Abstract
Steroidal glycoalkaloids (SGAs) are a class of nitrogen-containing glycosides occurring in several plant families and biosynthesized through a specific pathway. HMG-CoA reductase is the first enzyme of this pathway, and its transcription can be regulated by biotic and abiotic stressors and even in a tissue-specific manner. This study aimed to characterize the HMG genes family in a tuber-bearing potato species, Solanum commersonii, using transcriptional and functional approaches. Our results provided evidence that four ScHMGs with different tissue-specificities represent the HMG gene family in S. commersonii and that they originated from ScHMG1 through segmental duplications. Phylogenetic analysis suggests that ScHMG1 is the direct ortholog of AtHMG1, which is associated with SGAs accumulation in plants. Its overexpression in S. commersonii revealed that this gene plays a key role in the accumulation of glycoalkaloids regulating the production of dehydrocommersonine.
1. Introduction
Steroidal glycoalkaloids (SGAs) are a class of nitrogen-containing glycosides occurring in several members of the plant family Solanaceae, such as tomato, eggplant and potato. They are toxic to humans and have putative roles in plant defense against environmental stressors [1]. SGAs can be produced in all parts of the plants, including the edible ones, such as eggplant and tomato fruits and potato tubers, as well as in leaves, flowers and roots. Nowadays, over 50 SGAs have been identified. Among them, trisaccharides α-solamargine and α-solasonine predominate in eggplants; tetrasaccharides α-tomatine and dehydrotomatine in tomatoes; and trisaccharides α-chaconine and α-solanine in potatoes [2]. In particular, α-chaconine and α-solanine comprise more than 90% of the total SGA content in the tuber [3]. It has been demonstrated that wild tuber-bearing potatoes can produce additional glycoalkaloids, and it has been postulated that, during domestication, Solanaceae selected only two predominant SGAs, in order to have a lower number of SGAs, while maintaining resistance against phytopathogens [2]. Indeed, depending on the amount and type, SGAs can trigger various effects on plant physiology, and their role can be critical for plant resistance to biotic stressors, including fungi, bacteria, viruses and insects [2]. For example, SGAs in potatoes cause toxicity to the Guatemalan potato moth and the Colorado potato beetle, some of the worst potato pests [4,5]. Plant SGAs are biosynthesized by a specific pathway, where the major player is the 3-hydroxy-3-methylglutaryl coenzyme A reductase (HMG-CoA), which catalyzes the first committed step of sterol precursors biosynthesis. Once formed, the precursors undergo several biochemical processes, such as hydroxylation, oxidation, transamination and glycosylation, before they produce SGAs [6]. The HMG-coding genes are highly regulated by both abiotic and biotic stressors, like light [7], hormones [8], wounding [9] and pathogens [10]. At the genome level, HMG is encoded by small gene families, with two genes characterized in Arabidopsis thaliana [11] and radish [12], three in Hevea [13] and four in tomato [14]. In the cultivated potato Solanum tuberosum, three HMGs genes have been characterized [15]. It has been demonstrated that they are differentially expressed in response to wounding (StHMG1) or pathogens (StHMG2 and StHMG3) [7]. Krits et al. [16] showed that there is a correlation between transcript levels of StHMG1 and total SGAs. However, to the authors’ best knowledge, no studies have examined HMG gene family members in wild potato relatives, which can biosynthesize a greater amount of glycoalkaloids when compared to the cultivated form [17,18]. Such a lack of studies contrasts with the interest in efficiently using wild potatoes in breeding programs to obtain varieties with improved traits. Among wild tuber-bearing species, Solanum commersonii has become a model for molecular biology, genetics and breeding, garnering significant research interest. Indeed, this species possesses several desired resistances (to nematodes, bacteria and viruses) lacking in the cultivated potato [19,20,21,22]. It is particularly attractive for its freezing tolerance and capacity to cold acclimate [23]. Furthermore, it is the first wild potato whose genome sequence has been released [23], paving the way to different studies aimed to gain insights into genomic evolution at the sequence level from the wild to the cultivated form [24,25]. S. commersonii possesses a diverse mixture of glycoalkaloids, such as dehydrotomatine, dehydrodemissine and dehydrocommersonine, which are among the most poisonous compounds against different potato pests [26]. In this species, Vazquez et al. [27] also reported the presence of demissine, commersonine and a-tomatine. All of these characteristics make S. commersonii a suitable material for the elucidation of the role of HMGs in glycoalkaloid synthesis in potatoes and for studying the evolution of the HMG gene family from wild to cultivated species. Here, we report for the first time, in a wild potato, a study on the HMG gene family and the functional analysis of the gene ScHMG1 encoding HMG-CoA reductase.
