Identification of Differentially Expressed Gene Transcripts in Porcine Endometrium during Early Stages of Pregnancy
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Statement and Animal Materials
2.1.1. Ethical Statement
2.1.2. Animal Material
2.2. RNA Isolation
2.3. Laboratory Procedure of Microarray Experiment
2.4. Bioinformatics Analysis of Gene Expression Data Generated by Microarrays
2.4.1. Processing of Microarray Raw Data Analysis
2.4.2. Stringent Criteria of Excel-filtering of DEG-transcript-probes Microarray Data
2.4.3. Visualization of Upregulated and Downregulated DEG-transcript-probes of Microarray Data using Venn Diagrams, Heatmaps and ClueGO
2.5. Real time PCR Gene Expression Validation
2.5.1. Laboratory Procedure of Real-time PCR
2.5.2. Reference Genes Panel in the Microarray Validation Experiment
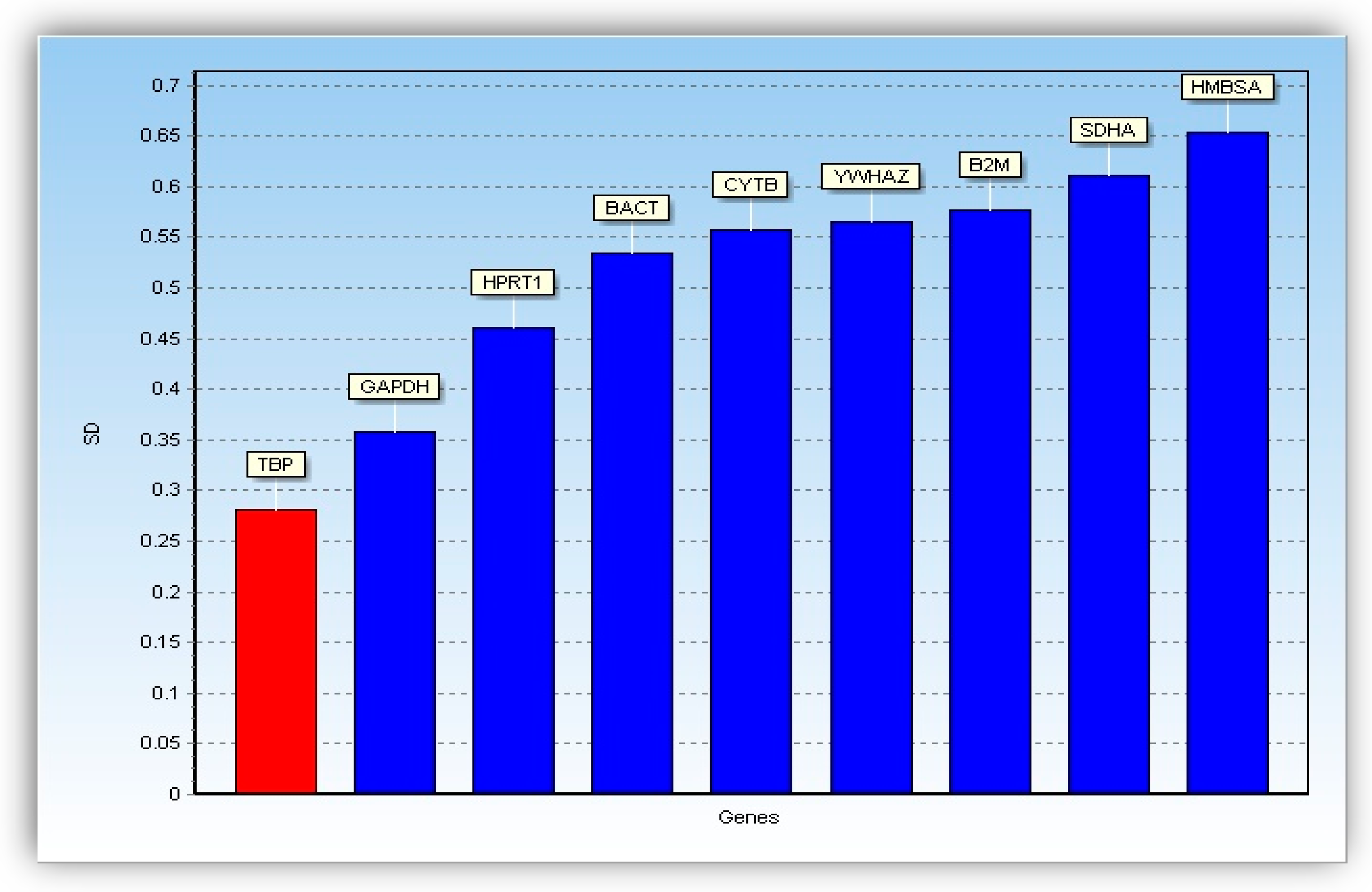
2.5.3. Analysis of Reference Genes Stability
2.5.4. Selected DEG-transcript-probes in the Microarray Validation Experiment
2.5.5. Statistics of Expression of Selected Genes in the Endometrium of Pregnant Gilts
3. Results
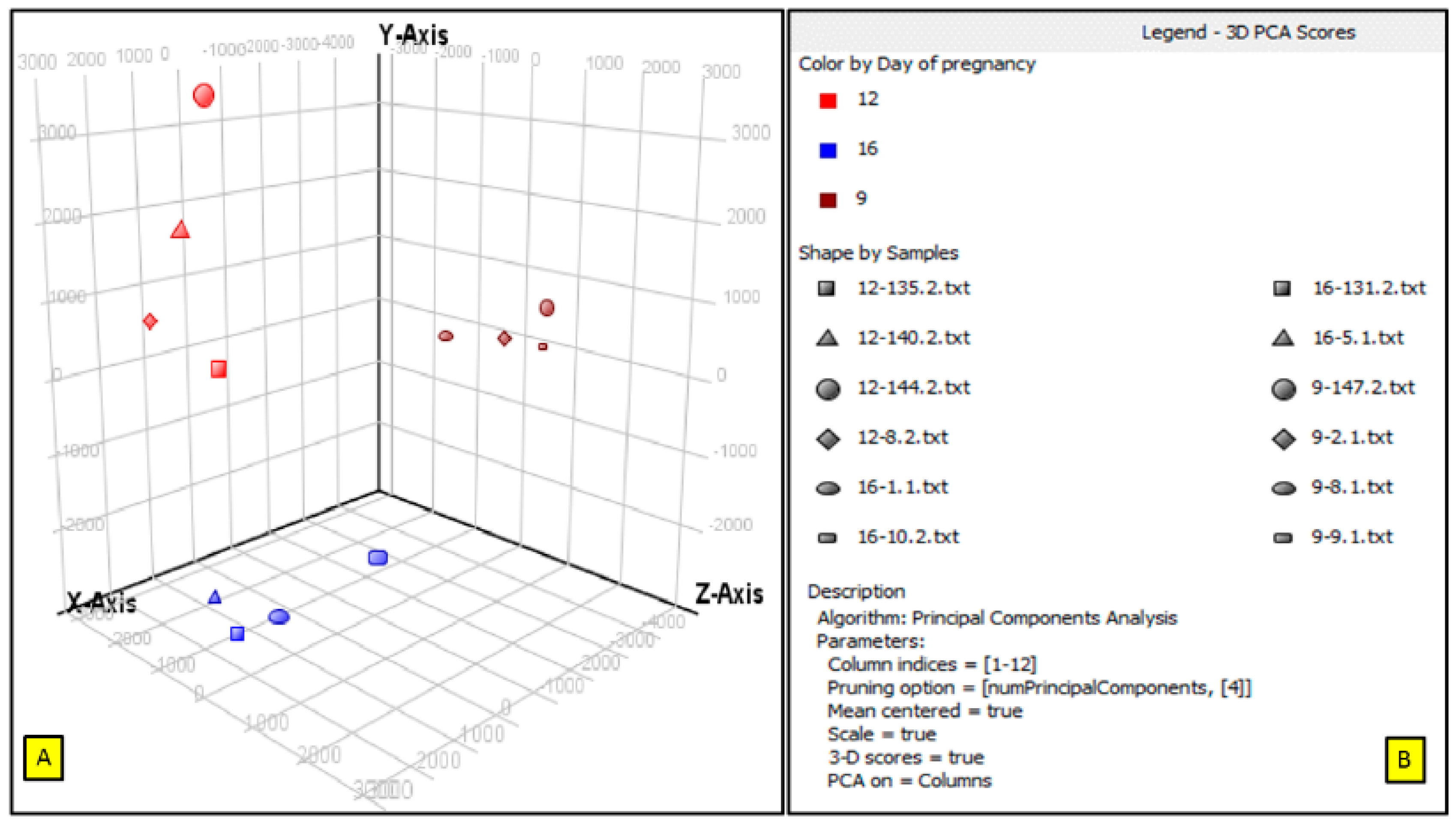
3.1. Principle Component Analysis (PCA)
3.2. Comparisons of Porcine Endometrium at Three Early Pregnancy Periods in PLW Gilts
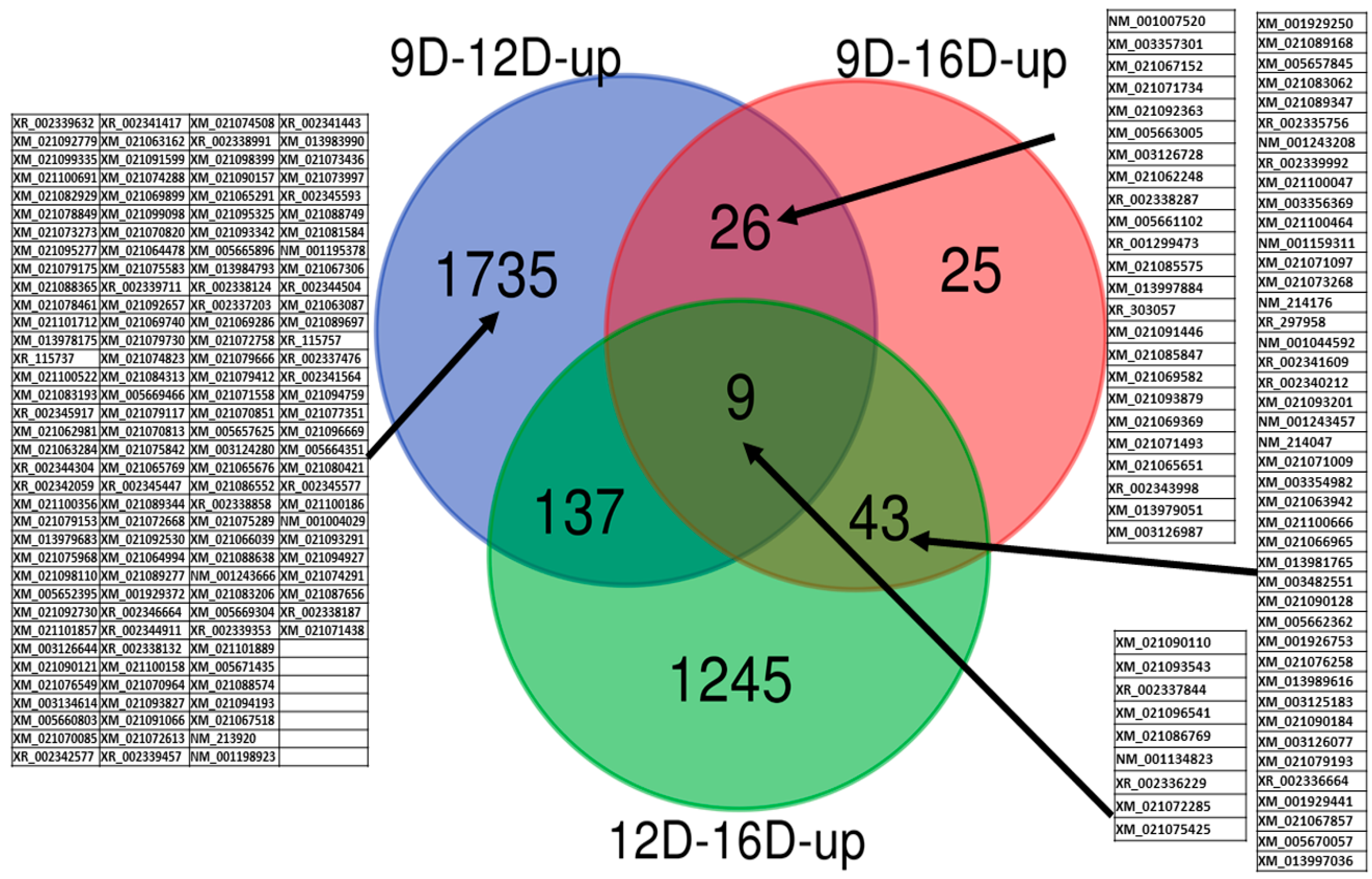
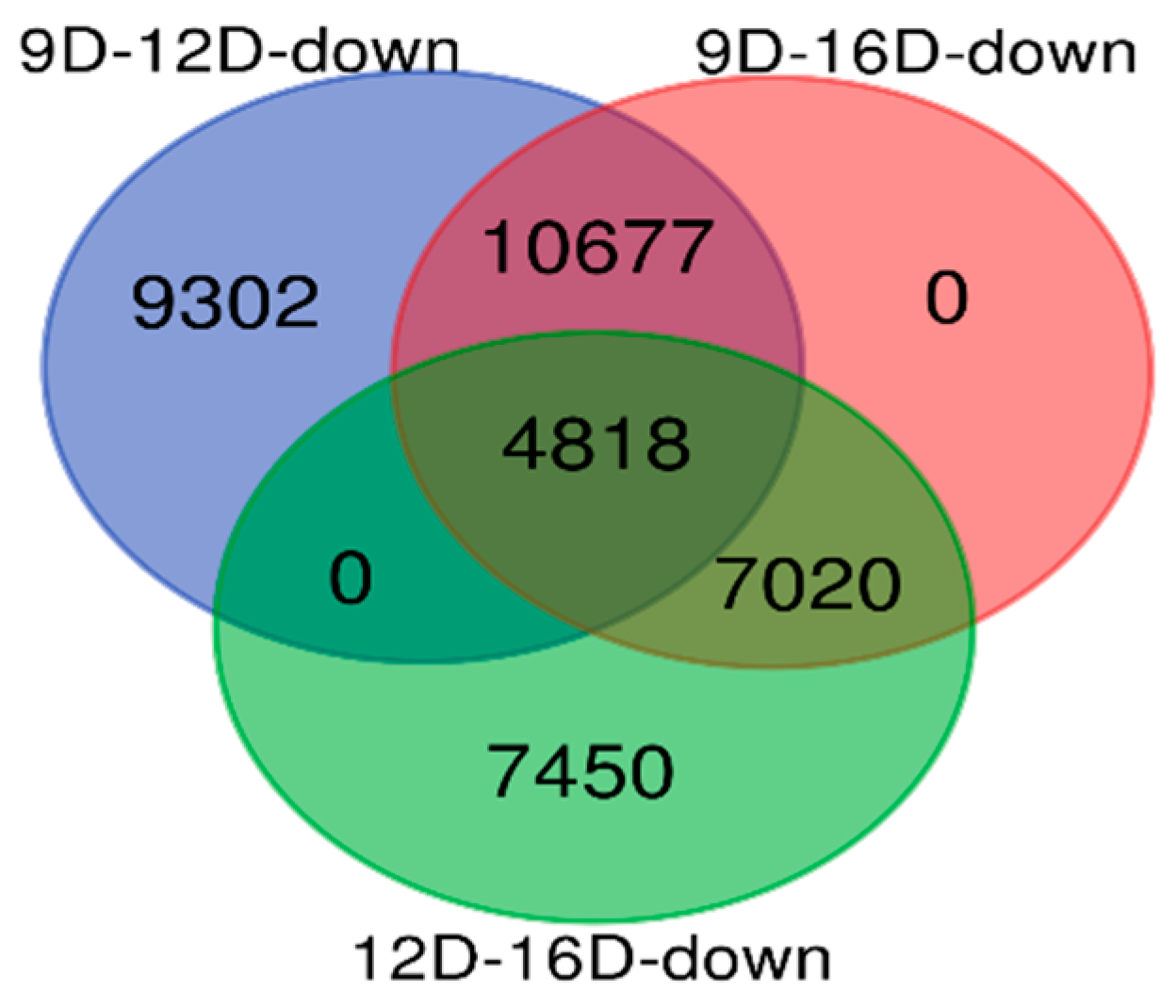
3.2.1. Comparisons of Porcine Endometrium 9D vs. 12D of the Early Pregnancy Period
3.2.2. Comparison of Porcine Endometrium 9D vs. 16D of the Early Pregnancy Period
3.2.3. Comparison of Porcine Endometrium 12D vs. 16D of the Early Pregnancy Period
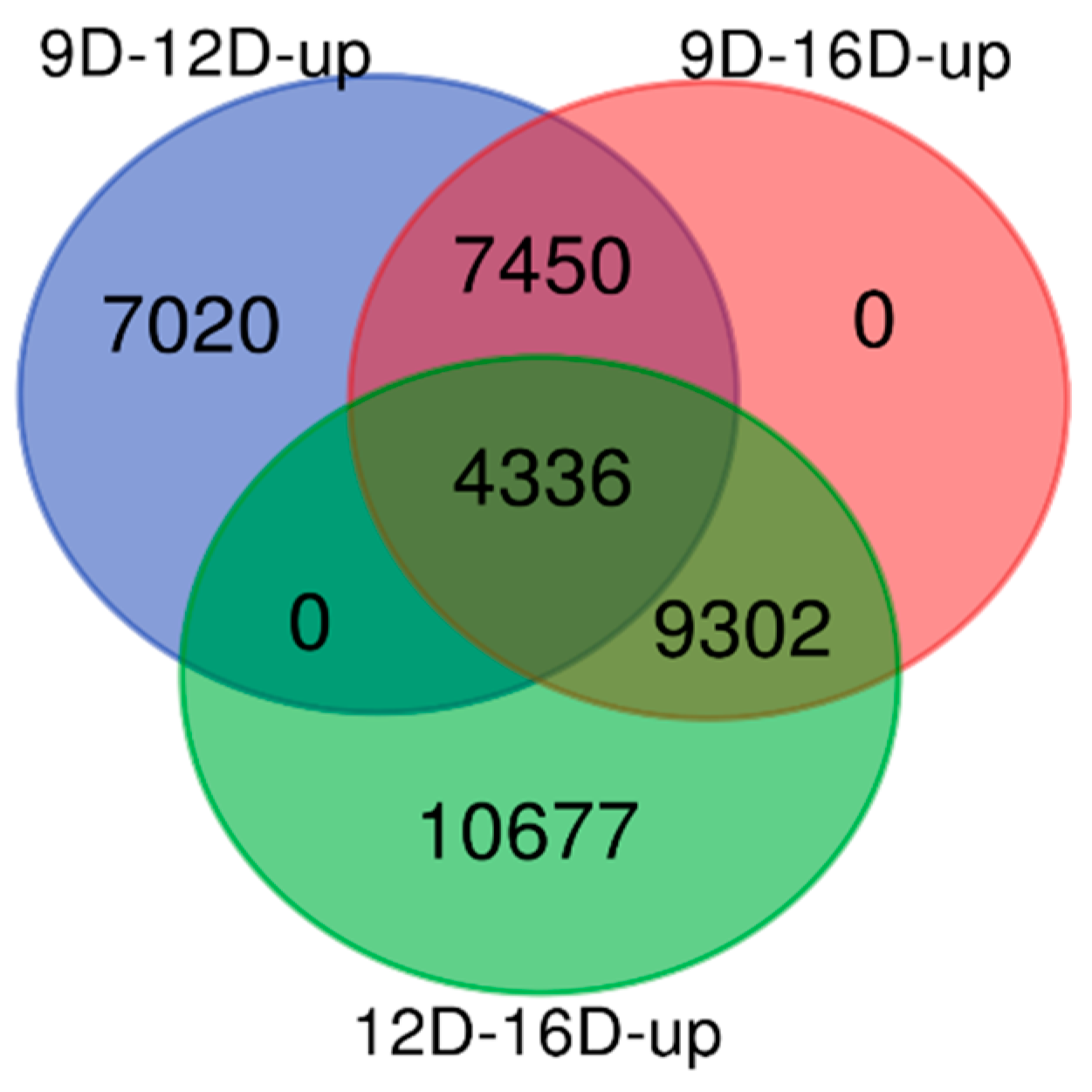
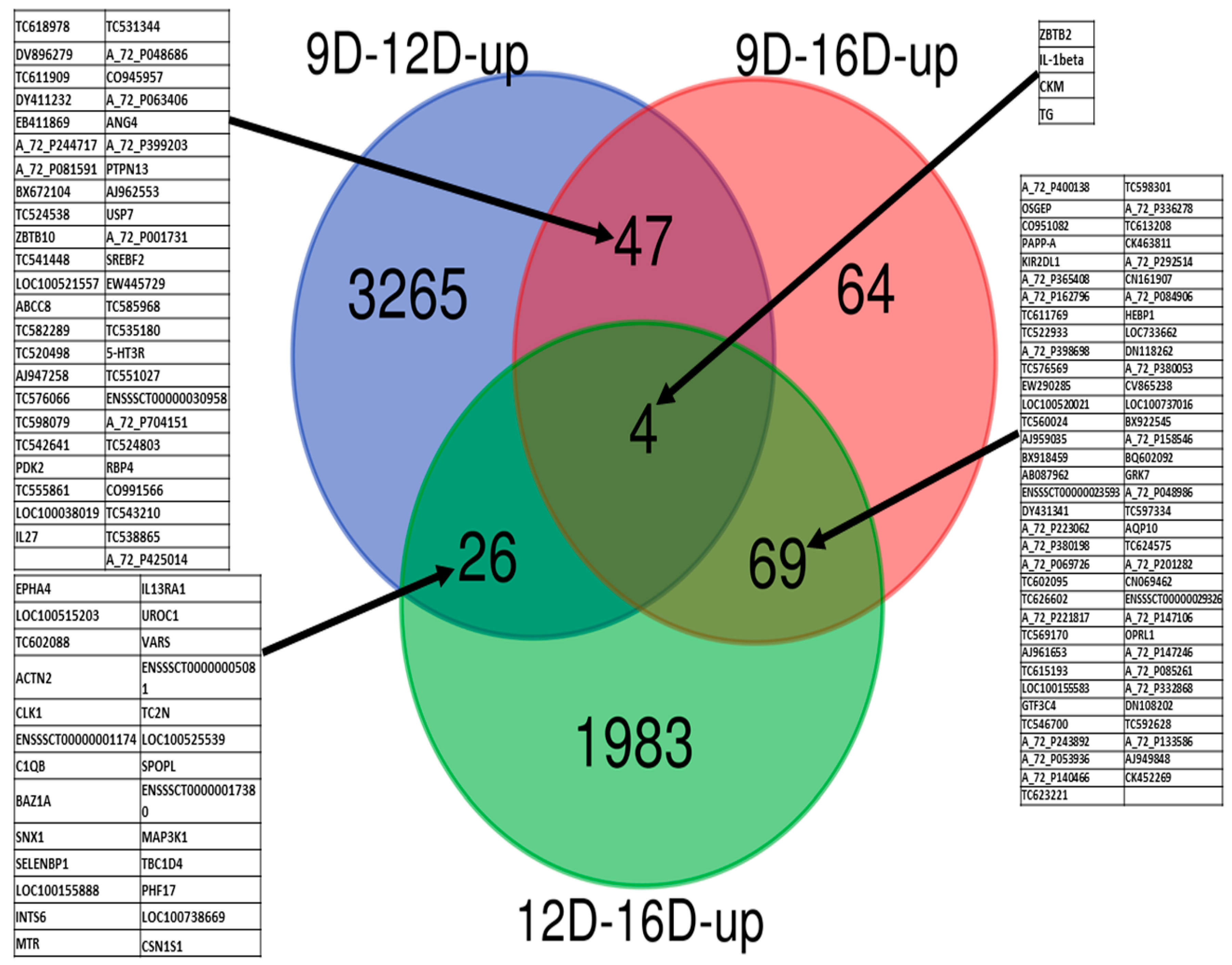
3.3. Comparison of Upregulated and Downregulated DEG-transcript-probes at Three Stages of Early Pregnancy in PLW Gilts
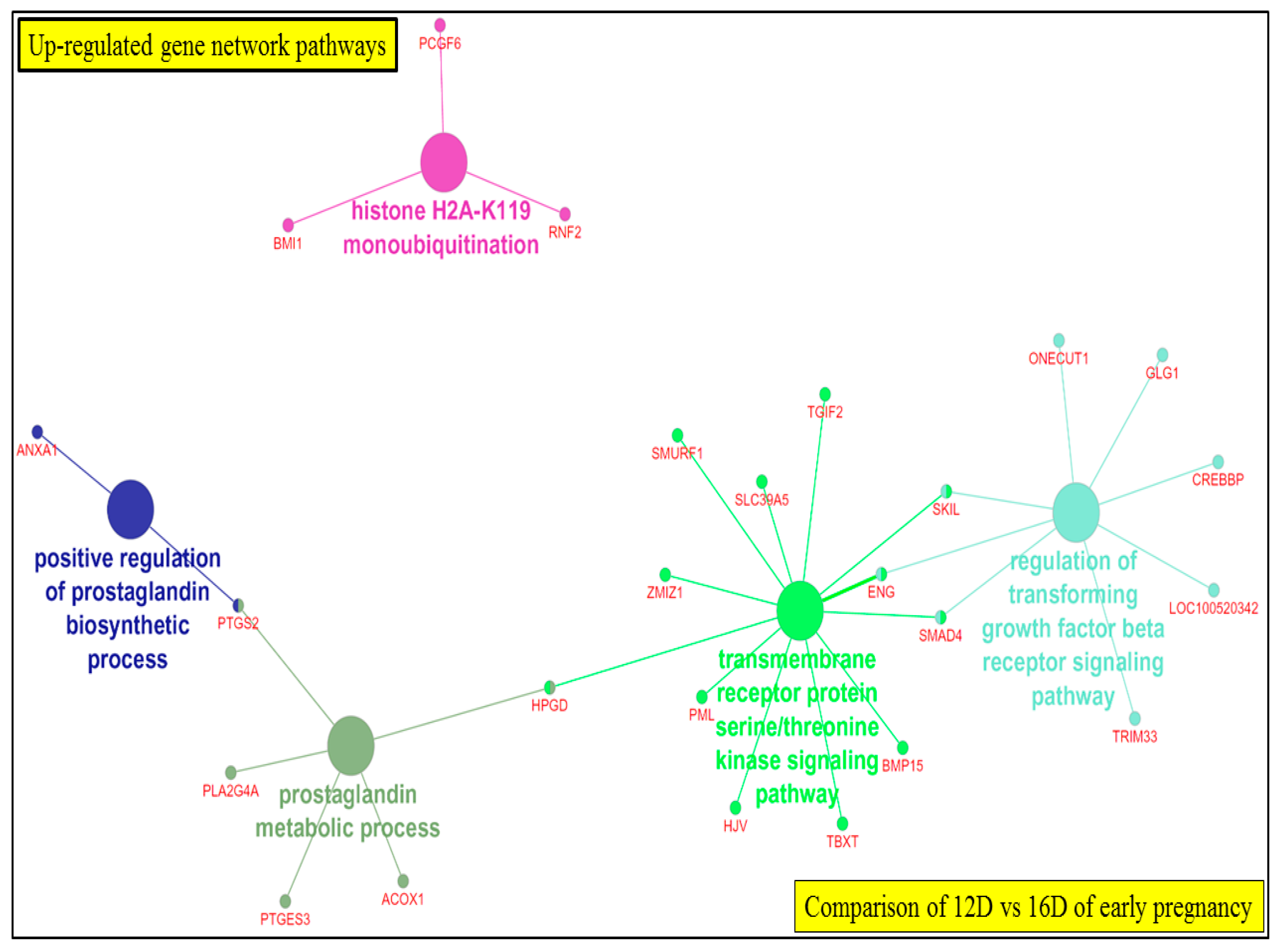
3.3.1. Comparison of Upregulated DEG-transcript-probes during Three Stages of Early Pregnancy
