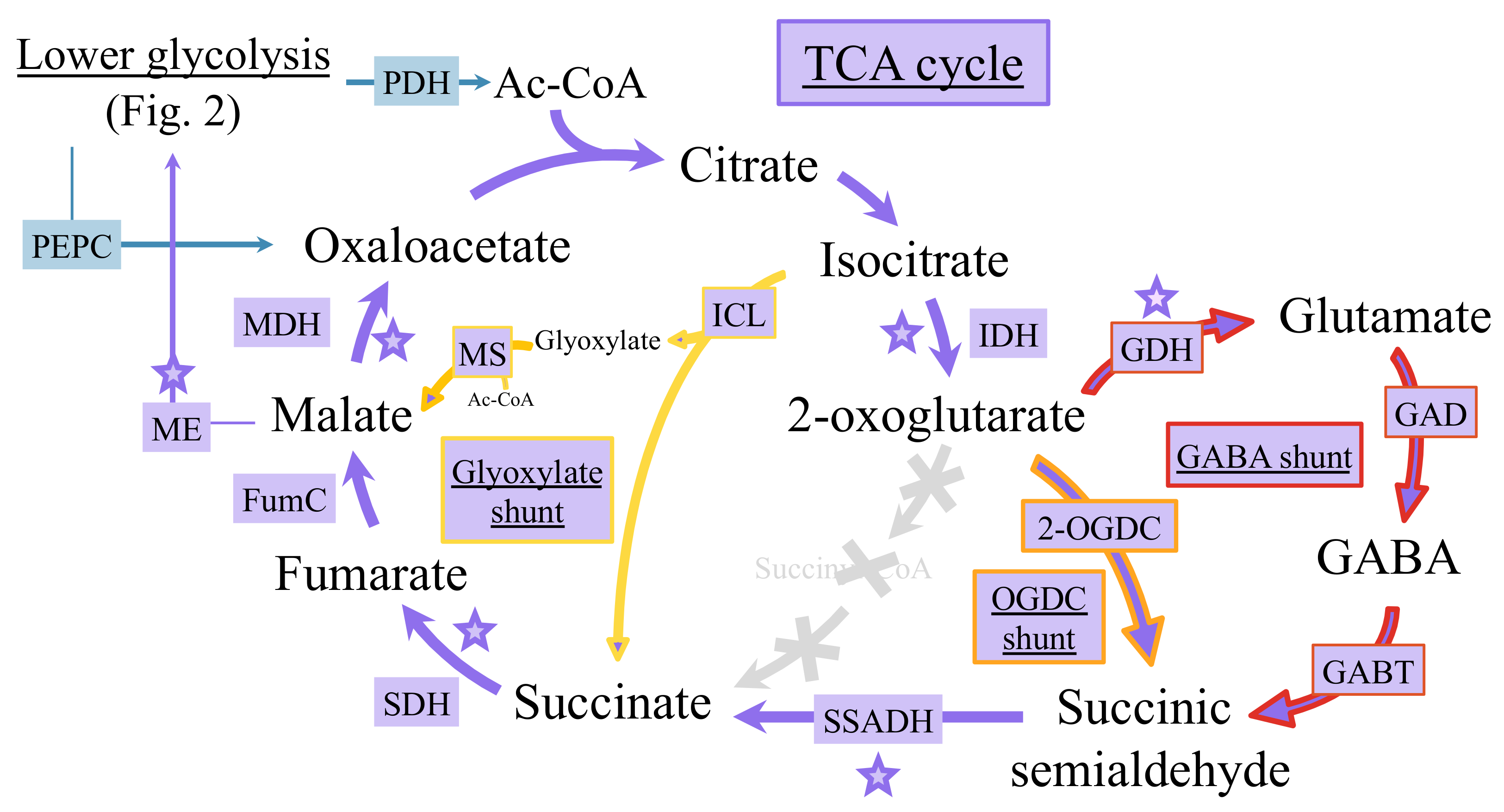
6.2. Occurrence of Multiple Tricarboxylic Acid Cycles
7. Nitrogen Fixation and Assimilation
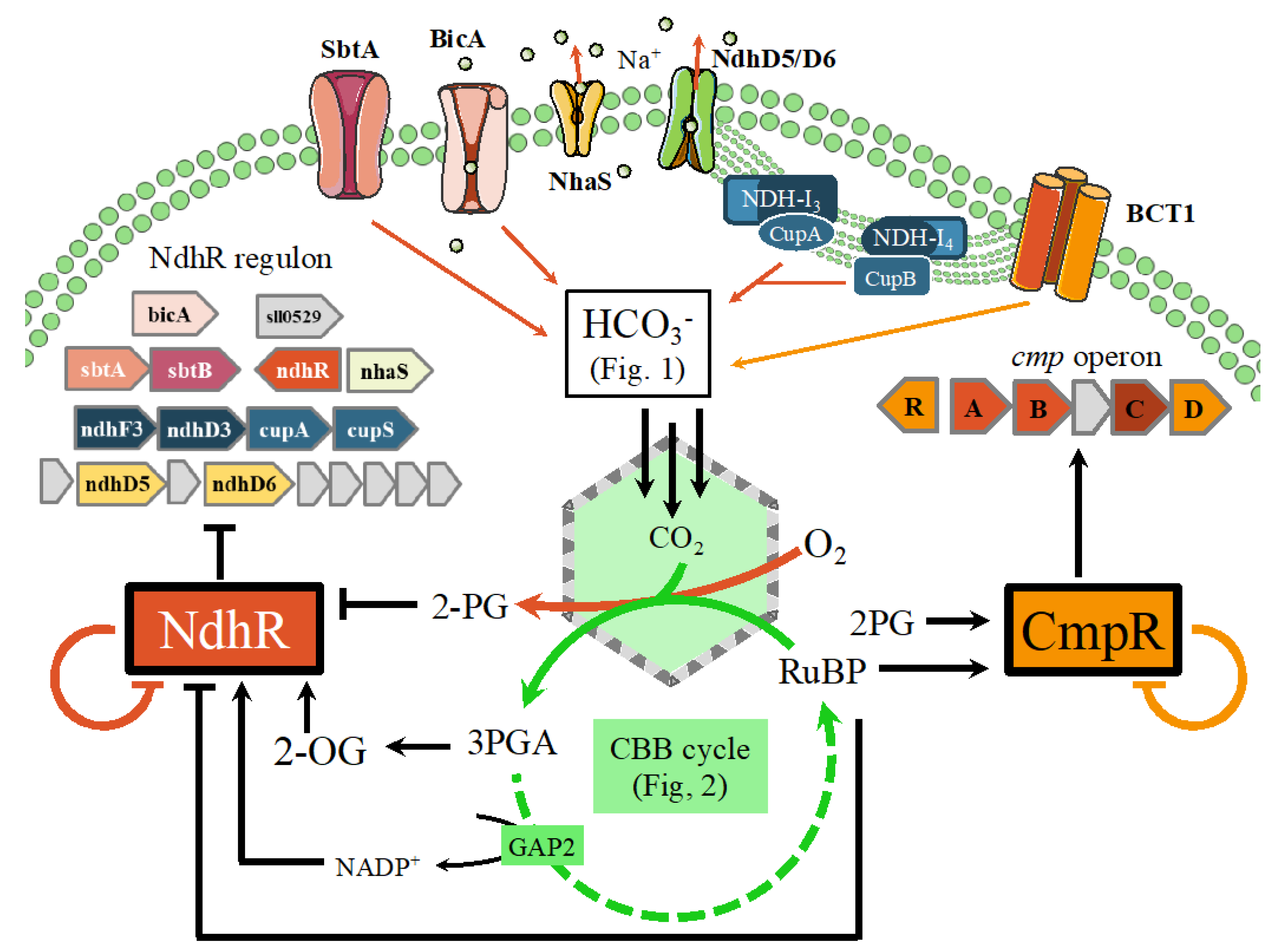
8. Coordination of the Carbon and Nitrogen Metabolisms
8.1. Role of Key Transcription Regulators (NdhR, CmpR, AbrB2, and LexA) and Associated Key Metabolites (2-oxoglutarate and 2-phosphoglycolate)
8.1.1. Role of the NdhR Transcription Factor
8.1.2. Role of the CmpR Transcription Factor
8.1.3. Roles of the AbrB2 Transcription Factor
8.1.4. Roles of the LexA Transcription Factor
8.2. Roles of the Key Transcription Factor NtcA and the Regulatory Proteins PII, PipX
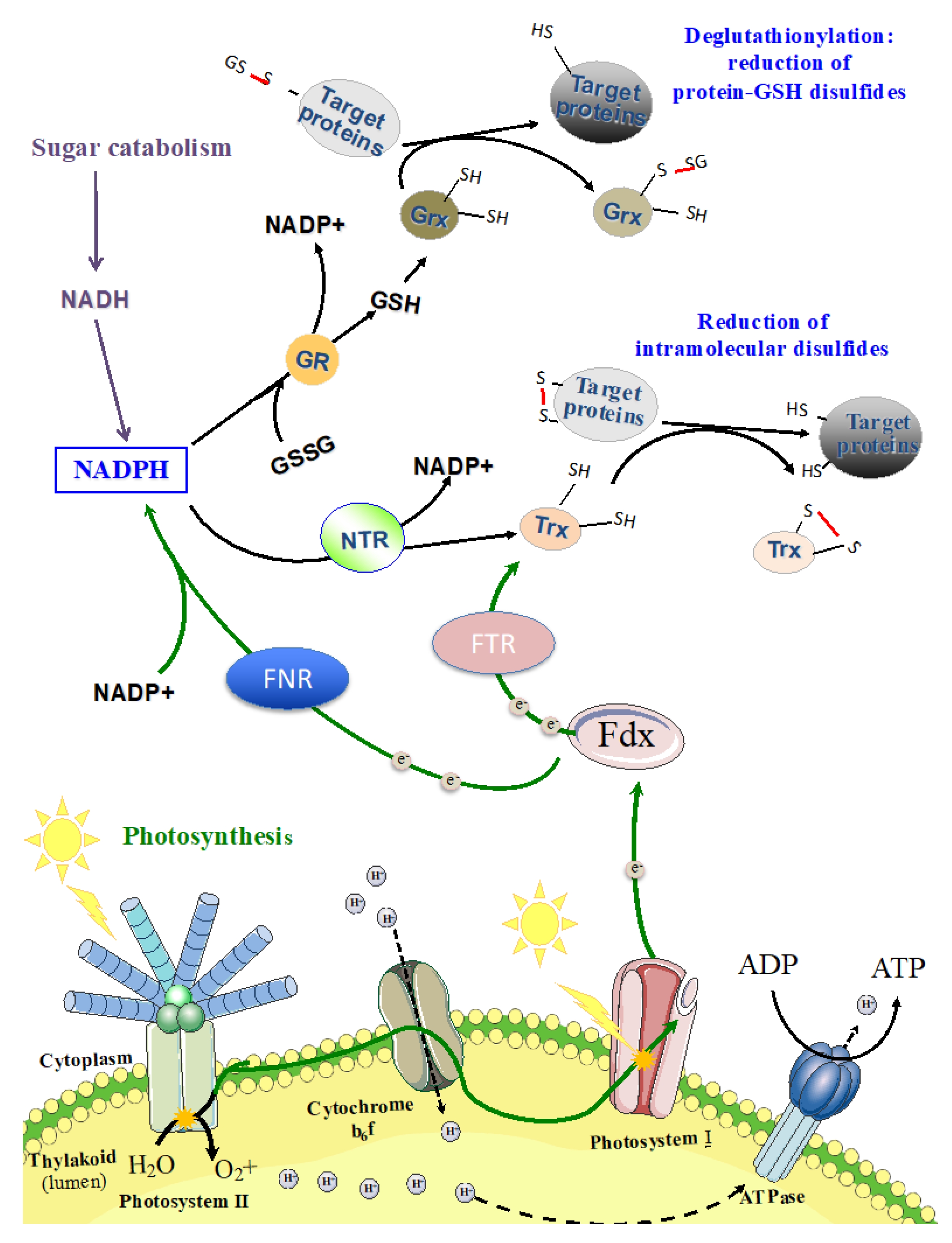
9. Redox Regulation of the Central Metabolism
9.1. Prominent Role of the Redox-Responsive CP12 Protein
9.2. Glutathionylation: An Overlooked Mechanism Operating in the Regulation of the Central Metabolism
10. Cyanobacteria Facing Carbon/Nitrogen Imbalance can Produce Exopolysaccharides and Biodegradable Plastics of Biotechnological Interest
11. Concluding Remarks
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Schopf, J.W. The paleobiological record of photosynthesis. Photosynth. Res. 2011, 107, 87–101. [Google Scholar] [CrossRef] [PubMed]
- Ponce-Toledo, R.I.; Deschamps, P.; López-García, P.; Zivanovic, Y.; Benzerara, K.; Moreira, D. An Early-Branching Freshwater Cyanobacterium at the Origin of Plastids. Curr. Biol. 2017, 27, 386–391. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, T.L.; Bryant, D.A.; Macalady, J.L. The role of biology in planetary evolution: Cyanobacterial primary production in low-oxygen Proterozoic oceans. Environ. Microbiol. 2016, 325–340. [Google Scholar] [CrossRef] [PubMed]
- Dai, W.; Chen, M.; Myers, C.; Ludtke, S.J.; Pettitt, B.M.; King, J.A.; Schmid, M.F.; Chiu, W. Visualizing Individual RuBisCO and Its Assembly into Carboxysomes in Marine Cyanobacteria by Cryo-Electron Tomography. J. Mol. Biol. 2018, 430, 4156–4167. [Google Scholar] [CrossRef]
- Singh, J.S.; Kumar, A.; Rai, A.N.; Singh, D.P. Cyanobacteria: A precious bio-resource in agriculture, ecosystem, and environmental sustainability. Front. Microbiol. 2016, 7. [Google Scholar] [CrossRef]
- Cassier-Chauvat, C.; Chauvat, F. Cyanobacteria: Wonderful Microorganisms for Basic and Applied Research. eLS 2018. [Google Scholar] [CrossRef]
- Nakamura, Y.; Kaneko, T.; Sato, S.; Mimuro, M.; Miyashita, H.; Tsuchiya, T.; Sasamoto, S.; Watanabe, A.; Kawashima, K.; Kishida, Y.; et al. Complete Genome Structure of Gloeobacter violaceus PCC 7421, a Cyanobacterium that Lacks Thylakoids. DNA Res. 2003, 10, 137–145. [Google Scholar] [CrossRef]
- Cassier-Chauvat, C.; Chauvat, F. Cell division in cyanobacteria. In The cell Biology of Cyanobacteria; Flores, E., Herrero, A., Eds.; Caister Academic Press: Norfolk, UK, 2014. [Google Scholar]
- Cassier-Chauvat, C.; Veaudor, T.; Chauvat, F. Comparative genomics of DNA recombination and repair in cyanobacteria: Biotechnological implications. Fronti. Microbiol. 2016, 7. [Google Scholar] [CrossRef]
- Narainsamy, K.; Farci, S.; Braun, E.; Junot, C.; Cassier-Chauvat, C.; Chauvat, F. Oxidative-stress detoxification and signalling in cyanobacteria: The crucial glutathione synthesis pathway supports the production of ergothioneine and ophthalmate. Mol. Microbiol. 2016, 100, 15–24. [Google Scholar] [CrossRef]
- Demay, J.; Bernard, C.; Reinhardt, A.; Marie, B. Natural products from cyanobacteria: Focus on beneficial activities. Mar. Drugs 2019, 17, 320. [Google Scholar] [CrossRef]
- Pearson, L.A.; Dittmann, E.; Mazmouz, R.; Ongley, S.E.; D’Agostino, P.M.; Neilan, B.A. The genetics, biosynthesis and regulation of toxic specialized metabolites of cyanobacteria. Harmful Algae 2016, 54, 98–111. [Google Scholar] [CrossRef] [PubMed]
- Panjiar, N.; Mishra, S.; Yadav, A.N.; Verma, P. Functional Foods from Cyanobacteria. In Microbial Functional Foods and Nutraceuticals; Gupta, V.K., Treichel, H., Shapaval, V.O., de Oliveira, L.A., Tuohy, M.G., Eds.; Wiley: New York, NY, USA, 2017. [Google Scholar]