2. Materials and Methods
2.1. Identification of Candidate HMG Genes and Phylogenetic Analysis
We used as queries the known HMG protein sequences of A. thaliana (downloaded from phytozome.jgi.doe.gov/pz/portal.html [11]) to search the orthologs in S. commersonii clone cmm1T of PI243503 (hereafter called cmm1T), S. tuberosum Group Phureja (DM1-3 516 R44) and S. lycopersicum, using Blastp with a threshold for significance of at least 103, in order to consider a match. Protein sequences of putative HMG in tomatoes and potatoes were confirmed through domain analysis by using the online software NCBI Search Domain [28]. All confirmed candidates were then aligned by using MUSCLE implemented in MEGAX [29], and the resulting matrix was used as input to identify the best fit for the substitution model for phylogeny (option find best DNA/Protein model). The phylogenetic analysis was finally performed, using the maximum likelihood method, with the best-predicted option JTT+G and 1000 bootstrap replicates. Our candidates were named based on the nomenclature of A. thaliana genes and the phylogenetic analysis. To further investigate the evolutionary history of our candidate proteins, the exon–intron organization was also studied by the online Spidey program [30]. The expression profiles of the HMG genes across four different tissues of S. commersonii and S. tuberosum (flower, leaf, root and stolon) were also studied. In particular, raw data for S. commersonii (deposited under the study SRP050412) and S. tuberosum (ERR029909, ERR029910, ERR029914 and ERR029917) were downloaded and analyzed, using Cufflinks–Cuffquant software (version 2.2.1); expression values for each gene were expressed as RPKM (Reads Per Kilobase of transcript per Million mapped reads), as described by Esposito et al. [24,31,32].
2.2. Determination of Gene Duplication Patterns
The patterns of HMG gene duplication were determined as segmental or tandem based on the position of paralogues on chromosomes, as reported by Zhao et al. [33] and Xia et al. [34]. The PGDD [35] was used to determine the genomic duplication blocks. The Ks values of paralogues in segmental duplication blocks were retrieved from the PGDD database computed by the KaKs calculator [36]. The timing of duplication events can be estimated by using the Ks value and a given clock-like rate, λ, through the formula T = Ks/2λ where, for tomato, λ = 1.5 × 10−8 substitutions per synonymous site per year [37].
2.3. Plant Material
Plantlets of S. commersonii were propagated in vitro on Murashige and Skoog (MS) medium (Sigma-Aldrich, St. Louis, MO, USA) with 1% (w/v) sucrose and 0.8% (w/v) agar. They were incubated at 24 °C, exposed to an irradiance of 200 mol m2 sec1 and under a 16 h/8 h (light/dark) photoperiod, as described by D’Amelia et al. [38].
2.4. Overexpression of ScHMG1 in S. commersonii
The ScHMG1 coding sequences were cloned in the 35SCaMV expression cassette of pGWB411 [39], using Gateway recombination technology (Life Technologies, Carlsbad, CA, USA). Agrobacterium tumefaciens cells ELECTRO MAX LBA4404 (Life Technologies, Carlsbad, CA, USA) transformed with each expression vector were used for co-cultivation of cmm1T leaf explants, according to the protocol of Cardi et al. [40]. A single plant was taken from a single callus. Transgenic plantlets were verified by genomic PCR on kanamycin gene-specific primers (Table S1) as reported by Brulè et al. [41].
2.5. RNA Extraction and Gene Expression Analyses
Total RNA was extracted from transformed plantlets. RNA extraction was performed by using the Spectrum Plant Total RNA Kit (Sigma-Aldrich, St. Louis, MO, USA) and the On-Column DNase I Digestion Set (Sigma-Aldrich, St. Louis, MO, USA). Then, 1 μg of RNA was reverse transcribed, using Oligo-Dt (20) and the SuperScript III reverse transcriptase (Invitrogen, www.invitrogen.com) in 20 μL final reaction volume, according to the manufacturer’s instructions. The obtained cDNA was used to monitor the expression change of genes through RT-qPCR. The experiment was carried out by using the 2x QuantiFast SYBR Green PCR Master Mix (Qiagen, Valencia, CA, USA) with ABI PRISM 7900HT (Applied Biosystems, Foster City, CA, USA). Each 15 μL reaction contained 300 nM of each primer and 1:5 diluted cDNA. Cycle conditions indicated by QuantiFast SYBR Green PCR Kit handbook (Qiagen, Valencia, CA, USA) were used. The housekeeping gene used for normalization of expression data was adenine phosphoribosyltransferase (APRT) in transgenic plantlets expression analysis. Results were analyzed by using the ABI PRISM 7900HT Sequence Detection System Version 2.1 (Applied Biosystems, Foster City, CA, USA). The relative expression was estimated according to the ∆∆Ct method [42], as described by Villano et al. [43] and Di Meo et al. [44]. Three biological and three technical replicates were prepared for the analysis.