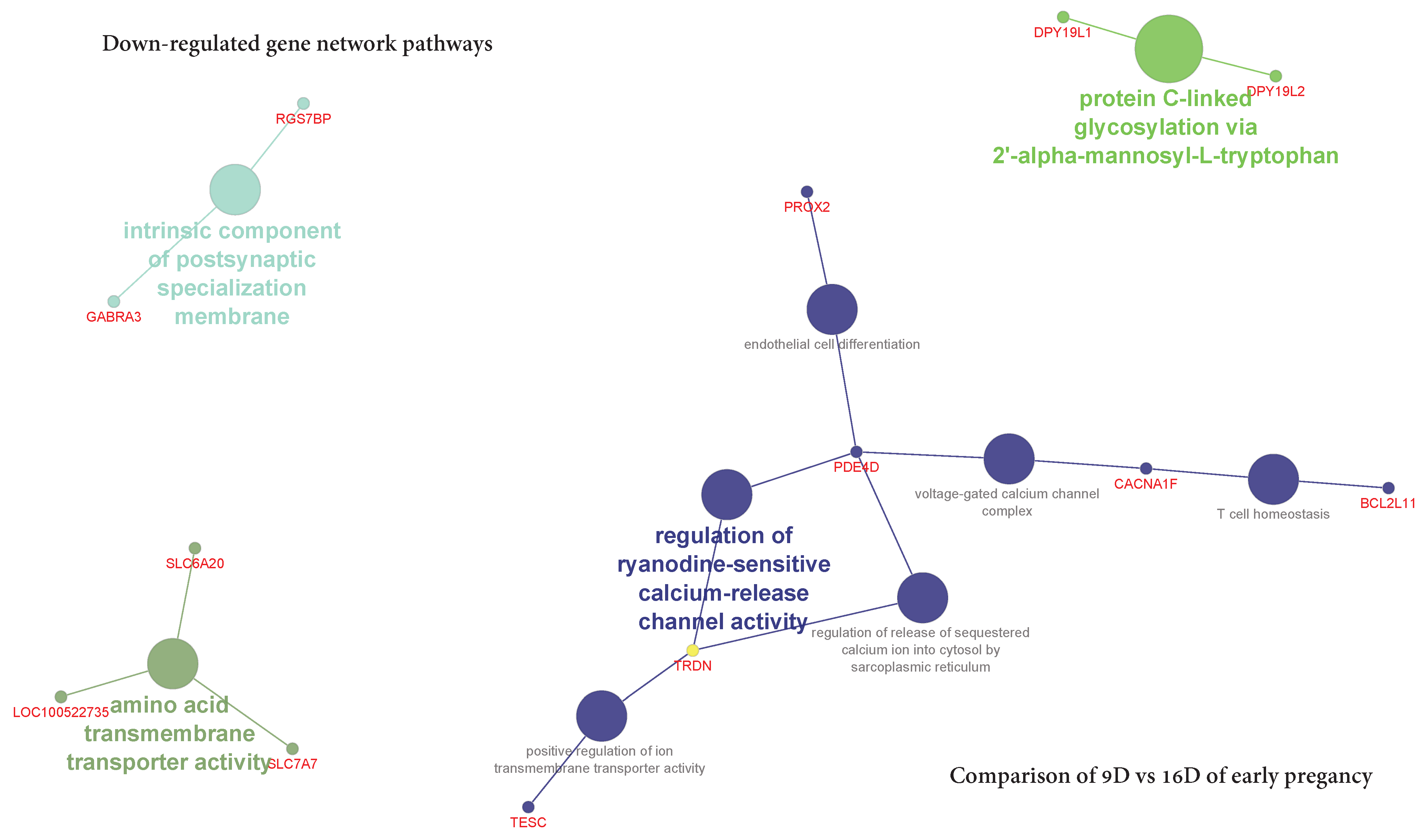
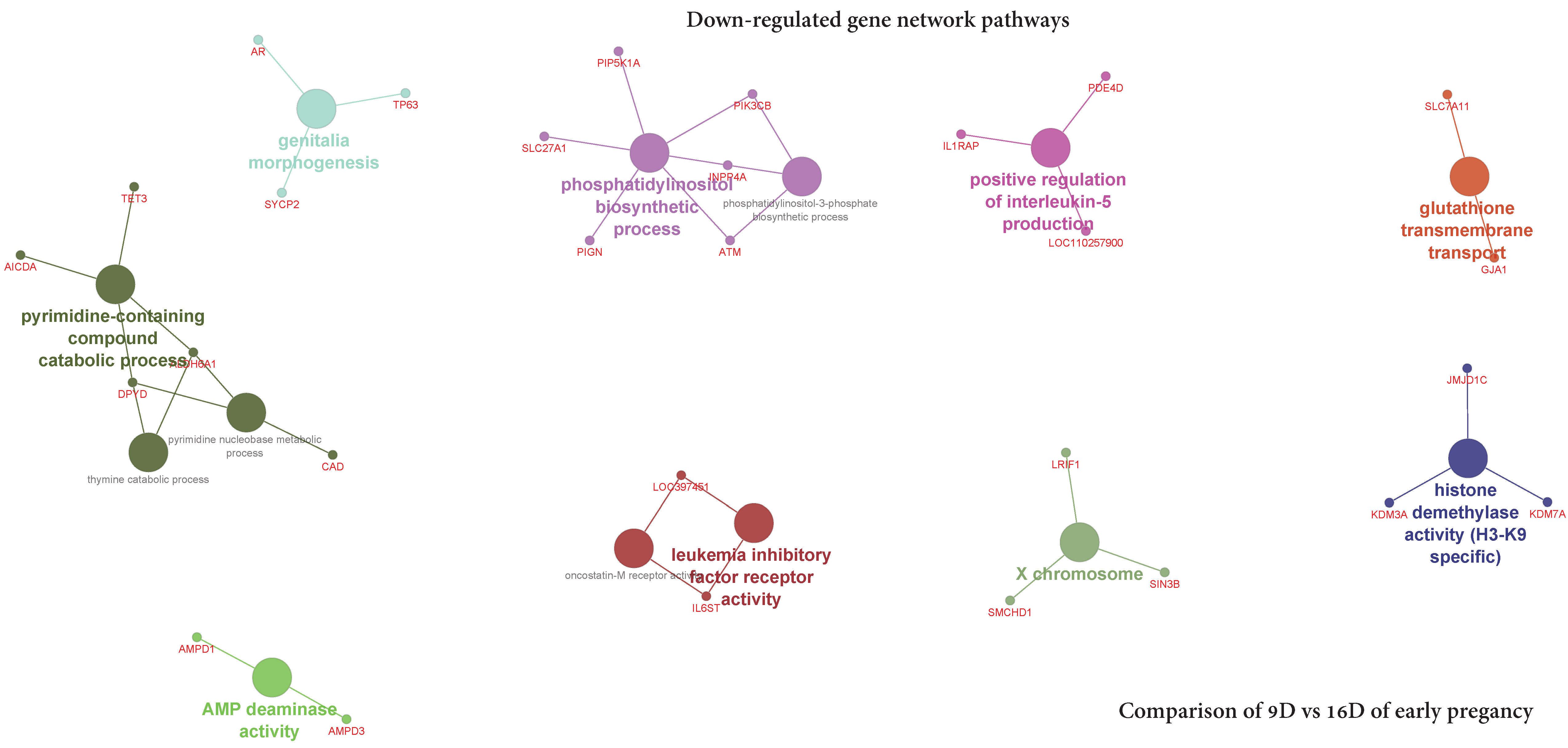
3.3.2. Comparison of Downregulated DEG-transcript-probes during Three Stages of Early Pregnancy
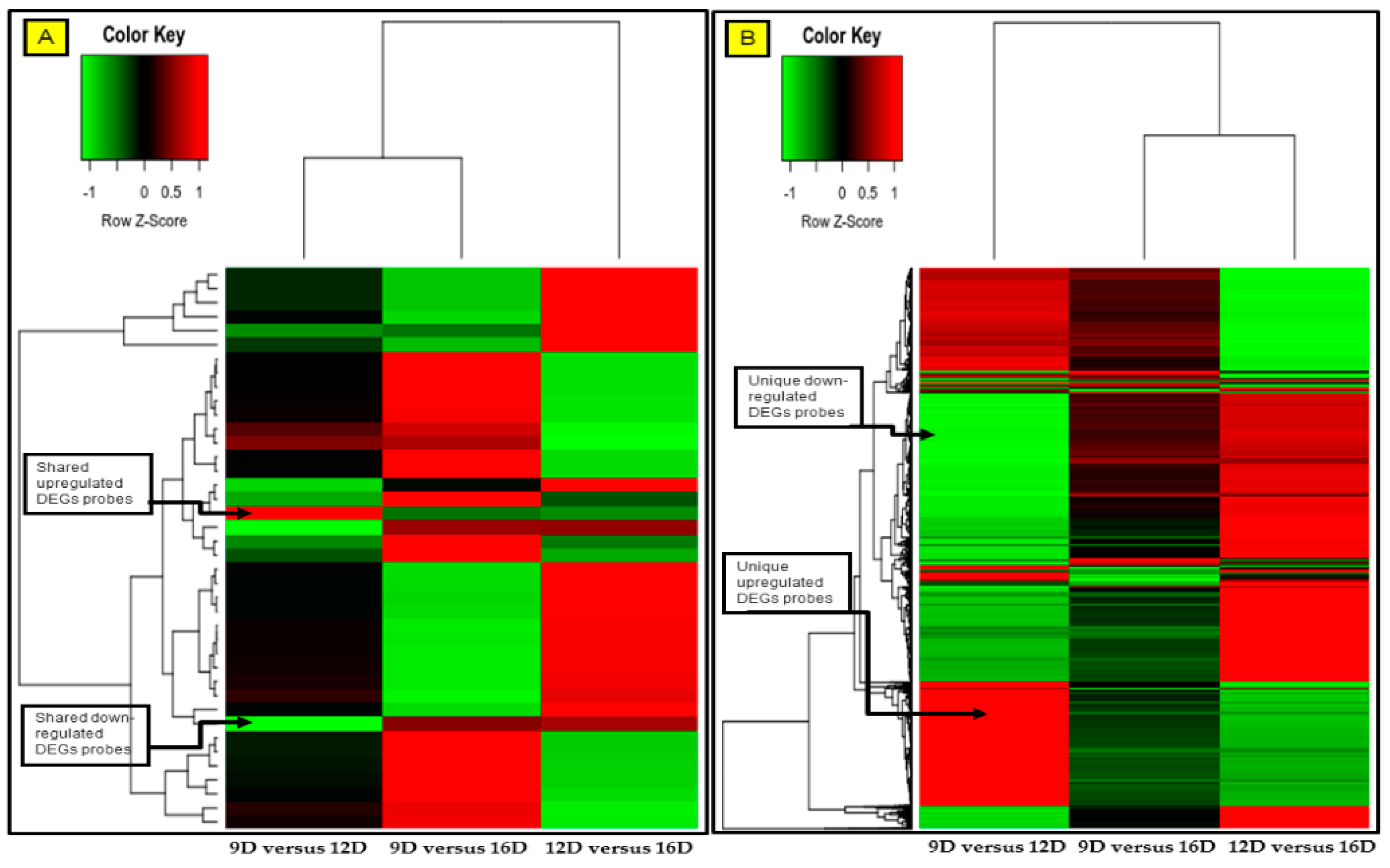
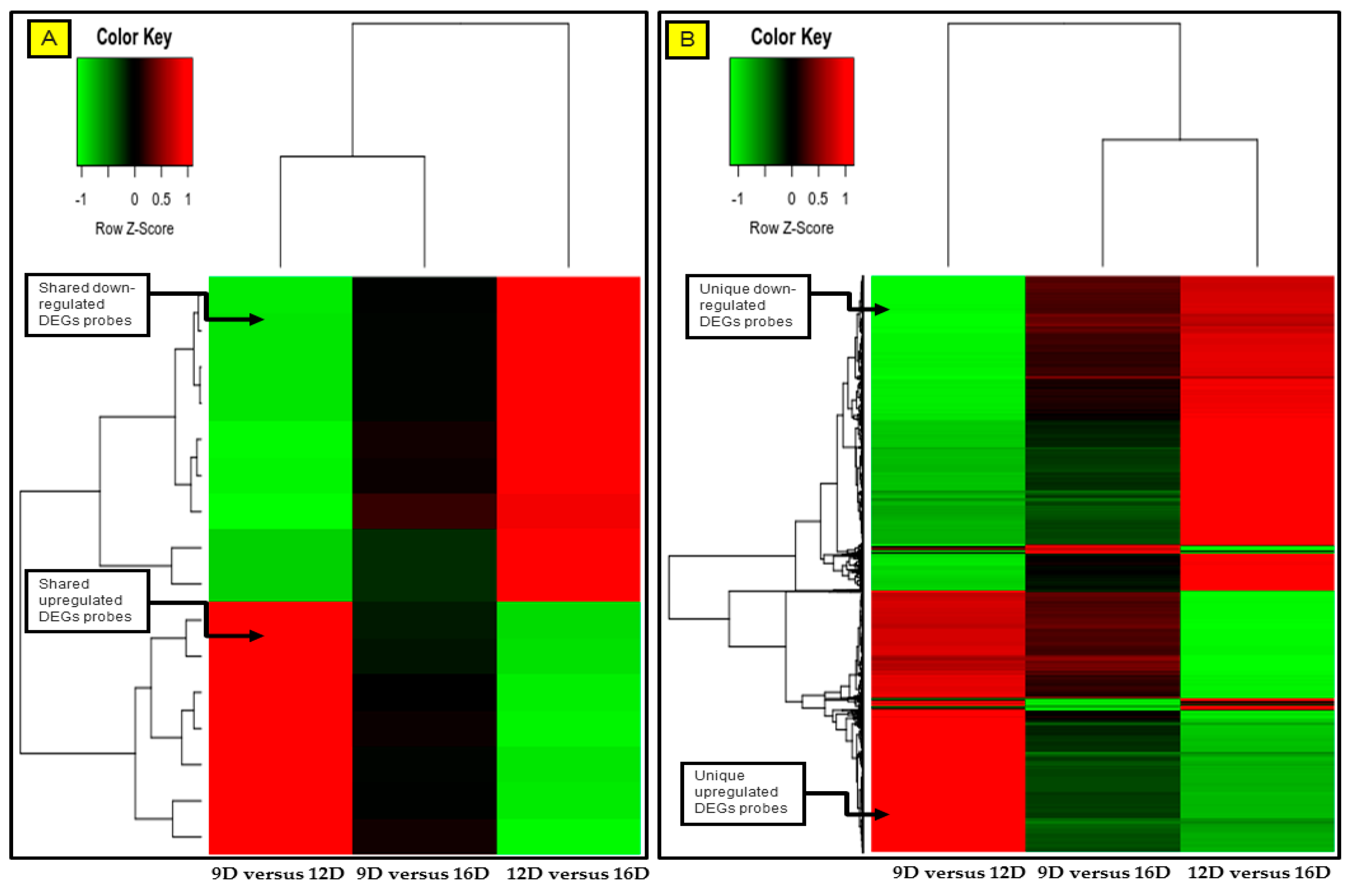
3.4. Comparison and the Heatmap Visualization of the Porcine Endometrium at 12D vs. 16D vs. 16D of early Pregnancy Period
3.5. Comparison and Visualization of Gene Network and Pathways using ClueGO of Porcine Endometrium at 12D vs. 16D vs. 16D of Early Pregnancy Period
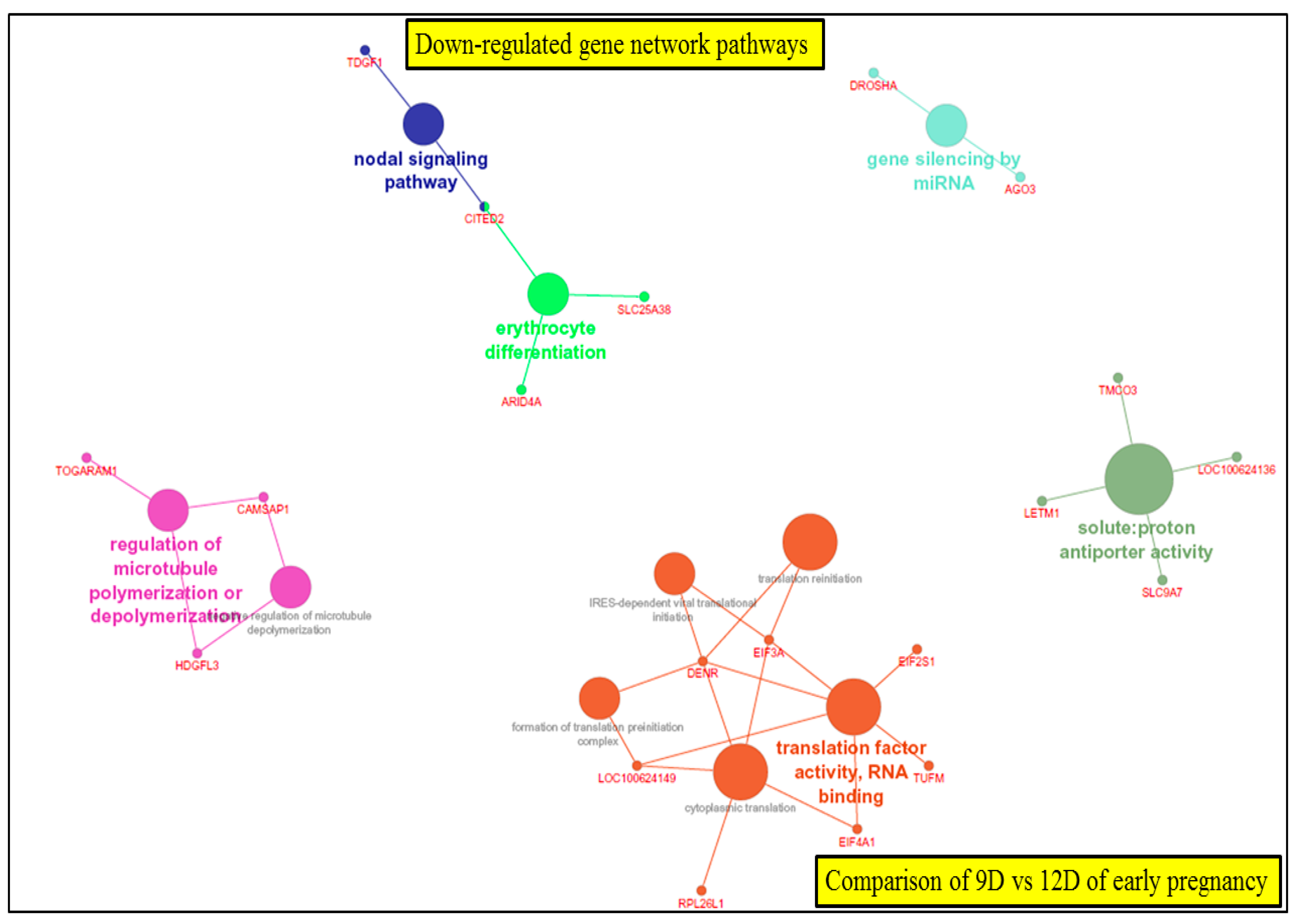
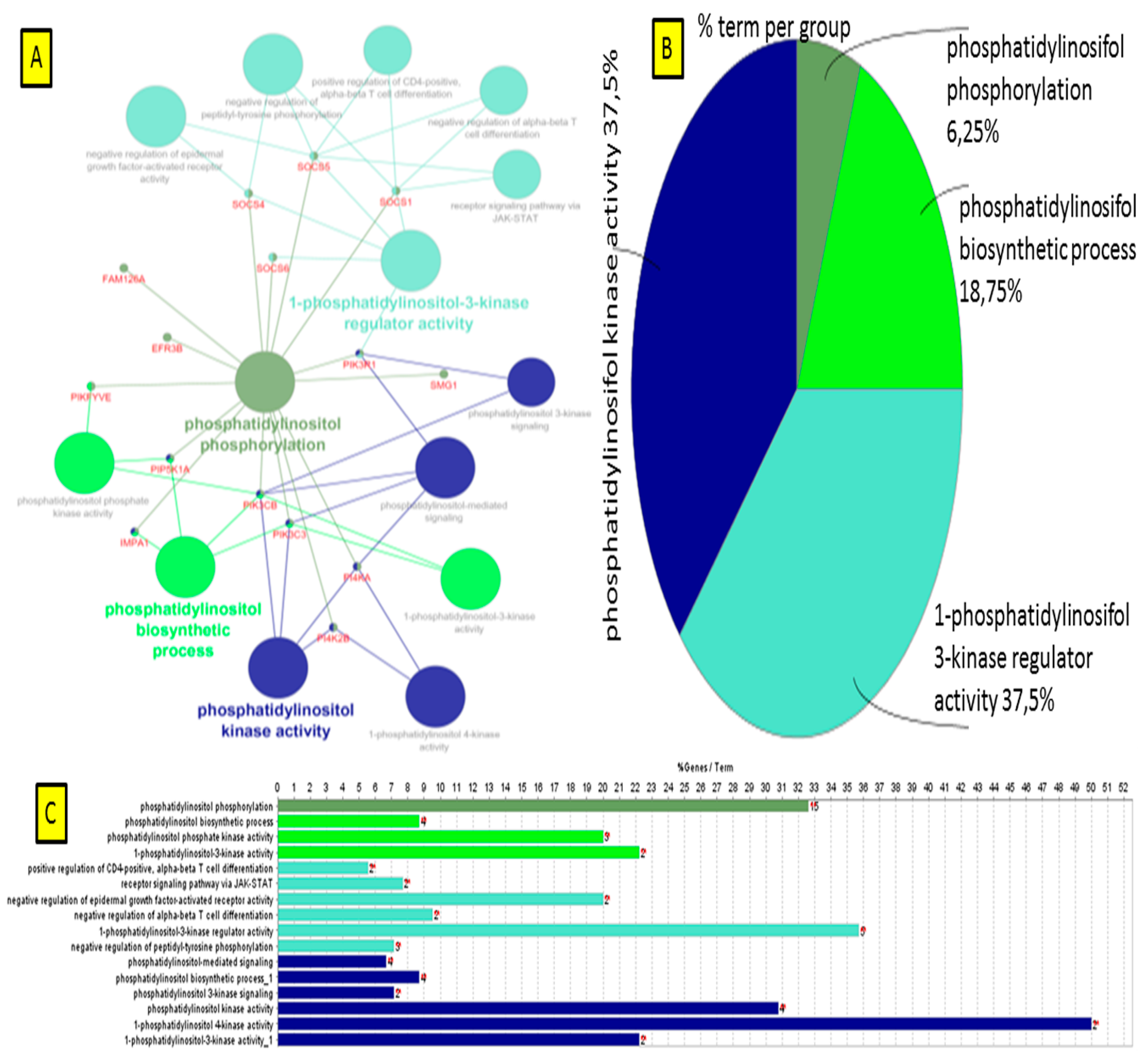
3.5.1. Visualization of Gene Network and Pathways of Porcine Endometrium at 9D vs. 12D of Early Pregnancy Period
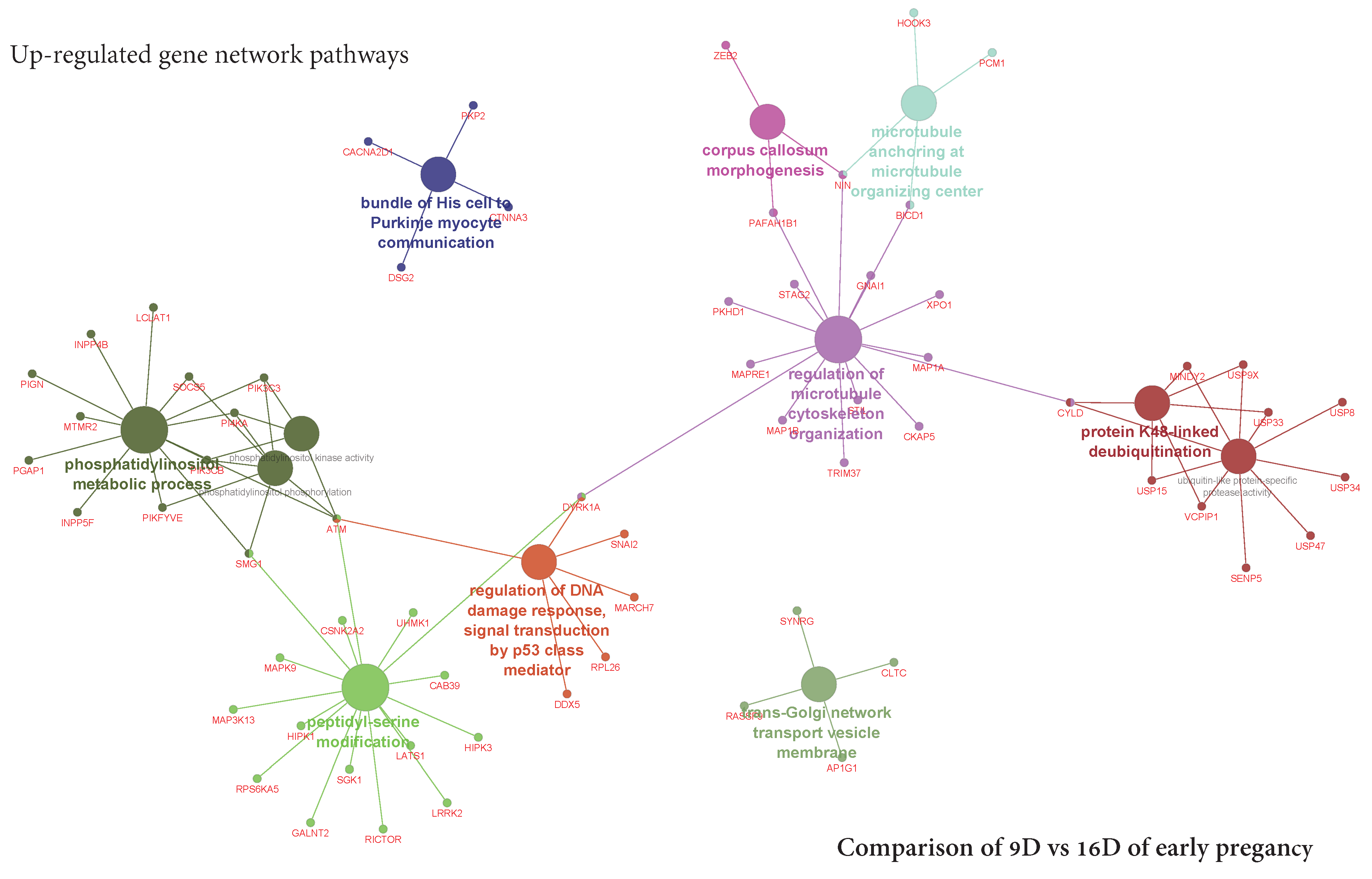
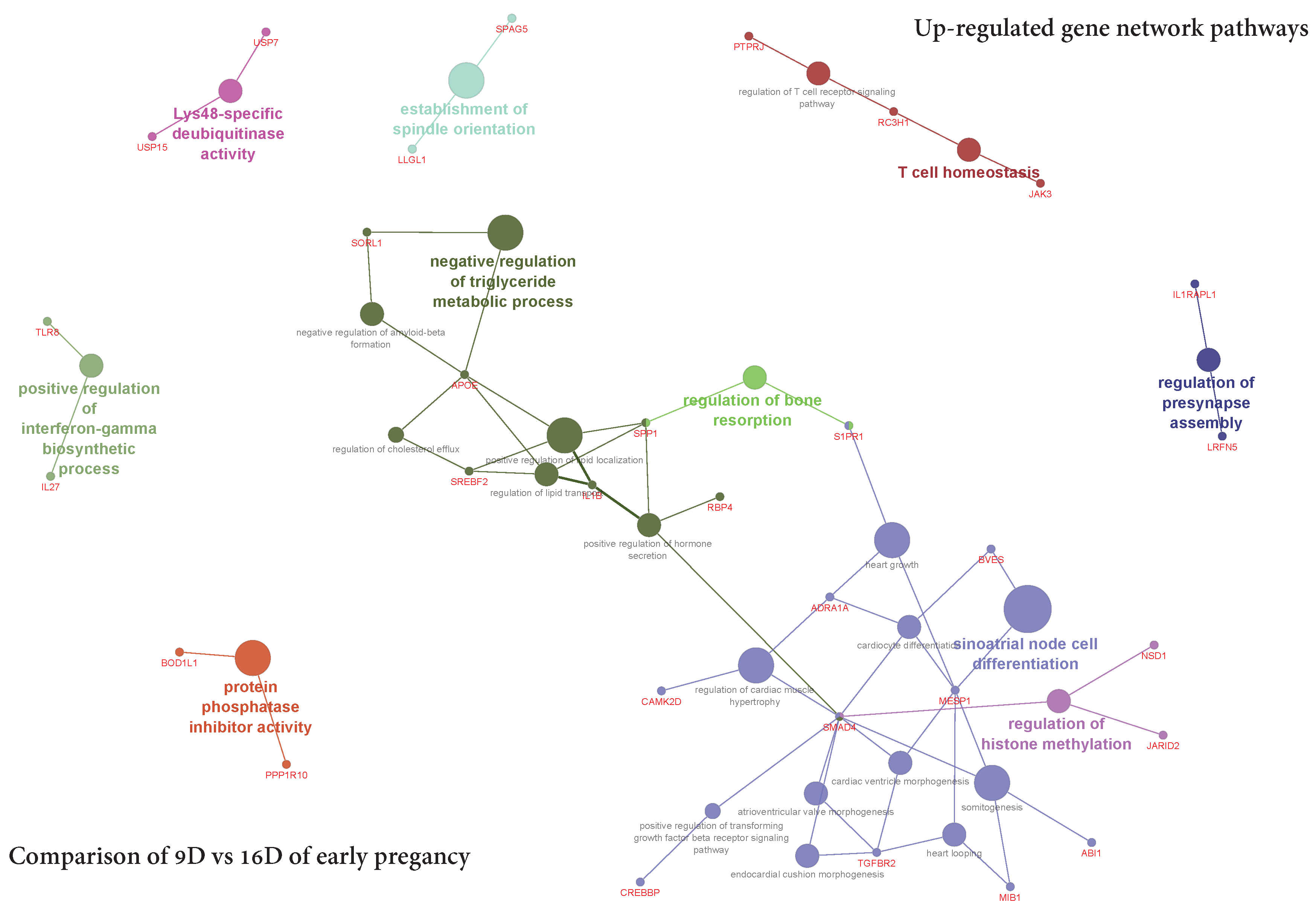
3.5.2. Visualization of Gene Network and Pathways of Porcine Endometrium at 9D vs. 16D of Early Pregnancy
3.5.3. Visualization of Gene Network and Pathways of Porcine Endometrium at 12D vs. 16D of Early Pregnancy Period
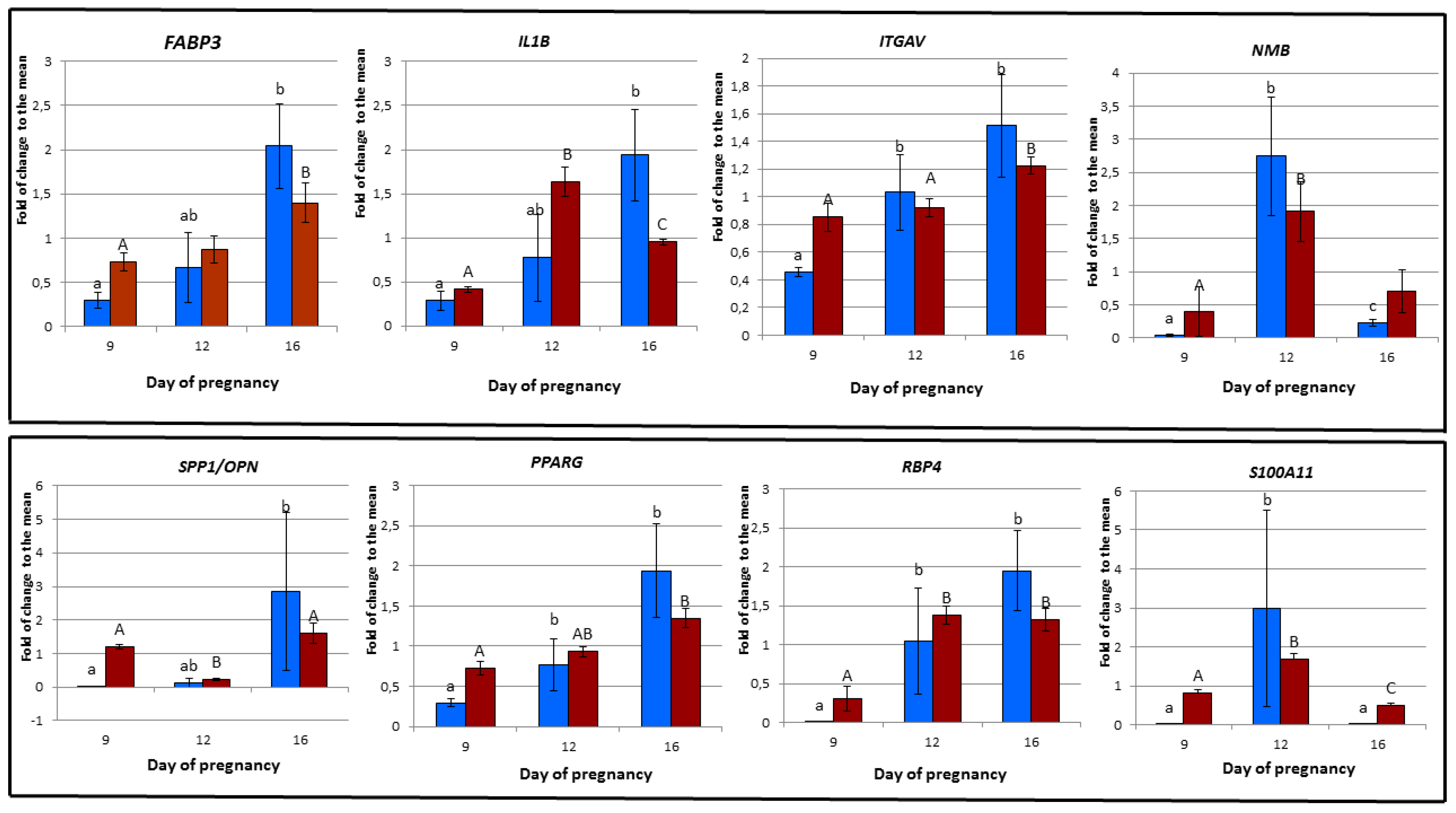
3.6. Validation of Microarray Experiment using RT-PCR
4. Discussion
4.1. Identification of DEG Transcripts in Porcine Endometrium during Early Stages of Pregnancy
4.2. Identification of Gene Networks and Pathways in Porcine Endometrium during Early Stages of Pregnancy
4.3. Genes Related to Signaling and Metabolism
4.4. Genes Involved in Growth Factors and Immune Response
4.5. Genes Involved in Epithelial Cell Differentiation and Development
4.6. Genes Involved in Ion and Nutrient Transport
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| DEG-transcript-probes | Differential Expressed Gene-transcript-probes |
| 9D | 9th day |
| 12D | 12th day |
| 16D | 16th day |
| PLW | Polish large-white |
| FC | Fold of change |
| TG | Thyroglobulin |
| ZBTB2 | Zinc finger and BTB domain containing-2 |
| CKM | Creatine kinase, M-type |