- Verseux, C.; Baqué, M.; Lehto, K.; De Vera, J.P.P.; Rothschild, L.J.; Billi, D. Sustainable life support on Mars-The potential roles of cyanobacteria. Int. J. Astrobiol. 2016, 15, 65–92. [Google Scholar] [CrossRef]
- Dittmann, E.; Gugger, M.; Sivonen, K.; Fewer, D.P. Natural Product Biosynthetic Diversity and Comparative Genomics of the Cyanobacteria. Trends Microbiol. 2015, 23, 642–652. [Google Scholar] [CrossRef] [PubMed]
- Cassier-Chauvat, C.; Dive, V.; Chauvat, F. Cyanobacteria: Photosynthetic factories combining biodiversity, radiation resistance, and genetics to facilitate drug discovery. Appl. Microbiol. Biotechnol. 2017, 101, 1359–1364. [Google Scholar] [CrossRef] [PubMed]
- Knoot, C.J.; Ungerer, J.; Wangikar, P.P.; Pakrasi, H.B. Cyanobacteria: Promising biocatalysts for sustainable chemical production. J. Biol. Chem. 2018, 293, 5044–5052. [Google Scholar] [CrossRef]
- Lin, P.C.; Pakrasi, H.B. Engineering cyanobacteria for production of terpenoids. Planta 2019, 249, 145–154. [Google Scholar] [CrossRef]
- Ducat, D.C.; Way, J.C.; Silver, P.A. Engineering cyanobacteria to generate high-value products. Trends Biotechnol. 2011, 29, 95–103. [Google Scholar] [CrossRef]
- Jansson, C.; Northen, T. Calcifying cyanobacteria-the potential of biomineralization for carbon capture and storage. Curr. Opin. Biotechnol. 2010, 21, 365–371. [Google Scholar] [CrossRef]
- Cassier-Chauvat, C.; Veaudor, T.; Chauvat, F. Advances in the function and regulation of hydrogenase in the cyanobacterium Synechocystis PCC6803. Int. J. Mol. Sci. 2014, 15, 19938–19951. [Google Scholar] [CrossRef]
- Magnuson, A. Heterocyst thylakoid bioenergetics. Life 2019, 9, 13. [Google Scholar] [CrossRef]
- Santos-Merino, M.; Singh, A.K.; Ducat, D.C. New applications of synthetic biology tools for cyanobacterial metabolic engineering. Front. Bioeng. Biotechnol. 2019, 7. [Google Scholar] [CrossRef] [PubMed]
- Veaudor, T.; Cassier-Chauvat, C.; Chauvat, F. Genomics of Urea Transport and Catabolism in Cyanobacteria: Biotechnological Implications. Front. Microbiol. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Lambert, D.H.; Stevens, S.E. Photoheterotrophic growth of Agmenellum quadruplicatum PR-6. J. Bacteriol. 1986, 165. [Google Scholar] [CrossRef] [PubMed]
- da Silva, G.P.; Mack, M.; Contiero, J. Glycerol: A promising and abundant carbon source for industrial microbiology. Biotechnol. Adv. 2009, 27, 30–39. [Google Scholar] [CrossRef] [PubMed]
- Rippka, R.; Deruelles, J.; Waterbury, J.B.; Herdman, M.; Stanier, R.Y. Generic assignments, strain histories and properties of pure cultures of cyano- bacteria. Journal of General Microbiology 11: 1-61. J. Gen. Microbiol. 1979, 111, 1–61. [Google Scholar] [CrossRef]
- Magdaong, N.C.M.; Blankenship, R.E. Photoprotective, excited-state quenching mechanisms in diverse photosynthetic organisms. J. Biol. Chem. 2018, 293, 5018–5025. [Google Scholar] [CrossRef] [PubMed]
- Barber, J. Photosynthetic water splitting by the Mn4Ca2+OX catalyst of photosystem II: Its structure, robustness and mechanism. Q. Rev. Biophys. 2017, 50. [Google Scholar] [CrossRef]
- Gao, J.; Wang, H.; Yuan, Q.; Feng, Y. Structure and function of the photosystem supercomplexes. Front. Plant Sci. 2018, 9. [Google Scholar] [CrossRef]
- Klatt, J.M.; Al-Najjar, M.A.A.; Yilmaz, P.; Lavik, G.; de Beer, D.; Polerecky, L. Anoxygenic photosynthesis controls oxygenic photosynthesis in a cyanobacterium from a sulfidic spring. Appl. Environ. Microbiol. 2015, 81, 2025–2031. [Google Scholar] [CrossRef]
- Bracher, A.; Whitney, S.M.; Hartl, F.U.; Hayer-Hartl, M. Biogenesis and Metabolic Maintenance of Rubisco. Annu. Rev. Plant Biol. 2017, 68, 29–60. [Google Scholar] [CrossRef]
- Zhu, B.; Chen, G.; Cao, X.; Wei, D. Molecular characterization of CO2 sequestration and assimilation in microalgae and its biotechnological applications. Bioresour. Technol. 2017, 244, 1207–1215. [Google Scholar] [CrossRef] [PubMed]