2.6. Copy Number Determination
Quantitative real-time PCR (qPCR) was used to estimate transgene copy numbers in transgenic plants [45]. Genomic DNA was extracted from transformed plantlets, using the DNeasy Plant Mini Kit (Qiagen, Valencia, CA, USA), following the manufacturer’s guidelines. Then 10 ng of genomic DNA was used in a 12.5 μL final reaction volume, according to the manufacturer’s instructions. The experiment was carried out in triplicates, using the 2x QuantiFast SYBR Green PCR Master Mix (Qiagen, Valencia, CA, USA) with ABI PRISM 7900HT (Applied Biosystems, Foster City, CA, USA). Relative gene copy numbers were obtained by using the ΔCt method of relative quantification, since the genes were amplified with similar efficiency in each sample. The relative gene copy number was calculated as 2−ΔCt, where ΔCt = Cttarget − Ctreference. The single-copy gene GBSS1 (granule-bound starch synthase 1) was used as a reference gene.
2.7. SGA Determination
SGA determination was performed by Liquid Chromatography–High Resolution Mass Spectrometry (LC–HRMS) analysis. Freeze-dried leaves (250 mg) were extracted with 20 mL of 2% acetic acid solution, using an Ultra Ika T18 basic Ultraturrax (Staufen, Germany) for 1 min. The extract was centrifuged at 14,800 rpm for 10 min and directly used for HMRS analysis. LC–HRMS data were acquired on an Accela U-HPLC system coupled to an Exactive Orbitrap mass spectrometer equipped with a heated electrospray interface (HESI) (Thermo Fisher Scientific, San Jose, CA). Chromatographic separation was performed, using a Gemini C18-110Å 5 μm column (150 mm × 2.0 mm) (Phenomenex, Torrance, CA) at 30 °C. The mobile phase consisted of 0.1% formic acid water (A) and 0.1% formic acid acetonitrile (B). Gradient elution was linearly programmed as follows: 10% B (1 min), 10%–90% B (7 min), constant to 90% B (2 min), 90%–10% B (2 min). The solvent flow rate was 200 μL/min, and the injection volume was 10 μL. The acquisition was performed in positive ionization mode in the mass range of m/z 100–1200. The resolving power was set to 50,000 full width at half-maximum (FWHM, m/z 200), resulting in a scan time of 1 s. The automatic gain control was used in balanced mode (1 × 106 ions); the maximum injection time was 100 ms. The spray voltage was at 3 kV, the capillary voltage was 30 V, the capillary temperature was at 275 °C and a sheath and auxiliary gas flow of 30 and 15 arbitrary units were used. The instrument was externally calibrated in the positive ion mode, following the Thermo Exactive calibration procedure. Chromatographic data acquisition and peak integration were performed by using Xcalibur software (Thermo Fisher Scientific, San Jose, USA). Compounds’ identification was performed by using exact mass values up to the fifth decimal digit, with mass tolerance ± 5 ppm. All analyzed compounds were expressed as equivalents of α-solanine. Molecular formula, theoretical mass, experimental mass and mass accuracy are reported in Table S2.
3. Results and Discussion
3.1. Comparative and Phylogenetic Analysis of the HMG Gene Families in Solanaceae
Several lines of evidence support that HMG-CoA reductase (HMG) has a key role in the regulation of the metabolic flux through the plant sterol biosynthetic pathway. Here, a genome-wide study of the HMG family has been carried out in three Solanaceae species, and one of these genes, ScHMG1, has been functionally characterized in wild tuber-bearing S. commersonii. Our study began with the identification of the HMG gene family members in cultivated tomato S. lycopersicum, cultivated potato S. tuberosum and S. commersonii, starting from the genome of the model plant A. thaliana. To date, two differentially expressed HMG genes, AtHMG1 and AtHMG2, have been characterized in A. thaliana [11,46]. Their contrasting role in SGA biosynthesis and plant development has been largely demonstrated. For example, the analysis of a null AtHMG1 evidenced the essential role of this gene in controlling several plant developmental traits, such as dwarfism, senescence and sterility. By contrast, the silencing of AtHMG2 did not affect either the phenotype or the fertility of the plant under normal growth conditions [47].