| IL1B | Interleukin-1beta |
| PTPN11 | Protein tyrosine phosphatase non-receptor type 11 |
| SELENOP | Selenoprotein P |
| CDC42BPA | Cell-division cycle-42 binding protein kinase alpha |
| MINPP1 | Multiple inositol-polyphosphate phosphatase 1 |
| VAV3 | Guanine nucleotide exchange factor-3 |
| CASC4 | Cancer susceptibility candidate gene 4 protein |
| COBLL1 | Cordon-bleu WH2 repeat protein like 1 |
| ACAP2 | ArfGAP with coiled-coil, ankyrin repeat and PH domains-2 |
| TMEM33 | Transmembrane protein 33 |
| KLHL15 | Kelch-like family member 15 |
| S100A11 | S100 calcium binding protein A11 |
| FABP3 | Fatty acid binding protein 3 |
| PPARG | Peroxisome proliferator activated receptor gamma |
| ITGAV | Integrin subunit alpha V |
| OPN | Secreted phosphoprotein 1 |
| NMB | Neuromedin B |
| RBP4 | Retinol binding protein 4 |
| RT-PCR | Reverse transcriptase polymerase chain reaction |
| HSP values | High-scoring segment pair values |
| blastn | Nucleotide-nucleotide BLAST |
| ALPPL2 | Adaptor protein, phosphotyrosine interacting with PH domain and leucine zipper 2 |
| RANBP17 | RAN binding protein 17 |
| NF1B | Nuclear factor I B |
| SPP1 | Secreted phosphoprotein 1 |
| CST6 | Cystatin E/M |
| TGF β1 | Transforming growth factor beta1 |
| FGF2 | Fibroblast growth factor 2 |
| EGF | Epidermal growth factor |
| ABPs | Actin-binding proteins |
| RIN | RNA integrity number |
| TBP | TATA-box binding protein |
| GAPDH | Glyceraldehyde 3-phosphate dehydrogenase |
| HPRT1 | Hypoxanthinine guanine phosphoribosyltransferase 1 |
| BACT | β-actin |
| CYTB | Mitochondrially encoded cytochrome B |
| YWHAZ | Tyrosine 3-Monooxygenase/Tryptophan 5-Monooxygenase Activation Protein zeta |
| B2 M | Beta-2-microglobulin |
| SDHA | Succinate dehydrogenase complex, subunit A |
| HMBSA | Hydroxymethylbilane synthase a |
| SD | Standard deviation |
| NCBI | National Center for Biotechnology Information |
| ACTB | Actin beta |