- Kolesinski, P.; Rydzy, M.; Szczepaniak, A. Is RAF1 protein from Synechocystis sp. PCC 6803 really needed in the cyanobacterial Rubisco assembly process? Photosynth. Res. 2017, 132, 135–148. [Google Scholar] [CrossRef] [PubMed]
- Hauser, T.; Bhat, J.Y.; Milicic, G.; Wendler, P.; Hartl, F.U.; Bracher, A.; Hayer-Hartl, M. Structure and mechanism of the Rubisco-assembly chaperone Raf1. Nat. Struct. Mol. Biol. 2015, 22, 720–728. [Google Scholar] [CrossRef] [PubMed]
- Forchhammer, K.; Selim, K.A. Carbon/nitrogen homeostasis control in cyanobacteria. FEMS Microbiol. Rev. 2019, 44, 33–53. [Google Scholar] [CrossRef]
- Wilson, R.H.; Martin-Avila, E.; Conlan, C.; Whitney, S.M. An improved Escherichia coli screen for Rubisco identifies a protein-protein interface that can enhance CO2-fixation kinetics. J. Biol. Chem. 2018, 293, 18–27. [Google Scholar] [CrossRef]
- Satagopan, S.; Huening, K.A.; Robert Tabita, F. Selection of Cyanobacterial (Synechococcus sp. Strain PCC 6301) RubisCO Variants with Improved Functional Properties That Confer Enhanced CO2-Dependent Growth of Rhodobacter capsulatus, a Photosynthetic Bacterium. mBio 2019, 10, e01537-19. [Google Scholar] [CrossRef]
- Zhang, A.; Carroll, A.L.; Atsumi, S. Carbon recycling by cyanobacteria: Improving CO2 fixation through chemical production. FEMS Microbiol. Lett. 2017, 364. [Google Scholar] [CrossRef]
- Liang, F.; Lindblad, P. Effects of overexpressing photosynthetic carbon flux control enzymes in the cyanobacterium Synechocystis PCC 6803. Metab. Eng. 2016, 38, 56–64. [Google Scholar] [CrossRef]
- Atsumi, S.; Higashide, W.; Liao, J.C. Direct photosynthetic recycling of carbon dioxide to isobutyraldehyde. Nat. Biotechnol. 2009, 27, 1177–1180. [Google Scholar] [CrossRef]
- Ruffing, A.M. Improved free fatty acid production in cyanobacteria with Synechococcus sp. PCC 7002 as host. Front. Bioeng. Biotechnol. 2014, 2, 1–10. [Google Scholar] [CrossRef]
- Shih, P.M.; Zarzycki, J.; Niyogi, K.K.; Kerfeld, C.A. Introduction of a synthetic CO2-fixing photorespiratory bypass into a cyanobacterium. J. Biol. Chem. 2014, 289, 9493–9500. [Google Scholar] [CrossRef] [PubMed]
- Sommer, M.; Sutter, M.; Gupta, S.; Kirst, H.; Turmo, A.; Lechno-Yossef, S.; Burton, R.L.; Saechao, C.; Sloan, N.B.; Cheng, X.; et al. Heterohexamers formed by CcmK3 and CcmK4 increase the complexity of beta carboxysome shells. Plant Physiol. 2019, 179, 156–167. [Google Scholar] [CrossRef] [PubMed]
- Kinney, J.N.; Axen, S.D.; Kerfeld, C.A. Comparative analysis of carboxysome shell proteins. Photosynth. Res. 2011, 109, 21–32. [Google Scholar] [CrossRef] [PubMed]
- Montgomery, B.L.; Lechno-Yossef, S.; Kerfeld, C.A. Interrelated modules in cyanobacterial photosynthesis: The carbon-concentrating mechanism, photorespiration, and light perception. J. Exp. Bot. 2016, 67, 2931–2940. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Cai, F.; Dou, Z.; Bernstein, S.L.; Leverenz, R.; Williams, E.B.; Heinhorst, S.; Shively, J.; Cannon, G.C.; Kerfeld, C.A. Advances in understanding carboxysome assembly in Prochlorococcus and Synechococcus implicate CsoS2 as a critical component. Life 2015, 50, 1141–1171. [Google Scholar] [CrossRef] [PubMed]
- Turmo, A.; Gonzalez-Esquer, C.R.; Kerfeld, C.A. Carboxysomes: Metabolic modules for CO2 fixation. FEMS Microbiol. Lett. 2017, 364. [Google Scholar] [CrossRef]
- Wang, H.; Yan, X.; Aigner, H.; Bracher, A.; Nguyen, N.D.; Hee, W.Y.; Long, B.M.; Price, G.D.; Hartl, F.U.; Hayer-Hartl, M. Rubisco condensate formation by CcmM in β-carboxysome biogenesis. Nature 2019, 566. [Google Scholar] [CrossRef]
- Mangan, N.M.; Flamholz, A.; Hood, R.D.; Milo, R.; Savage, D.F. PH determines the energetic efficiency of the cyanobacterial CO2 concentrating mechanism. Proc. Natl. Acad. Sci. USA 2016, 113, 5354–5362. [Google Scholar] [CrossRef]
- Sun, Y.; Wollman, A.J.M.; Huang, F.; Leake, M.C.; Liu, L.N. Single-organelle quantification reveals stoichiometric and structural variability of carboxysomes dependent on the environment. Plant Cell 2019, 31, 1648–1664. [Google Scholar] [CrossRef]