Within Solanaceae, recently, Zhang et al. [7] reported that the S. tuberosum genome holds two HMG genes (StHMG1 and StHMG2). However, in our study, we were able to identify four StHMGs, namely StHMG1, StHMG3, StHMG4 and StHMG5 (Figure 1A), and here we propose a new potato HMG gene family classification. Direct orthologs of AtHMG1 were found in S. tuberosum (PGSC0003DMP400017516) and S. commersonii (maker_scaffold1783_augustus_gene_0_28), but not in tomato. By contrast, no robust orthology relationship was found for AtHMG2, which did not cluster with any Solanum HMGs (Figure 1A). The remaining sequences were named HMG3, HMG4 and HMG5, using progressive numbering. It is noteworthy that StHMG5 has been previously annotated as StHMG2 [26,48]. However, our data rule out any orthologous relationship between StHMG5 and AtHMG2. Regarding StHMG4, we found two different isoforms in S. tuberosum annotated as PGSC0003DMP400024173 (which has a strong similarity with the N-terminal portion of SlHMG4 and ScHMG4) and PGSC0003DMP400024174 (strongly similar to the C-terminal portion of the same proteins). Therefore, we considered them to be a single protein. HMG3, HMG4 and HMG5 had one-to-one direct ortholog in S. commersonii, S. tuberosum and S. lycopersicum, suggesting a gene expansion occurred in Solanaceae since it last shared a common ancestor with the model species A. thaliana.
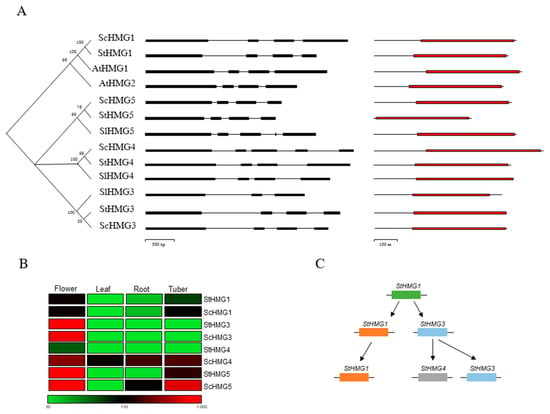
Figure 1.
(A) Phylogenetic analysis of the HMG gene family in Arabidopsis, Solanum lycopersicum, S. tuberosum and S. commersonii. Domain distribution (red boxes represent the HMG-CoA reductase class I domain) with NCBI search domain and exon–intron structure (black boxes represent the exons) of HMGs from Arabidopsis, S. lycopersicum, S. tuberosum and S. commersonii. (B) Expression patterns of ScHMGs and StHMGs in flowers, leaves, roots and tubers. (C) Duplication events occurring in HMG genes in S. tuberosum.
We identified four HMG genes also in S. commersonii, with one-to-one orthologs to the StHMGs. They were therefore named ScHMG1, ScHMG3, ScHMG4 and ScHMG5. No direct orthologs to AtHMG2 were identified. To gain insights into the evolution of HMG genes between Arabidopsis, S. lycopersicum, S. tuberosum and S. commersonii, we examined their exon–intron structure (Figure 1A). Most of the genes showed four exons, with the only exception being SlHMG3 (three exons), SlHMG4 and ScHMG4 (five exons) (Figure 1A, Table S3). At the protein level, a single common HMG-CoA reductase class I domain was found in all orthologous sequences; this domain consists of five features: catalytic residue, NADP(H) binding site, substrate-binding pocket, inhibitor binding site and tetramerization interface. Both Arabidopsis and the two potato species’ HMGs showed all these features, whereas SlHMG3 contained only the NADP(H) binding site and tetramerization interface (Table S3). Recently, Zhang et al. [7] reported that the potato HMG genes (StHMG1 and StHMG2) could be differently regulated in potato tubers subjected to light exposure, demonstrating a different specialization of these genes in response to abiotic stressors. Even in Arabidopsis, a different specialization of HMGs in the induction of SGAs has been evidenced [49]. Hence, we explored the expression of the different HMG genes, using public RNA-seq data and also focused on the divergence between S. commersonii and S. tuberosum. First, the gene structure previously described was confirmed for all HMG genes, excluding the possibility of any pseudogene in the dataset. Then, HMGs of both species were expressed in flower, leaf, root and tuber tissues (Figure 1B). To some extent, a match of the HMG expression patterns between S. commersonii and S. tuberosum and across all the analyzed tissues was found. In particular, most of the HMGs were highly expressed in flower, while a higher specificity of HMG5 to tuber tissue was found in both species. HMG1 and HMG3 paralogs in S. commersonii and S. tuberosum showed a similar expression pattern. We observed a strong expression of ScHMG4 throughout all tissues, including in the leaf, where no other HMGs were particularly expressed. Overall, these results suggest a different plant-tissue-specificity and regulation of HMGs. HMG-CoA reductase 1 is regulated at the transcriptional, posttranscriptional and posttranslational levels, and reduced activity is associated with reduced mRNA levels [50,51]. A high abundance of mRNA transcripts was observed in flowers, where different terpenoids usually accumulate [52]. HMGs expressed in the tubers are particularly interesting, since they may drive the production of glycoalkaloids in the edible part of the potato. In terms of expression profile, S. commersonii has two tuber-specific HMGs (ScHMG4 and ScHMG5) compared to the cultivated potato, in which only StHMG5 is highly expressed in tubers. This outcome correlates with the higher level of terpenoids and general glycoalkaloids of wild potato tubers compared to the cultivated potatoes.