| HMBS | hydroxymethylbilane synthase |
| FABP3 | Fatty acid-binding protein 3 |
| IL1B | Interleukin 1B |
| ITGAV | Integrin AV |
| NMB | Neuromedin B |
| OPN | Osteopontin |
| PPARG | Peroxisome proliferator-activated gamma receptor |
| LC480 | Light cycler 480 |
References
- Wang, D.; Liu, K.; Yang, Y.; Wang, T.; Rao, Q.; Guo, W.; Zhang, Z. Prognostic value of leukemia inhibitory factor and its receptor in pancreatic adenocarcinoma. Futur. Oncol. 2019, 16, 4461–4473. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.; Jang, H.; Seo, H.; Yoo, I.; Han, J.; Kim, M.; Lee, S.; Ka, H. Changes in calcium levels in the endometrium throughout pregnancy and the role of calcium on endometrial gene expression at the time of conceptus implantation in pigs. Mol. Reprod. Dev. 2019, 86, 883–895. [Google Scholar] [CrossRef] [PubMed]
- King, G.J.; Atkinson, B.A.; Robertson, H.A. Implantation and early placentation in domestic ungulates. J. Reprod. Fertil. Suppl. 1982, 31, 87–95. [Google Scholar]
- Zhang, H.; Wang, S.; Liu, M.; Zhang, A.; Wu, Z.; Zhang, Z.; Li, J. Differential gene expression in the endometrium on gestation day 12 provides insight into sow prolificacy. BMC Genom. 2013, 14, 45. [Google Scholar] [CrossRef] [PubMed]
- Burton, G.J.; Fowden, A.L. The placenta: A multifaceted, transient organ. Philos. Trans. R. Soc. B Biol. Sci. 2015, 370, 20140066. [Google Scholar] [CrossRef]
- Gu, T.; Zhu, M.; Schroyen, M.; Qu, L.; Nettleton, D.; Kuhar, D.; Lunney, J.K.; Ross, J.W.; Zhao, S.; Tuggle, C.K. Endometrial gene expression profiling in pregnant Meishan and Yorkshire pigs on day 12 of gestation. BMC Genom. 2014, 15, 156. [Google Scholar] [CrossRef]
- Zeng, S.; Bick, J.; Ulbrich, S.E.; Bauersachs, S. Cell type-specific analysis of transcriptome changes in the porcine endometrium on Day 12 of pregnancy. BMC Genom. 2018, 19, 459. [Google Scholar] [CrossRef]