- Huang, F.; Vasieva, O.; Sun, Y.; Faulkner, M.; Dykes, G.F.; Zhao, Z.; Liu, L.N. Roles of RBcX in carboxysome biosynthesis in the cyanobacterium Synechococcus elongatus PCC7942. Plant Physiol. 2019, 179, 184–194. [Google Scholar] [CrossRef]
- Cameron, J.C.; Wilson, S.C.; Bernstein, S.L.; Kerfeld, C.A. Biogenesis of a bacterial organelle: The carboxysome assembly pathway. Cell 2013, 155, 1131–1140. [Google Scholar] [CrossRef] [PubMed]
- MacCready, J.S.; Basalla, J.L.; Vecchiarelli, A.G. Origin and Evolution of Carboxysome Positioning Systems in Cyanobacteria. Mol. Biol. Evol. 2020. [Google Scholar] [CrossRef]
- Takeya, M.; Hirai, M.Y.; Osanai, T. Allosteric inhibition of phosphoenolpyruvate carboxylases is determined by a single amino acid residue in cyanobacteria. Sci. Rep. 2017, 7. [Google Scholar] [CrossRef] [PubMed]
- Angermayr, S.A.; Van Der Woude, A.D.; Correddu, D.; Vreugdenhil, A.; Verrone, V.; Hellingwerf, K.J. Exploring metabolic engineering design principles for the photosynthetic production of lactic acid by Synechocystis sp. PCC6803. Biotechnol. Biofuels 2014, 7. [Google Scholar] [CrossRef] [PubMed]
- Luinenburg, I.; Coleman, J.R. A requirement for phosphoenolpyruvate carboxylase in the cyanobacterium Synechococcus PCC 7942. Arch. Microbiol. 1990, 154, 471–474. [Google Scholar] [CrossRef]
- Hasunuma, T.; Matsuda, M.; Kato, Y.; Vavricka, C.J.; Kondo, A. Temperature enhanced succinate production concurrent with increased central metabolism turnover in the cyanobacterium Synechocystis sp. PCC 6803. Metab. Eng. 2018, 48, 109–120. [Google Scholar] [CrossRef]
- Durall, C.; Lindberg, P.; Yu, J.; Lindblad, P. Increased ethylene production by overexpressing phosphoenolpyruvate carboxylase in the cyanobacterium Synechocystis PCC 6803. Biotechnol. Biofuels 2020, 13. [Google Scholar] [CrossRef]
- Cheah, Y.E.; Xu, Y.; Sacco, S.A.; Babele, P.K.; Zheng, A.O.; Johnson, C.H.; Young, J.D. Systematic identification and elimination of flux bottlenecks in the aldehyde production pathway of Synechococcus elongatus PCC 7942. Metab. Eng. 2020, 60, 56–65. [Google Scholar] [CrossRef]
- Chen, X.; Schreiber, K.; Appel, J.; Makowka, A.; Fähnrich, B.; Roettger, M.; Hajirezaei, M.R.; Sönnichsen, F.D.; Schönheit, P.; Martin, W.F.; et al. The Entner-Doudoroff pathway is an overlooked glycolytic route in cyanobacteria and plants. Proc. Natl. Acad. Sci. USA 2016, 113, 5441–5446. [Google Scholar] [CrossRef]
- Xiong, W.; Cano, M.; Wang, B.; Douchi, D.; Yu, J. The plasticity of cyanobacterial carbon metabolism. Curr. Opin. Chem. Biol. 2017, 41, 12–19. [Google Scholar] [CrossRef]
- Wan, N.; DeLorenzo, D.M.; He, L.; You, L.; Immethun, C.M.; Wang, G.; Baidoo, E.E.K.; Hollinshead, W.; Keasling, J.D.; Moon, T.S.; et al. Cyanobacterial carbon metabolism: Fluxome plasticity and oxygen dependence. Biotechnol. Bioeng. 2017, 114, 1593–1602. [Google Scholar] [CrossRef] [PubMed]
- Kanwal, S.; Incharoensakdi, A. GABA synthesis mediated by γ-aminobutanal dehydrogenase in Synechocystis sp. PCC6803 with disrupted glutamate and α-ketoglutarate decarboxylase genes. Plant Sci. 2020, 290. [Google Scholar] [CrossRef] [PubMed]
- Gründel, M.; Knoop, H.; Steuer, R. Activity and functional properties of the isocitrate lyase in the cyanobacterium Cyanothece sp. PCC 7424. Microbiology (United Kingdom) 2017, 163. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Bryant, D.A. Biochemical validation of the glyoxylate cycle in the cyanobacterium Chlorogloeopsis fritschii strain PCC 9212. J. Biol. Chem. 2015, 290, 14019–14030. [Google Scholar] [CrossRef]
- Esteves-Ferreira, A.A.; Inaba, M.; Fort, A.; Araújo, W.L.; Sulpice, R. Nitrogen metabolism in cyanobacteria: Metabolic and molecular control, growth consequences and biotechnological applications. Crit. Rev. Microbiol. 2018, 44, 541–560. [Google Scholar] [CrossRef]
- Watzer, B.; Spät, P.; Neumann, N.; Koch, M.; Sobotka, R.; MacEk, B.; Hennrich, O.; Forchhammer, K. The signal transduction protein PII controls ammonium, nitrate and urea uptake in cyanobacteria. Front. Microbiol. 2019, 10. [Google Scholar] [CrossRef]