In this study, the relative contributions of tandem and segmental duplications of ScHMGs were investigated. For the former, we based the analysis on the chromosomal localization of HMG orthologs in S. commersonii, while for the latter, the Plant Genome Duplication Database (PGDD) was interrogated. However, since a specific repository for S. commersonii at the PGDD is not yet available, we retrieved the gene HMG IDs of S. tuberosum, along with their ks values. We hypothesized that the results harvested in potato reflect what occurs in S. commersonii. Even though tandem duplications were not found, two segmental duplications were identified. In particular, StHMG3 was present both on segmental block_35 (ks 0,72; ks/ka 0,07) and on block_88 (ks 1; ks/ka 0,21), suggesting that it amplified twice in potato. Notably, we found that StHMG3 localized, along with StHMG4 on block_35 and with StHMG1 on block_88. Using ks values retrieved from the database and the molecular clock for potato, we showed that StHMG3 originated from StHMG1 by a segmental duplication event that occurred around 33 million years ago (MYA), whereas StHMG4 originated later, from ScHMG3 (~25 MYA) (Figure 1C). Our findings underly the idea that the number of HMG copies expanded through segmental duplication rather than other mechanisms, such as tandem duplication or retrotransposition.
3.2. ScHMG1 Overexpressing Plants and Their Expression Pattern
Based on our findings, among the four candidates identified in this study, we decided to use ScHMG1 to test its role in the biosynthesis of SGAs in S. commersonii. Previous papers on tobacco demonstrated that the constitutive expression of HMG resulted in phytosterol overproduction, indicating a major role of HMG in the upregulation of sterol abundance [53,54]. Carpintero et al. [26] and Ginzberg et al. [51] provided evidence that StHMG1 and StHMG2 were the primary metabolism genes directly involved in SGA biosynthesis, and that overexpression of StHMG1 in potato plants increased SGA content in the leaves. No papers have been published on the role of their orthologs in wild potato species, a strategic source of stress resistances and tuber quality traits. In the present study, we set up a constitutive overexpression of ScHMG1 in S. commersonii and investigated both its expression and the abundance of SGAs in positive clones. Among all the HMGs found in S. commersonii, ScHMG1 is the direct ortholog of the Arabidopsis AtHMG1, which has been found to play important physiological roles for plant growth and development [47,49,55]. After transformation, eight explants of ScHMG1 were positive to genomic PCR, with a transformation efficiency of 8.9% (not shown). The transgene copy number was efficiently identified through qPCR. It revealed that four ScHMG1 transgenic plants (hmg1.3, hmg1.4, hmg1.6 and hmg1.7) contained a relative gene copy number higher than 1 (data not shown). ScHMG1 overexpression in leaves of transgenic plants, compared to the control plants, varied from 0.7 in hmg1.7 to 3.4 in hmg1.1 (Figure 2). Based on a threshold of 1.5 log2FC, we selected hmg1.1, hmg1.4, hmg1.6 and hmg1.8 transgenic lines for further analysis.
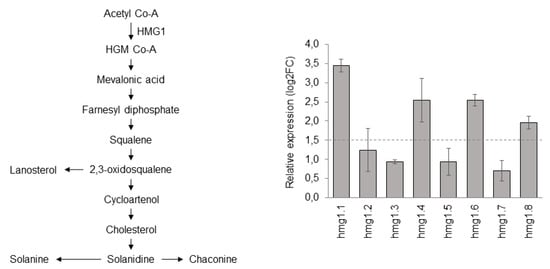
Figure 2.
Schematic representation of the steroidal glycoalkaloids pathway and expression of ScHMG1 in positive transgenic lines.
The levels of major (dehydrodemissine and dehydrocommersonine) and minor (solanidadienol triose, solanidenediol triose, solanidadienol lycotetraose, solanidenol lycotetraose and solanidenediol tetraose) glycoalkaloids were determined in transgenic clones and in the untransformed control (cmm1T) (Figure 3A,B, respectively).
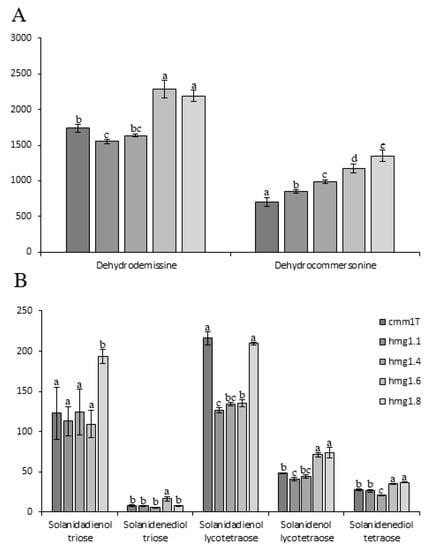
Figure 3.