- Zhang, S.; Lin, H.; Kong, S.; Wang, S.; Wang, H.; Wang, H.; Armant, D.R. Physiological and molecular determinants of embryo implantation. Mol. Asp. Med. 2013, 34, 939–980. [Google Scholar] [CrossRef]
- Waclawik, A.; Kaczmarek, M.M.; Blitek, A.; Kaczyński, P.; Ziecik, A.J. Embryo-maternal dialogue during pregnancy establishment and implantation in the pig. Mol. Reprod. Dev. 2017, 84, 842–855. [Google Scholar] [CrossRef]
- Ziecik, A.J.; Waclawik, A.; Kaczmarek, M.M.; Blitek, A.; Jalali, B.M.; Andronowska, A. Mechanisms for the Establishment of Pregnancy in the Pig. Reprod. Domest. Anim. 2011, 46, 31–41. [Google Scholar] [CrossRef]
- Soede, N.M.; Langendijk, P.; Kemp, B. Reproductive cycles in pigs. Anim. Reprod. Sci. 2011, 124, 251–258. [Google Scholar] [CrossRef] [PubMed]
- Franczak, A.; Wojciechowicz, B.; Kotwica, G. Transcriptomic analysis of the porcine endometrium during early pregnancy and the estrous cycle. Reprod. Biol. 2013, 13, 229–237. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Seo, H.; Choi, Y.; Shim, J.; Kim, H.; Lee, C.-K.; Ka, H. Microarray Analysis of Gene Expression in the Uterine Endometrium during the Implantation Period in Pigs. Asian-Australasian J. Anim. Sci. 2012, 25, 1102–1116. [Google Scholar] [CrossRef] [PubMed]
- Bazer, F.W.; Burghardt, R.; Johnson, G.A.; Spencer, T.E.; Wu, G. Mechanisms for the establishment and maintenance of pregnancy: Synergies from scientific collaborations. Biol. Reprod. 2018, 99, 225–241. [Google Scholar] [CrossRef]
- Bazer, F.W.; Spencer, T.E.; Johnson, G.A.; Burghardt, R.; Wu, G. Comparative aspects of implantation. Reproduction 2009, 138, 195–209. [Google Scholar] [CrossRef]
- Lin, H.; Wang, H.; Wang, Y.; Liu, C.; Wang, C.; Guo, J. Transcriptomic Analysis of the Porcine Endometrium during Embryo Implantation. Genes 2015, 6, 1330–1346. [Google Scholar] [CrossRef]
- Geisert, R.D.; Schmitt, R.A.M. Early embryonic survival in the pig: Can it be improved? J. Anim. Sci. 2002, 80, E54–E65. [Google Scholar]