- Giner-Lamia, J.; Robles-Rengel, R.; Hernández-Prieto, M.A.; Isabel Muro-Pastor, M.; Florencio, F.J.; Futschik, M.E. Identification of the direct regulon of NtcA during early acclimation to nitrogen starvation in the cyanobacterium Synechocystis sp. PCC 6803. Nucleic Acids Res. 2017, 45, 11800–11820. [Google Scholar] [CrossRef]
- Veaudor, T.; Cassier-Chauvat, C.; Chauvat, F. Overproduction of the cyanobacterial hydrogenase and selection of a mutant thriving on urea, as a possible step towards the future production of hydrogen coupled with water treatment. PLoS ONE 2018, 13, e0198836. [Google Scholar] [CrossRef]
- Herrero, A.; Stavans, J.; Flores, E. The multicellular nature of filamentous heterocyst-forming cyanobacteria. FEMS Microbiol. Rev. 2016, 40, 831–854. [Google Scholar] [CrossRef]
- Avilan, L.; Roumezi, B.; Risoul, V.; Bernard, C.S.; Kpebe, A.; Belhadjhassine, M.; Rousset, M.; Brugna, M.; Latifi, A. Phototrophic hydrogen production from a clostridial [FeFe] hydrogenase expressed in the heterocysts of the cyanobacterium Nostoc PCC 7120. Appl. Microbiol. Biotechnol. 2018, 102, 5775–5783. [Google Scholar] [CrossRef]
- Robles-Rengel, R.; Florencio, F.J.; Muro-Pastor, M.I. Redox interference in nitrogen status via oxidative stress is mediated by 2-oxoglutarate in cyanobacteria. New Phytol. 2019, 224, 216–228. [Google Scholar] [CrossRef] [PubMed]
- García-Domínguez, M.; Reyes, J.C.; Florencio, F.J. NtcA represses transcription of gifA and gifB, genes that encode inhibitors of glutamine synthetase type I from Synechocystis sp. PCC 6803. Mol. Microbiol. 2000, 35, 1192–1201. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.C.; Zhou, C.Z.; Burnap, R.L.; Peng, L. Carbon/Nitrogen Metabolic Balance: Lessons from Cyanobacteria. Trends Plant Sci. 2018, 23, 1116–1130. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.L.; Wang, X.P.; Sun, H.; Han, S.J.; Li, W.F.; Cui, N.; Lin, G.M.; Zhang, J.Y.; Cheng, W.; Cao, D.D.; et al. Coordinating carbon and nitrogen metabolic signaling through the cyanobacterial global repressor NdhR. Proc. Natl. Acad. Sci. USA 2017, 115, 403–408. [Google Scholar] [CrossRef]
- Domain, F.; Houot, L.; Chauvat, F.; Cassier-Chauvat, C. Function and regulation of the cyanobacterial genes lexA, recA and ruvB: LexA is critical to the survival of cells facing inorganic carbon starvation. Mol. Microbiol. 2004, 53, 65–80. [Google Scholar] [CrossRef]
- Kizawa, A.; Kawahara, A.; Takashima, K.; Takimura, Y.; Nishiyama, Y.; Hihara, Y. The LexA transcription factor regulates fatty acid biosynthetic genes in the cyanobacterium Synechocystis sp. PCC 6803. Plant J. 2017, 92, 89–98. [Google Scholar] [CrossRef]
- Muro-Pastor, A.M.; Hess, W.R. Regulatory RNA at the crossroads of carbon and nitrogen metabolism in photosynthetic cyanobacteria. Biochimica et Biophysica Acta-Gene Regulatory Mechanisms 2020, 1863, 194477. [Google Scholar] [CrossRef]
- Figge, R.M.; Cassier-Chauvat, C.; Chauvat, F.; Cerff, R. Characterization and analysis of an NAD(P)H dehydrogenase transcriptional regulator critical for the survival of cyanobacteria facing inorganic carbon starvation and osmotic stress. Mol. Microbiol. 2001, 39, 455–468. [Google Scholar] [CrossRef]
- Takahashi, Y.; Yamaguchi, O.; Omata, T. Roles of CmpR, a LysR family transcriptional regulator, in acclimation of the cyanobacterium Synechococcus sp. strain PCC 7942 to low-CO2 and high-light conditions. Mol. Microbiol. 2004, 52, 837–845. [Google Scholar] [CrossRef]
- Ishii, A.; Hihara, Y. An AbrB-like transcriptional regulator, Sll0822, is essential for the activation of nitrogen-regulated genes in Synechocystis sp. PCC 6803. Plant Physiol. 2008, 148, 660–670. [Google Scholar] [CrossRef]
- Wang, H.L.; Postier, B.L.; Burnap, R.L. Alterations in Global Patterns of Gene Expression in Synechocystis sp. PCC 6803 in Response to Inorganic Carbon Limitation and the Inactivation of ndhR, a LysR Family Regulator. J. Biol. Chem. 2004, 279, 5739–5751. [Google Scholar] [CrossRef] [PubMed]