Major (A) and minor (B) glycoalkaloids content detected in leaves of wild type (cmm1T) and selected transgenic plants, hmg1.1, hmg1.4, hmg1.6 and hmg1.8. Each value represents the mean of three replicates (±SD). Means denoted by the same letter did not differ significantly at p ≤ 0.05, according to Duncan’s multiple range test.
Solanidenediol triose was the less abundant SGA in all samples, with an average of 9.2 (±3.7) mg/100 gr DM. Concerning major SGAs, dehydrocommersonine varied from 706.2 (cmm1T) to 1351.3 (hmg1.8) mg/100 gr DM, while dehydrodemissine ranged from 1551.9 (hmg1.1) to 2280.4 (hmg1.6) mg/100 gr DM. All transgenic lines had a dehydrocommersonine content significantly higher than that of the untransformed cmm1T. A different behavior was observed for minor SGAs, which did not exhibit a clear accumulation trend. Solanidadienol and solanidenediol triose content were significantly different than the control only in hmg1.8 and hmg1.6, respectively. Conversely, solanidadienol lycotetraose, solanidenol lycotetraose and solanidenediol tetraose contents were significantly different than the control in all transgenic lines, except for hmg1.8, hmg1.4 and hmg1.1, respectively. Our results are similar to those of Ginzberg et al. [51], who overexpressed HMG1 from S. chacoense in cultivated potatoes and did not find a consistent increase of SGA in transgenic lines. Furthermore, the expression levels of the ectopic HMG1 reported by Ginzberg et al. [51] in potato were much higher than those of our study (about 20 times to 700 times higher). It can confer the additional hypothesis that the effect can be masked by the possibility of coordinated regulation of the downstream pathway. Krits et al. [16] and Ginzberg et al. [51] showed correlations between transcript levels of StHMG1 and SGA production in S. tuberosum genotypes. In our work, we did not detect a consistent correlation between ScHMG1 and total SGAs in leaf, except for dehydrocommersonine, which significantly increased in all our transgenic lines compared to cmm1T. Our results suggest two hypotheses: (1) In S. commersonii leaves, there is a more complex regulation which blocks the contribution of terpenoid precursors (produced by the activity of HMG-CoA) in SGA accumulation; (2) the HMG1-coded enzyme is weakly active, and consequently other HMG genes here identified can be stronger activators. This latter hypothesis is an important issue for future research aimed to investigate the contribution of the different HMG genes in SGA production.
4. Conclusions
SGAs are secondary metabolites that are associated with resistance to various environmental stresses in Solanaceae. Among the key genes of the SGA pathway, HMG plays a critical role and has been investigated in several cultivated plants. By contrast, no information is available on the behavior of HMG genes in wild species. In the present study, we proposed a new potato HMG gene family classification and showed that both cultivated S. tuberosum and its wild relative S. commersonii harbor four HMG genes. Our research suggests that HMG homologs arose from HMG1 through segmental duplication events rather than other mechanisms of gene duplication. ScHMG1 is also the direct ortholog of AtHMG1, known to be a key gene in SGA regulation in Arabidopsis. ScHMG1 was successfully overexpressed in S. commersonii, allowing the identification of a correlation between this gene and dehydrocommersonine accumulation.
Supplementary Materials
The following are available online at https://www.mdpi.com/2075-1729/10/4/37/s1, Table S1. Name and sequence of primers used in the present study. Table S2: Molecular formula, theoretical mass, experimental mass and mass accuracy used in the HRMS analysis. Table S3. Characteristics of the HMG genes identified in the present study. For each gene, Gene and Sequence ID, protein sequence and length, gene structure (exon number and length and intron length) and the features of HMG-CoA reductase class I domain are reported.
Author Contributions
Conceptualization, M.G.A. and R.A.; data curation, C.V., M.G.A. and F.C.; formal analysis, M.G.A.; investigation, M.G.A. and F.C.; methodology, R.F. and P.V.; project administration, D.C.; software, S.E.; supervision, R.A. and D.C.; writing—original draft, C.V.; writing—review and editing, C.V., V.D., S.E., R.A., P.V. and D.C. All authors have read and agreed to the published version of the manuscript.