- Kaplon, M.J.; Rothschild, M.F.; Berger, P.J.; Healey, M. Genetic and phenotypic trends in Polish large white nucleus swine herds. J. Anim. Sci. 1991, 69, 551. [Google Scholar] [CrossRef]
- Gurgul, A.; Jasielczuk, I.; Ropka-Molik, K.; Semik-Gurgul, E.; Pawlina-Tyszko, K.; Szmatoła, T.; Szyndler-Nędza, M.; Bugno-Poniewierska, M.; Blicharski, T.; Szulc, K.; et al. A genome-wide detection of selection signatures in conserved and commercial pig breeds maintained in Poland. BMC Genet. 2018, 19, 95. [Google Scholar] [CrossRef]
- Chomczynski, P.; Sacchi, N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 1987, 162, 156–159. [Google Scholar] [CrossRef]
- Dobbin, K.; Shih, J.H.; Simon, R. Questions and answers on design of dual-label microarrays for identifying differentially expressed genes. J. Natl. Cancer Inst. 2003, 95, 1362–1369. [Google Scholar] [CrossRef] [PubMed]
- Two-Color Microarray Based Gene Expression Analysis. Protocol Version 6.9.1. Available online: https://www.agilent.com/cs/Library/usermanuals/public/G4140-90050_GeneExpression_TwoColor_6.9.pdf (accessed on 28 August 2015).
- Jolliffe, I.T.; Cadima, J. Principal component analysis: A review and recent developments. Philos. Trans. R. Soc. A Math. Phys. Eng. Sci. 2016, 374, 20150202. [Google Scholar] [CrossRef] [PubMed]
- Qian, H.-R.; Huang, S.-G. Comparison of false discovery rate methods in identifying genes with differential expression. Genomics 2005, 86, 495–503. [Google Scholar] [CrossRef] [PubMed]
- Glickman, M.; Rao, S.R.; Schultz, M.R. False discovery rate control is a recommended alternative to Bonferroni-type adjustments in health studies. J. Clin. Epidemiol. 2014, 67, 850–857. [Google Scholar] [CrossRef] [PubMed]
- Sus Scrofa (Pig) Genome Assembly and Annotation Reprot. Available online: https://www.ncbi.nlm.nih.gov/genome/84?genome_assembly_id=317145 (accessed on 11 December 2019).