- Dutheil, J.; Saenkham, P.; Sakr, S.; Leplat, C.; Ortega-Ramos, M.; Bottin, H.; Cournac, L.; Cassier-Chauvat, C.; Chauvat, F. The AbrB2 autorepressor, expressed from an atypical promoter, represses the hydrogenase operon to regulate hydrogen production in Synechocystis strain PCC6803. J. Bacteriol. 2012, 194, 5423–5433. [Google Scholar] [CrossRef] [PubMed]
- Orf, I.; Timm, S.; Bauwe, H.; Fernie, A.R.; Hagemann, M.; Kopka, J.; Nikoloski, Z. Can cyanobacteria serve as a model of plant photorespiration?-A comparative meta-analysis of metabolite profiles. J. Exp. Bot. 2016, 67, 2941–2952. [Google Scholar] [CrossRef] [PubMed]
- Leplat, C.; Champeimont, R.; Saenkham, P.; Cassier-Chauvat, C.; Jean-Christophe, A.; Chauvat, F. Genome-wide transcriptome analysis of hydrogen production in the cyanobacterium Synechocystis: Towards the identification of new players. Int. J. Hydrog. Energy 2013, 38, 1866–1872. [Google Scholar] [CrossRef]
- Gutekunst, K.; Phunpruch, S.; Schwarz, C.; Schuchardt, S.; Schulz-Friedrich, R.; Appel, J. LexA regulates the bidirectional hydrogenase in the cyanobacterium Synechocystis sp. PCC 6803 as a transcription activator. Mol. Microbiol. 2005, 58, 810–823. [Google Scholar] [CrossRef]
- Oliveira, P.; Lindblad, P. LexA, a transcription regulator binding in the promoter region of the bidirectional hydrogenase in the cyanobacterium Synechocystis sp. PCC 6803. FEMS Microbiol. Lett. 2005, 251, 59–66. [Google Scholar] [CrossRef]
- Patterson-Fortin, L.M.; Colvin, K.R.; Owttrim, G.W. A LexA-related protein regulates redox-sensitive expression of the cyanobacterial RNA helicase, crhR. Nucleic Acids Res. 2006, 34, 3446–3454. [Google Scholar] [CrossRef]
- Pimentel, J.S.M.; Giani, A. Microcystin production and regulation under nutrient stress conditions in toxic Microcystis strains. Appl. Environ. Microbiol. 2014, 80, 5836–5843. [Google Scholar] [CrossRef]
- Mo, H.; Xie, X.; Zhu, T.; Lu, X. Effects of global transcription factor NtcA on photosynthetic production of ethylene in recombinant Synechocystis sp. PCC 6803. Biotechnol. Biofuels 2017, 10. [Google Scholar] [CrossRef]
- Scholl, J.; Dengler, L.; Bader, L.; Forchhammer, K. Phosphoenolpyruvate carboxylase from the cyanobacterium Synechocystis sp. PCC 6803 is under global metabolic control by P II signaling. Mol. Microbiol. 2020. [Google Scholar] [CrossRef]
- Lüddecke, J.; Forchhammer, K. From PII signaling to metabolite sensing: A novel 2-oxoglutarate sensor that details PII-NAGK complex formation. PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Nguyen, A.Y.; Dai, Z.; Su, D.; Gaffrey, M.J.; Moore, R.J.; Jacobs, J.M.; Monroe, M.E.; Smith, R.D.; Koppenaal, D.W.; et al. Proteome-wide light/dark modulation of thiol oxidation in cyanobacteria revealed by quantitative site-specific redox proteomics. Mol. Cell Proteom. 2014, 13, 3270–3285. [Google Scholar] [CrossRef] [PubMed]
- Florencio, F.J.; Pérez-Pérez, M.E.; López-Maury, L.; Mata-Cabana, A.; Lindahl, M. The diversity and complexity of the cyanobacterial thioredoxin systems. Photosynth. Res. 2006, 89, 157–171. [Google Scholar] [CrossRef]
- López-Calcagno, P.E.; Howard, T.P.; Raines, C.A. The CP12 protein family: A thioredoxin-mediated metabolic switch? Front. Plant Sci. 2014, 5. [Google Scholar] [CrossRef]
- Hackenberg, C.; Hakanpäa, J.; Cai, F.; Antonyuk, S.; Eigner, C.; Meissner, S.; Laitaoja, M.; Jänis, J.; Kerfeld, C.A.; Dittmann, E.; et al. Structural and functional insights into the unique CBS–CP12 fusion protein family in cyanobacteria. Proc. Natl. Acad. Sci. USA 2018, 115, 7141–7146. [Google Scholar] [CrossRef] [PubMed]
- McFarlane, C.R.; Shah, N.R.; Kabasakal, B.V.; Echeverria, B.; Cotton, C.A.R.; Bubeck, D.; Murray, J.W. Structural basis of light-induced redox regulation in the Calvin–Benson cycle in cyanobacteria. Proc. Natl. Acad. Sci. USA 2019, 116, 20984–20990. [Google Scholar] [CrossRef] [PubMed]
- Stanley, D.N.; Raines, C.A.; Kerfeld, C.A. Comparative analysis of 126 cyanobacterial genomes reveals evidence of functional diversity among homologs of the redox-regulated CP12 protein. Plant Physiol. 2013, 161, 824–835. [Google Scholar] [CrossRef] [PubMed]