Acknowledgments
We thank Raffaele Garramone (University of Naples) for technical support.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Uluwaduge, D.I. Glycoalkaloids, bitter tasting toxicants in potatoes: A review. Int. Food Sci. Nutr. 2018, 3, 188–193. [Google Scholar]
- Friedman, M. Chemistry and anticarcinogenic mechanisms of glycoalkaloids produced by eggplants, potatoes, and tomatoes. J. Agric. Food Chem. 2015, 63, 3323–3337. [Google Scholar] [CrossRef] [PubMed]
- Itkin, M.; Heinig, U.; Tzfadia, O.; Bhide, A.J.; Shinde, B.; Cardenas, P.D.; Bocobza, S.E.; Unger, T.; Malitsky, S.; Finkers, R. Biosynthesis of antinutritional alkaloids in solanaceous crops is mediated by clustered genes. Science 2013, 341, 175–179. [Google Scholar] [CrossRef] [PubMed]
- Karlsson, M.F.; Birgersson, G.; Witzgall, P.; Lekfeldt, J.D.S.; Punyasiri, P.N.; Bengtsson, M. Guatemalan potato moth Tecia solanivora distinguish odour profiles from qualitatively different potatoes Solanum tuberosum L. Phytochemistry 2013, 85, 72–81. [Google Scholar] [CrossRef] [PubMed]
- Paudel, J.R.; Davidson, C.; Song, J.; Maxim, I.; Aharoni, A.; Tai, H.H. Pathogen and pest responses are altered due to RNAi-mediated knockdown of GLYCOALKALOID METABOLISM 4 in Solanum tuberosum. Mol. Plant-Microbe Interact. 2017, 30, 876–885. [Google Scholar] [CrossRef]
- Wang, C.C.; Meng, L.H.; Gao, Y.; Grierson, D.; Fu, D.Q. Manipulation of light signal transduction factors as a means of modifying steroidal glycoalkaloids accumulation in tomato leaves. Front. Plant Sci. 2018, 9, 437. [Google Scholar] [CrossRef]
- Zhang, W.; Zuo, C.; Chen, Z.; Kang, Y.; Qin, S. RNA sequencing reveals that both abiotic and biotic stress-responsive genes are induced during expression of steroidal glycoalkaloid in potato tuber subjected to light exposure. Genes 2019, 10, 920. [Google Scholar] [CrossRef]
- Moore, K.B.; Oishi, K.K. 3-Hydroxy-3-methylglutaryl coenzyme A reductase activity in the endosperm of maize vivipary mutants. Plant Physiol. 1994, 105, 119–125. [Google Scholar] [CrossRef]
- Alex, D.; Bach, T.J.; Chye, M.L. Expression of Brassica juncea 3-hydroxy-3-methylglutaryl CoA synthase is developmentally regulated and stress-responsive. Plant J. 2000, 22, 415–426. [Google Scholar] [CrossRef]
- Ha, S.H.; Kim, J.B.; Hwang, Y.S.; Lee, S.W. Molecular characterization of three 3-hydroxy-3-methylglutaryl-CoA reductase genes including pathogen-induced Hmg2 from pepper (Capsicum annuum). Biochim. Biophys. Acta Gene Struct. Expr. 2000, 1625, 253–260. [Google Scholar] [CrossRef]
- Enjuto, M.; Balcells, L.; Campos, N.; Caelles, C.; Arro, M.; Boronat, A. Arabidopsis thaliana contains two differentially expressed 3-hydroxy-3-methylglutaryl-CoA reductase genes, which encode microsomal forms of the enzyme. Proc. Natl. Acad. Sci. USA 1994, 91, 927–931. [Google Scholar] [CrossRef] [PubMed]
- Vollack, K.U.; Dittrich, B.; Ferrer, A.; Boronat, A.; Bach, T.J. Two radish genes for 3-hydroxy-3-methylglutaryl-CoA reductase isozymes complement mevalonate auxotrophy in a yeast mutant and yield membrane-bound active enzyme. J. Plant Physiol. 1994, 143, 479–487. [Google Scholar] [CrossRef]
- Chye, M.-L.; Tan, C.-T.; Chua, N.-H. Three genes encode 3-hydroxy-3-methylglutaryl-coenzyme A reductase in Hevea brasiliensir: Hmgl and hmg3 are differentially expressed. Plant Mol. Biol. 1992, 19, 473–484. [Google Scholar] [CrossRef]
- Bach, T.J.; Wettstein, A.; Boronat, A.; Enjuto, M.; Gruissem, W.; Narita, J.O. Properties and molecular cloning of plant HMG-CoA reductase. In Physiology and Biochemistry of Sterols; Patterson, G.W., Nes, W.D., Eds.; American Oil Chemists Society: Washington, DC, USA, 1991; pp. 29–49. [Google Scholar]
- Yang, Z.; Park, H.; Lacy, G.H.; Cramer, C.L. Differential activation of potato 3-hydroxy-3-methylglutaryl coenzyme: A reductase gene by wounding and pathogen challenge. Plant Cell 1991, 3, 397–405. [Google Scholar] [PubMed]