- Fassler, J.; Cooper, P. BLAST Glossary. NCBI Help Manual. Available online: https://www.ncbi.nlm.nih.gov/books/NBK62051/ (accessed on 14 July 2011).
- Oliveros, J.C. Venny. An Interactive Tool for Comparing Lists with Venn’s Diagrams (2007–2015). Available online: http://bioinfogp.cnb.csic.es/tools/venny/index.html (accessed on 1 July 2007).
- Warnes, G.; Bolker, B.; Bonebakker, L.; Gentleman, R.; Huber, W.; Liaw, A.; Lumley, T.; Mächler, M.; Magnusson, A.; Möller, S. Gplots: Various r Programming Tools for Plotting Data. 2005. Available online: https://cran.r-project.org/web/packages/gplots/index.html (accessed on 27 September 2019).
- Wilkinson, L.; Friendly, M. The History of the Cluster Heat Map. Am. Stat. 2009, 63, 179–184. [Google Scholar] [CrossRef]
- Bindea, G.; Galon, J.; Mlecnik, B. CluePedia Cytoscape plugin: Pathway insights using integrated experimental and in silico data. Bioinformatics 2013, 29, 661–663. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Thallinger, G.G.; Obermayr, E.; Charoentong, P.; Tong, D.; Trajanoski, Z.; Zeillinger, R. A Sequence Based Validation of Gene Expression Microarray Data. Am. J. Bioinform. 2012, 1, 1–9. [Google Scholar] [CrossRef]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate snormalisation of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, research0034-1. [Google Scholar] [CrossRef]
- De Spiegelaere, W.; Dern-Wieloch, J.; Weigel, R.; Schumacher, V.; Schorle, H.; Nettersheim, D.; Bergmann, M.; Brehm, R.; Kliesch, S.; Vandekerckhove, L.; et al. Reference Gene Validation for RT-qPCR, a Note on Different Available Software Packages. PLoS ONE 2015, 10, e0122515. [Google Scholar] [CrossRef]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T. Primer-BLAST: A tool to design target specific primers for polymerase chain reaction. BMC Bioinform. 2012, 13, 134. Available online: http://www.ncbi.nlm.nih.gov/tools/primer-blast/ (accessed on 23 August 2012). [CrossRef]
- OligoAnalyzer: Users Guide and Protocols, Integrated DNA Technologies, Inc, Iowa, USA. Available online: https://eu.idtdna.com/analyzer/Applications/OligoAnalyzer/ (accessed on 31 December 2019).
- Ruijter, J.M.; Ramakers, C.; Hoogaars, W.M.H.; Karlen, Y.; Bakker, O.; Hoff, M.J.B.V.D.; Moorman, A.F.M. Amplification efficiency: Linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res. 2009, 37, e45. [Google Scholar] [CrossRef] [PubMed]
- Calculate and Draw Custom Venn Diagrams. VIB/ UGent Bioinformatics and Evolutionary Genomics, Gent, Belgium. Available online: http://bioinformatics.psb.ugent.be/webtools/Venn/ (accessed on 31 December 2019).
- Ka, H.; Seo, H.; Kim, M.; Choi, Y.; Lee, C.-K. Identification of differentially expressed genes in the uterine endometrium on day 12 of the estrous cycle and pregnancy in pigs. Mol. Reprod. Dev. 2009, 76, 75–84. [Google Scholar] [CrossRef] [PubMed]
- Samborski, A.; Graf, A.; Krebs, S.; Kessler, B.; Bauersachs, S. Deep Sequencing of the Porcine Endometrial Transcriptome on Day 14 of Pregnancy1. Biol. Reprod. 2013, 88, 84. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Li, A.; Chen, W.; Wei, J.; Fu, J.; Wang, A. Differential Gene Expression in Uterine Endometrium During Implantation in Pigs1. Biol. Reprod. 2015, 92, 52. [Google Scholar] [CrossRef]
- Kim, M.; Seo, H.; Choi, Y.; Yoo, I.; Seo, M.; Lee, C.-K.; Kim, H.; Ka, H. Analysis of Stage-Specific Gene Expression Profiles in the Uterine Endometrium during Pregnancy in Pigs. PLoS ONE 2015, 10, e0143436. [Google Scholar] [CrossRef]
- Østrup, E.; Bauersachs, S.; Blum, H.; Wolf, E.; Hyttel, P. Differential endometrial gene expression in pregnant and non-pregnant sows. Biol. Reprod. 2010, 83, 277–285. [Google Scholar] [CrossRef]
- Kiewisz, J.; Krawczynski, K.; Lisowski, P.; Blitek, A.; Zwierzchowski, L.; Ziecik, A.J.; Kaczmarek, M.M. Global gene expression profiling of porcine endometria on Days 12 and 16 of the estrous cycle and pregnancy. Theriogenology 2014, 82, 897–909. [Google Scholar] [CrossRef]
- Huang, J.; Liu, R.; Su, L.; Xiao, Q.; Yu, M. Transcriptome Analysis Revealed the Embryo-Induced Gene Expression Patterns in the Endometrium from Meishan and Yorkshire Pigs. Int. J. Mol. Sci. 2015, 16, 22692–22710. [Google Scholar] [CrossRef]
- Altimimi, H.; Schnetkamp, P.P. Na+/Ca2+-K+ exchangers (NCKX): Functional properties and physiological roles. Channels 2007, 1, 62–69. [Google Scholar] [CrossRef]
- Choi, Y.; Seo, H.; Shim, J.; Yoo, I.; Ka, H. Calcium extrusion regulatory molecules: Differential expression during pregnancy in the porcine uterus. Domest. Anim. Endocrinol. 2014, 47, 1–10. [Google Scholar] [CrossRef]
- Whitaker, M. Calcium at sfertilisation and in early development. Physiol. Rev. 2006, 86, 25–88. [Google Scholar] [CrossRef]
- Geisert, R.D.; Brookbank, J.W.; Roberts, R.M.; Bazer, F.W. Establishment of pregnancy in the pig: II. Cellular remodeling of the porcine blastocyst during elongation on day 12 of pregnancy. Biol. Reprod. 1982, 27, 941–955. [Google Scholar] [CrossRef] [PubMed]
- Ruan, Y.C.; Chen, H.; Chan, H.C. Ion channels in the endometrium: Regulation of endometrial receptivity and embryo implantation. Hum. Reprod. Updat. 2014, 20, 517–529. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Li, P.; Zhang, Q.; He, L.; Su, G.; Huang, Y.; Lu, Z.; Hu, W.; Ding, H.; Huang, R. Transcriptome analysis of the endometrium from Chinese Erhualian sows that differ in calcium ion concentration and litter size. Anim. Genet. 2019, 50, 326–333. [Google Scholar] [CrossRef] [PubMed]
- Jalali, B.M.; Likszo, P.; Andronowska, A.; Skarzynski, D.J. Alterations in the distribution of actin and its binding proteins in the porcine endometrium during early pregnancy: Possible role in epithelial remodeling and embryo adhesion. Theriogenology 2018, 116, 17–27. [Google Scholar] [CrossRef]
- Galton, V.A.; Martinez, E.; Hernandez, A.; St Germain, E.A.; Bates, J.M.; St Germain, D.L. The type 2 iodothyronine deiodinase is expressed in the rat uterus and induced during pregnancy. Endocrin 2001, 142, 2123–2128. [Google Scholar] [CrossRef]
- Kasik, J. Selenoprotein p expression in liver, uterus and placenta during late pregnancy. Placenta 1995, 16, 67–74. [Google Scholar] [CrossRef]
- Ceko, M.J.; Hummitzsch, K.; Hatzirodos, N.; Bonner, W.M.; Aitken, J.B.; Russell, D.L.; Lane, M.; Rodgers, R.; Harris, H.H. X-Ray fluorescence imaging and other analyses identify selenium and GPX1 as important in female reproductive function. Metallomics 2015, 7, 71–82. [Google Scholar] [CrossRef]
- Boesch, M.; Sopper, S.; Marth, C.; Fiegl, H.; Wiedemair, A.; Rössler, J.; Hatina, J.; Wolf, D.; Reimer, D.; Zeimet, A.G. Evaluation of Vav3.1 as prognostic marker in endometrial cancer. J. Cancer Res. Clin. Oncol. 2018, 144, 2067–2076. [Google Scholar] [CrossRef]
- Anczuków, O.; Akerman, M.; Cléry, A.; Wu, J.; Shen, C.; Shirole, N.H.; Raimer, A.; Sun, S.; Jensen, M.A.; Hua, Y.; et al. SRSF1-Regulated Alternative Splicing in Breast Cancer. Mol. Cell 2015, 60, 105–117. [Google Scholar] [CrossRef] [PubMed]
- Dziuk, P. Effect of migration, distribution and spacing of pig embryos on pregnancy and fetal survival. J. Reprod. Fertil. Suppl. 1985, 33, 57–63. [Google Scholar] [PubMed]
- Geisert, R.D.; Zavy, M.T.; Moffatt, R.J.; Blair, R.M.; Yellin, T. Embryonic steroids and the establishment of pregnancy in pigs. J. Reprod. Fertil. Suppl. 1990, 40, 293–305. [Google Scholar] [PubMed]
- Modi, D.; Bhartiya, P. Physiology of embryo-endometrial cross talk. Biomed. Res. J. 2015, 2, 83. [Google Scholar] [CrossRef]
- Su, Y.; Zhang, J.-J.; He, J.-L.; Liu, X.-Q.; Chen, X.-M.; Ding, Y.-B.; Tong, C.; Peng, C.; Geng, Y.-Q.; Wang, Y.-X.; et al. Endometrial autophagy is essential for embryo implantation during early pregnancy. J. Mol. Med. 2020, 98, 555–567. [Google Scholar] [CrossRef]
- Smolinska, N.; Szeszko, K.; Dobrzyn, K.; Kiezun, M.; Rytelewska, E.; Kisielewska, K.; Gudelska, M.; Bors, K.; Wyrebek, J.; Kopij, G.; et al. Transcriptomic Analysis of Porcine Endometrium during Implantation after In Vitro Stimulation by Adiponectin. Int. J. Mol. Sci. 2019, 20, 1335. [Google Scholar] [CrossRef]
- Samborski, A.; Graf, A.; Krebs, S.; Kessler, B.; Reichenbach, M.; Reichenbach, H.-D.; Ulbrich, S.E.; Bauersachs, S. Transcriptome Changes in the Porcine Endometrium During the Preattachment Phase1. Biol. Reprod. 2013, 89, 134. [Google Scholar] [CrossRef]
- Dimova, T.; Mihaylova, A.; Spassova, P.; Georgieva, R. Establishment of the Porcine Epitheliochorial Placenta Is Associated with Endometrial T-Cell Recruitment. Am. J. Reprod. Immunol. 2007, 57, 250–261. [Google Scholar] [CrossRef]
- Wollenhaupt, K.; Kettler, A.; Brüssow, K.-P.; Schneider, F.; Kanitz, W.; Einspanier, R. Regulation of the expression and bioactivation of the epidermal growth factor receptor system by estradiol in pig oviduct and endometrium. Reprod. Fertil. Dev. 2001, 13, 167. [Google Scholar] [CrossRef]