- Gontero, B.; Maberly, S.C. An intrinsically disordered protein, CP12: Jack of all trades and master of the Calvin cycle. Biochem. Soc. Trans. 2012, 40, 995–999. [Google Scholar] [CrossRef] [PubMed]
- Kanno, M.; Carroll, A.L.; Atsumi, S. Global metabolic rewiring for improved CO2 fixation and chemical production in cyanobacteria. Nat. Commun. 2017, 8. [Google Scholar] [CrossRef]
- Marteyn, B.; Domain, F.; Legrain, P.; Chauvat, F.; Cassier-Chauvat, C. The thioredoxin reductase-glutaredoxins-ferredoxin crossroad pathway for selenate tolerance in Synechocystis PCC6803. Mol. Microbiol. 2009, 71, 520–532. [Google Scholar] [CrossRef]
- Narainsamy, K.; Marteyn, B.; Sakr, S.; Cassier-Chauvat, C.; Chauvat, F. Genomics of the Pleïotropic Glutathione System in Cyanobacteria. In Advances in Botanical Research; Elsevier Academic Press: San Diego, CA, USA, 2013; ISBN 9780123943132. [Google Scholar]
- Carvalho, A.N.; Marques, C.; Guedes, R.C.; Castro-Caldas, M.; Rodrigues, E.; Van Horssen, J.; Gama, M.J. S-Glutathionylation of Keap1: A new role for glutathione S-transferase pi in neuronal protection. FEBS Lett. 2016, 590, 1455–1466. [Google Scholar] [CrossRef] [PubMed]
- Sakr, S.; Dutheil, J.; Saenkham, P.; Bottin, H.; Leplat, C.; Ortega-Ramos, M.; Aude, J.C.; Chapuis, V.; Guedeney, G.; Decottignies, P.; et al. The activity of the Synechocystis PCC6803 AbrB2 regulator of hydrogen production can be post-translationally controlled through glutathionylation. Int. J. Hydrog. Energy 2013, 38, 13547–13555. [Google Scholar] [CrossRef]
- Marteyn, B.; Sakr, S.; Farci, S.; Bedhomme, M.; Chardonnet, S.; Decottignies, P.; Lemaire, S.D.; Cassier-Chauvat, C.; Chauvat, F. The Synechocystis PCC6803 MerA-like enzyme operates in the reduction of both mercury and uranium under the control of the glutaredoxin 1 enzyme. J. Bacteriol. 2013, 195, 4138–4145. [Google Scholar] [CrossRef] [PubMed]
- Chardonnet, S.; Sakr, S.; Cassier-Chauvat, C.; Le Maréchal, P.; Chauvat, F.; Lemaire, S.D.; Decottignies, P. First proteomic study of S-glutathionylation in cyanobacteria. J. Proteome Res. 2015, 14, 59–71. [Google Scholar] [CrossRef]
- Gao, X.H.; Bedhomme, M.; Veyel, D.; Zaffagnini, M.; Lemaire, S.D. Methods for analysis of protein glutathionylation and their application to photosynthetic organisms. Mol. Plant 2009, 2, 218–235. [Google Scholar] [CrossRef]
- Pereira, S.; Zille, A.; Micheletti, E.; Moradas-Ferreira, P.; De Philippis, R.; Tamagnini, P. Complexity of cyanobacterial exopolysaccharides: Composition, structures, inducing factors and putative genes involved in their biosynthesis and assembly. FEMS Microbiol. Rev. 2009, 33, 917–941. [Google Scholar] [CrossRef]
- Jittawuttipoka, T.; Planchon, M.; Spalla, O.; Benzerara, K.; Guyot, F.; Cassier-Chauvat, C.; Chauvat, F. Multidisciplinary Evidences that Synechocystis PCC6803 Exopolysaccharides Operate in Cell Sedimentation and Protection against Salt and Metal Stresses. PLoS ONE 2013, 8. [Google Scholar] [CrossRef]
- Kamennaya, N.A.; Zemla, M.; Mahoney, L.; Chen, L.; Holman, E.; Holman, H.Y.; Auer, M.; Ajo-Franklin, C.M.; Jansson, C. High pCO2-induced exopolysaccharide-rich ballasted aggregates of planktonic cyanobacteria could explain Paleoproterozoic carbon burial. Nat. Commun. 2018, 9, 216. [Google Scholar] [CrossRef]
- Stuart, R.K.; Mayali, X.; Boaro, A.A.; Zemla, A.; Everroad, R.C.; Nilson, D.; Weber, P.K.; Lipton, M.; Bebout, B.M.; Pett-Ridge, J.; et al. Light regimes shape utilization of extracellular organic C and N in a cyanobacterial biofilm. mBio 2016, 7, e00650-16. [Google Scholar] [CrossRef] [PubMed]
- Planchon, M.; Jittawuttipoka, T.; Cassier-Chauvat, C.; Guyot, F.; Gelabert, A.; Benedetti, M.F.; Chauvat, F.; Spalla, O. Exopolysaccharides protect Synechocystis against the deleterious effects of Titanium dioxide nanoparticles in natural and artificial waters. J. Colloid Interface Sci. 2013, 405, 35–43. [Google Scholar] [CrossRef]
- Rossi, F.; De Philippis, R. Role of cyanobacterial exopolysaccharides in phototrophic biofilms and in complex microbial mats. Life 2015, 5, 1218–1238. [Google Scholar] [CrossRef] [PubMed]
- Bernaerts, T.M.M.; Gheysen, L.; Foubert, I.; Hendrickx, M.E.; Van Loey, A.M. The potential of microalgae and their biopolymers as structuring ingredients in food: A review. Biotechnol. Adv. 2019, 37. [Google Scholar] [CrossRef] [PubMed]
- Zampieri, R.M.; Adessi, A.; Caldara, F.; Codato, A.; Furlan, M.; Rampazzo, C.; De Philippis, R.; La Rocca, N.; Dalla Valle, L. Anti-Inflammatory Activity of Exopolysaccharides from Phormidium sp. ETS05, the Most Abundant Cyanobacterium of the Therapeutic Euganean Thermal Muds, Using the Zebrafish Model. Biomolecules 2020, 10, 582. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Esquer, C.R.; Smarda, J.; Rippka, R.; Axen, S.D.; Guglielmi, G.; Gugger, M.; Kerfeld, C.A. Cyanobacterial ultrastructure in light of genomic sequence data. Photosynth. Res. 2016, 192, 147–157. [Google Scholar] [CrossRef]
- Balaji, S.; Gopi, K.; Muthuvelan, B. A review on production of poly β hydroxybutyrates from cyanobacteria for the production of bio plastics. Algal Res. 2013, 2, 278–285. [Google Scholar] [CrossRef]
- Kamravamanesh, D.; Pflügl, S.; Nischkauer, W.; Limbeck, A.; Lackner, M.; Herwig, C. Photosynthetic poly-β-hydroxybutyrate accumulation in unicellular cyanobacterium Synechocystis sp. PCC 6714. AMB Express 2017, 7. [Google Scholar] [CrossRef] [PubMed]
- Ansari, S.; Fatma, T. Cyanobacterial polyhydroxybutyrate (PHB): Screening, optimization and characterization. PLoS ONE 2016, 11. [Google Scholar] [CrossRef] [PubMed]
- Kaewbai-ngam, A.; Incharoensakdi, A.; Monshupanee, T. Increased accumulation of polyhydroxybutyrate in divergent cyanobacteria under nutrient-deprived photoautotrophy: An efficient conversion of solar energy and carbon dioxide to polyhydroxybutyrate by Calothrix scytonemicola TISTR 8095. Bioresour. Technol. 2016, 212. [Google Scholar] [CrossRef]
- Koch, M.; Berendzen, K.W.; Forchhammer, K. On the role and production of polyhydroxybutyrate (Phb) in the cyanobacterium Synechocystis sp. pcc 6803. Life 2020, 10, 47. [Google Scholar] [CrossRef]
- Carpine, R.; Du, W.; Olivieri, G.; Pollio, A.; Hellingwerf, K.J.; Marzocchella, A.; Branco dos Santos, F. Genetic engineering of Synechocystis sp. PCC6803 for poly-β-hydroxybutyrate overproduction. Algal Res. 2017, 27, 117–127. [Google Scholar] [CrossRef]
- Courtene-Jones, W.; Quinn, B.; Ewins, C.; Gary, S.F.; Narayanaswamy, B.E. Microplastic accumulation in deep-sea sediments from the Rockall Trough. Mar. Pollut. Bull. 2020, 154. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Wang, X.; Su, M.; Zou, X.; Duan, L.; Zhang, H. Characteristics of Plastic Pollution in the Environment: A Review. Bull. Environ. Contam. Toxicol. 2020. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Veaudor, T.; Blanc-Garin, V.; Chenebault, C.; Diaz-Santos, E.; Sassi, J.-F.; Cassier-Chauvat, C.; Chauvat, F. Recent Advances in the Photoautotrophic Metabolism of Cyanobacteria: Biotechnological Implications. Life 2020, 10, 71. https://doi.org/10.3390/life10050071
Veaudor T, Blanc-Garin V, Chenebault C, Diaz-Santos E, Sassi J-F, Cassier-Chauvat C, Chauvat F. Recent Advances in the Photoautotrophic Metabolism of Cyanobacteria: Biotechnological Implications. Life. 2020; 10(5):71. https://doi.org/10.3390/life10050071
Chicago/Turabian StyleVeaudor, Théo, Victoire Blanc-Garin, Célia Chenebault, Encarnación Diaz-Santos, Jean-François Sassi, Corinne Cassier-Chauvat, and Franck Chauvat. 2020. "Recent Advances in the Photoautotrophic Metabolism of Cyanobacteria: Biotechnological Implications" Life 10, no. 5: 71. https://doi.org/10.3390/life10050071
APA StyleVeaudor, T., Blanc-Garin, V., Chenebault, C., Diaz-Santos, E., Sassi, J.-F., Cassier-Chauvat, C., & Chauvat, F. (2020). Recent Advances in the Photoautotrophic Metabolism of Cyanobacteria: Biotechnological Implications. Life, 10(5), 71. https://doi.org/10.3390/life10050071