- Jeong, W.; Kim, J.; Bazer, F.W.; Song, G. Epidermal growth factor stimulates proliferation and migration of porcine trophectoderm cells through protooncogenic protein kinase 1 and extracellular-signal-regulated kinases 1/2 mitogen-activated protein kinase signal transduction cascades during early pregnancy. Mol. Cell Endocrinol. 2013, 381, 302–311. [Google Scholar]
- Choi, Y.; Seo, H.; Kim, M.; Ka, H. Dynamic Expression of Calcium-Regulatory Molecules, TRPV6 and S100G, in the Uterine Endometrium During Pregnancy in Pigs1. Biol. Reprod. 2009, 81, 1122–1130. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.-M.; Ding, G.-L.; Jiang, Y.; Pan, H.-J.; Zhang, D.; Wang, T.-T.; Zhang, R.-J.; Shu, J.; Sheng, J.-Z.; Huang, H.-F. Down-regulation of S100A11, a calcium-binding protein, in human endometrium may cause reproductive failure. J. Clin. Endocrinol. Metab. 2012, 97, 3672–3683. [Google Scholar] [CrossRef] [PubMed]
- Geisert, R.D.; Johnson, G.A.; Burghardt, R. Implantation and Establishment of Pregnancy in the Pig. Regulation of Implantation and Establishment of Pregnancy in Mammals; Springer: Cham, Switzerland, 2015; Volume 216, pp. 137–163. [Google Scholar] [CrossRef]
- Zeng, S.; Ulbrich, S.E.; Bauersachs, S. Spatial sorganisation of endometrial gene expression at the onset of embryo attachment in pigs. BMC Genom. 2019, 20, 895. [Google Scholar] [CrossRef] [PubMed]
- Frank, J.W.; Seo, H.; Burghardt, R.; Bayless, K.J.; Johnson, G.A. ITGAV (alpha v integrins) bind SPP1 (osteopontin) to support trophoblast cell adhesion. Reproduction 2017, 153, 695–706. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xue, S.; Liu, X.; Liu, H.; Hu, T.; Qiu, X.; Zhang, J.; Lei, M. Analyses of Long Non-Coding RNA and mRNA profiling using RNA sequencing during the pre-implantation phases in pig endometrium. Sci. Rep. 2016, 6, 20238. [Google Scholar] [CrossRef]



| Number of Animals/sample ID | Day of Pregnancy | Number of Corpus Luteum in Right Ovary | Number of Corpus Luteum in Left Ovary |
|---|---|---|---|
| 2.1 | 9 | 5 | 5 |
| 9.1 | 9 | 6 | 6 |
| 8.1 | 9 | 7 | 6 |
| 147.2 | 9 | 9 | 4 |
| 135.2 | 12 | 4 | 9 |
| 140.2 | 12 | 11 | 5 |
| 8.2 | 12 | 9 | 9 |
| 144.2 | 12 | 7 | 10 |
| 131.2 | 16 | 8 | 11 |
| 5.1 | 16 | 4 | 12 |
| 10.2 | 16 | 7 | 7 |
| 1.1 | 16 | 7 | 8 |
| Gene | Full-Name | Primer Sequence | NCBI Access-Code |
|---|---|---|---|
| TBP | TATA-Box Binding Protein | F: 5’ - GATGGACGTTCGGTTTAGG- 3’ R: 5’ - AGCAGCACAGTACGAGCAA- 3’ | DQ178122 |
| GAPDH | Glyceraldehyde-3-Phosphate Dehydrogenase | F:5’-ACTCACTCTTCTACCTTTGATGCT-3’ R: 5’ - TGTTGCTGTAGCCAAATTCA - 3’ | NM_001206359 |
| ACTB | Actin Beta | F: 5’ - GGACTTCGAGCAGGAGATGG - 3’ R: 5’ - GCACCGTGTTGGCGTAGAGG - 3’ | XM_003357928 |
| SDHA | Succinate Dehydrogenase Complex Flavoprotein Subunit A | F: 5’ - GAACCGAAGATGGCAAGA - 3’ R: 5’ - CAGGAGATCCAAGGCAAA - 3’ | XM_005659031 |
| HPRT1 | Hypoxanthine Phosphoribosyltransferase 1 | F: 5’ - CCGAGGATTTGGAAAAGGT- 3’ R: 5’ - CTATTTCTGTTCAGTGCTTTG- 3’ | NM_001032376 |
| CYTB | mitochondrially encoded cytochrome b | F: 5’ - CACATCCAAACAACGAAGCA- 3’ R: 5’ -GTTCTACGGGTTGTCCTCCA - 3’ | AY830188.1 |
| B2 M | beta-2-microglobulin | F: 5’ - AAACGGAAAGCCAAATTACC- 3’ R: 5’ -ATCCACAGCGTTAGGAGTGA - 3’ | XM_005659651 |
| YWHAZ | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein zeta | F: 5’ - ATGCAACCAACACATCCTATC- 3’ R: 5’ - GCATTATTAGCGTGCTGTCTT - 3’ | XM_005662949 |
| HMBS | hydroxymethylbilane synthase | F: 5’ - CTGTTTACCCAAGGAGCTGGA- 3’ R: 5’ - TGAAGCCAGGAGGAAGCA- 3’ | NM_001097412 |
| Gene | Primer Sequence | NCBI Access-Code |
|---|---|---|
| FABP3 | F: 5’ - GGCCAACATGACCAAGCCTA - 3’ R: 5’ - CTGCCATGGGTGAGTGTCAG - 3’ | NM_001099931 |
| RBP4 | F: 5’ - CCCCGAGGGACTCTTTCTGC - 3’ R: 5’ - TCTTGAACTTGGCAGGGTCC - 3’ | NM_214057 |
| PPARG | F: 5’ - TTAGATGACAGCGACCTGGC - 3’ R: 5’ - GTGAAGGCTCATGTCCGTCT - 3’ | NM_214379 |
| IL1B | F: 5’ - GTACATGGTTGCTGCCTGAA - 3’ R: 5’ - TGGCACACTCACCCCAAAG - 3’ | NM_214055 |
| SPP1 | F: 5’ - CACATTGTAGCGAGGTGGGA - 3’ R: 5’ - GTGACGGCTTGTATTTCTTATACGG - 3’ | NM_214023 |
| ITGAV | F: 5’ - GACTTCACTGCTGATGGTGC - 3’ R: 5’ - ACAAAAGATGTGGATAGTAAGGTCT - 3’ | NM_001083932 |
| NMB | F: 5’ - TGTGATTTCTGGTTGAGTTGCC - 3’ R: 5’ - CAAGACATACAGCAGGGACG - 3’ | NM_001123145 |
| S100A8 | F: 5’ - CCGACATGGCAAAAAGACCC- 3’ R: 5’ - ATAAAGGAGTCATGGCAAGCTA- 3’ | NM_001004045 |
| RT-PCR | FABP3 | IL1B | ITGAV | NMB | OPN | PPARG | RBP4 | S100A11 |
| 9D vs. 12D | 0.248 | 0.083 | 0.021 | 0.021 | 0.083 | 0.043 | 0.021 | 0.021 |
| 9D vs. 16D | 0.021 | 0.021 | 0.021 | 0.043 | 0.021 | 0.021 | 0.021 | 0.083 |
| 12 vs. 16D | 0.083 | 0.083 | 0.564 | 0.0021 | 0.083 | 0.149 | 0.149 | 0.021 |
| Microarray | FABP3/A_72_P424889 | IL1B/A_72_P463511 | ITGAV/A_72_P051521 | NMB/A_72_P441888 | OPN/SPP1/A_72_P145586 | PPARG/A_72_P441339 | RBP4/A_72_P146486 | S100A11/A_72_P041606 |
| 9D vs. 12D | 0.574 | 0.00000012 | 0.658 | 0.0432 | 0.0000000754 | 0.123 | 0.00456 | 0.000144 |
| 9D vs. 16D | 0.0284 | 0.0000105 | 0.0572 | 0.578 | 0.274 | 0.00671 | 0.0189 | 0.0217 |
| 12D vs. 16D | 0.105 | 0.000364 | 0.00836 | 0.0639 | 0.00000132 | 0.00865 | 0.807 | 0.000144 |
| Stages | RT-PCR | FABP3 | IL1B | ITGAV | NMB | OPN | PPARG | RBP4 | S100A11 |
| 9D | Mean | 0.29678507 | 0.2862616 | 0.45462323 | 0.039134517 | 0.022101078 | 0.293498944 | 0.004475395 | 0.002163842 |
| 9D | SD | 0.181874213 | 0.217897791 | 0.066866002 | 0.025376539 | 0.011917908 | 0.102534468 | 0.003005485 | 0.00106232 |
| 9D | SEM | 0.090937107 | 0.108948895 | 0.033433001 | 0.01268827 | 0.005958954 | 0.051267234 | 0.001502743 | 0.00053116 |
| 12D | Mean | 0.661363926 | 0.772976768 | 1.032295435 | 2.742343022 | 0.128210738 | 0.768243071 | 1.045137864 | 2.988849196 |
| 12D | SD | 0.792157361 | 0.991360659 | 0.550797913 | 1.79142117 | 0.258761542 | 0.657077116 | 1.372147219 | 5.060744924 |
| 12D | SEM | 0.39607868 | 0.49568033 | 0.275398957 | 0.895710585 | 0.129380771 | 0.328538558 | 0.68607361 | 2.530372462 |
| 16D | Mean | 2.041851004 | 1.940761632 | 1.513081335 | 0.218522461 | 2.849688183 | 1.938257985 | 1.950386742 | 0.008986962 |
| 16D | SD | 0.952351557 | 1.040282257 | 0.745442463 | 0.103526723 | 4.720728783 | 1.162845646 | 1.020400836 | 0.006955678 |
| 16D | SEM | 0.476175779 | 0.520141128 | 0.372721232 | 0.051763361 | 2.360364391 | 0.581422823 | 0.510200418 | 0.003477839 |
| Stages | Microarray | FABP3/A_72_P424889 | IL1B/A_72_P463511 | ITGAV/A_72_P051521 | NMB/A_72_P441888 | OPN/SPP1/A_72_P145586 | PPARG/A_72_P441339 | RBP4/A_72_P146486 | S100A11/A_72_P041606 |
| 9D | Mean | 2439.201 | 48.485 | 15496.08 | 23200.24 | 344.068 | 238.932 | 25301.21 | 9645.699 |
| 9D | SD | 1008.043083 | 10.89272496 | 5371.666 | 63055.29 | 55.352831 | 76.57238 | 36255.42 | 2418.796989 |
| 9D | SEM | 356.3970497 | 3.851159844 | 1899.171 | 22293.41 | 19.570181 | 27.07242 | 12818.23 | 855.1738765 |
| 12D | Mean | 2906.12 | 192.019 | 16750.34 | 113595 | 61.806 | 307.028 | 114344.7 | 19990.117 |
| 12D | SD | 1492.354766 | 54.20298132 | 3277.125 | 76325.14 | 21.970376 | 63.24315 | 26856.04 | 4844.695652 |
| 12D | SEM | 527.6270877 | 19.16364783 | 1158.639 | 26985.01 | 7.767701 | 22.35983 | 9495.044 | 1712.858574 |
| 16D | Mean | 4662.605 | 111.572 | 22283.82 | 42044.74 | 459.502 | 445.37 | 109625.8 | 5752.108 |
| 16D | SD | 2097.077529 | 11.64962418 | 3102.399 | 54008.59 | 246.97822 | 115.78 | 33988.1 | 1899.866899 |
| 16D | SEM | 741.4288708 | 4.118764126 | 1096.864 | 19094.92 | 87.319986 | 40.93442 | 12016.61 | 671.7043838 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pierzchała, M.; Pierzchała, D.; Ogłuszka, M.; Poławska, E.; Blicharski, T.; Roszczyk, A.; Nawrocka, A.; Urbański, P.; Stepanow, K.; Ciepłoch, A.; et al. Identification of Differentially Expressed Gene Transcripts in Porcine Endometrium during Early Stages of Pregnancy. Life 2020, 10, 68. https://doi.org/10.3390/life10050068
Pierzchała M, Pierzchała D, Ogłuszka M, Poławska E, Blicharski T, Roszczyk A, Nawrocka A, Urbański P, Stepanow K, Ciepłoch A, et al. Identification of Differentially Expressed Gene Transcripts in Porcine Endometrium during Early Stages of Pregnancy. Life. 2020; 10(5):68. https://doi.org/10.3390/life10050068
Chicago/Turabian StylePierzchała, Mariusz, Dorota Pierzchała, Magdalena Ogłuszka, Ewa Poławska, Tadeusz Blicharski, Agnieszka Roszczyk, Agata Nawrocka, Pawel Urbański, Kamila Stepanow, Aleksandra Ciepłoch, and et al. 2020. "Identification of Differentially Expressed Gene Transcripts in Porcine Endometrium during Early Stages of Pregnancy" Life 10, no. 5: 68. https://doi.org/10.3390/life10050068
APA StylePierzchała, M., Pierzchała, D., Ogłuszka, M., Poławska, E., Blicharski, T., Roszczyk, A., Nawrocka, A., Urbański, P., Stepanow, K., Ciepłoch, A., Korwin-Kossakowska, A., te Pas, M. F. W., Slaska, B., Buszewska-Forajta, M., Jaśkowski, J. M., Sachajko, M., Herudzińska, M., Jaśkowski, B. M., Niżański, W., ... Pareek, C. S. (2020). Identification of Differentially Expressed Gene Transcripts in Porcine Endometrium during Early Stages of Pregnancy. Life, 10(5), 68. https://doi.org/10.3390/life10050068