- Krits, P.; Fogelman, E.; Ginzberg, I. Potato steroidal glycoalkaloid levels and the expression of key isoprenoid metabolic genes. Planta 2007, 227, 143–150. [Google Scholar] [CrossRef]
- Deahl, K.L.; Cantelo, W.W.; Sinden, S.L.; Sanford, L.L. The effect of light intensity on Colorado potato beetle resistance and foliar glycoalkaloid concentration of four Solanum chacoense clones. Am. J. Potato 1991, 68, 659–666. [Google Scholar] [CrossRef]
- Kozukue, N.; Yoon, K.S.; Byun, G.I.; Misoo, S.; Levin, C.E.; Friedman, M. Distribution of glycoalkaloids in potato tubers of 59 accessions of two wild and five cultivated Solanum species. J. Agric. Food Chem. 2008, 56, 11920–11928. [Google Scholar] [CrossRef]
- Hanneman, R.E. The potato germplasm resource. Am. Potato J. 1989, 66, 655–667. [Google Scholar] [CrossRef]
- Hawkes, J.G. The Potato: Evolution, Biodiversity and Genetic Resources; Belhaven Press: Birmingham, UK, 1990. [Google Scholar]
- Micheletto, S.; Boland, R.; Huarte, M. Argentinian wild diploid Solanum species as sources of quantitative late blight resistance. Theor. Appl. Genet. 2000, 101, 902–906. [Google Scholar] [CrossRef]
- D’Amelia, V.; Villano, C.; Aversano, R. Emerging genetic technologies to improve crop productivity. Encycl. Food Secur. Sustain. 2019, 3, 152–158. [Google Scholar]
- Aversano, R.; Contaldi, F.; Ercolano, M.R.; Grosso, V.; Iorizzo, M.; Tatino, F.; Xumerle, L.; Dal Molin, A.; Avanzato, C.; Ferrarini, A. The Solanum commersonii genome sequence provides insights into adaptation to stress conditions and genome evolution of wild potato relatives. Plant Cell 2015, 27, 954–968. [Google Scholar] [CrossRef] [PubMed]
- Esposito, S.; Aversano, R.; D’Amelia, V.; Villano, C.; Alioto, D.; Mirouze, M.; Carputo, D. Dicer-like and RNA-dependent RNA polymerase gene family identification and annotation in the cultivated Solanum tuberosum and its wild relative S. commersonii. Planta 2018, 248, 729–743. [Google Scholar] [CrossRef] [PubMed]
- Villano, C.; Miraglia, V.; Iorizzo, M.; Aversano, R.; Carputo, D. Combined use of molecular markers and high-resolution melting (HRM) to assess chromosome dosage in potato hybrids. J. Hered. 2016, 107, 187–192. [Google Scholar] [CrossRef] [PubMed]
- Manrique-Carpintero, N.C.; Tokuhisa, J.G.; Ginzberg, I.; Holliday, J.A.; Veilleux, R.E. Sequence diversity in coding regions of candidate genes in the glycoalkaloid biosynthetic pathway of wild potato species. G3 Genes Genomes Genet. 2013, 3, 1467–1479. [Google Scholar] [CrossRef] [PubMed]
- Vázquez, A.; González, G.; Ferreira, F.; Moyna, P.; Kenne, L. Glycoalkaloids of Solanum commersonii Dun. Ex Poir. Euphytica 1997, 95, 195–201. [Google Scholar] [CrossRef]
- NCBI Search Domain. Available online: https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi. (accessed on 4 March 2018).
- Tamura, K.; Dudley, J.; Nei, M.; Kumar, S. MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Boil. Evol. 2007, 24, 1596–1599. [Google Scholar] [CrossRef]
- Wheelan, S.J.; Church, D.M.; Ostell, J.M. Spidey: A tool for mRNA-to-genomic alignments. Genome Res. 2001, 11, 1952–1957. [Google Scholar] [CrossRef]
- Esposito, S.; Aversano, R.; Bradeen, J.M.; Di Matteo, A.; Villano, C.; Carputo, D. Deep-sequencing of Solanum commersonii small RNA libraries reveals riboregulators involved in cold stress response. Plant Biol. 2020, 22, 133–142. [Google Scholar] [CrossRef]
- Esposito, S.; D’Amelia, V.; Carputo, D.; Aversano, R. Genes involved in stress signals: The CBLs-CIPKs network in cold tolerant Solanum commersonii. Biol. Plant. 2019, 63, 699–709. [Google Scholar] [CrossRef]
- Zhao, P.; Zhu, K.Y.; Jiang, H. Heterologous expression, purification, and biochemical characterization of a greenbug (Schizaphis graminum) acetylcholinesterase encoded by a paralogous gene (ace-1). J. Biochem. Mol. Toxicol. 2010, 24, 51–59. [Google Scholar] [CrossRef]
- Xia, K.; Liu, T.; Ouyang, J.; Wang, R.; Fan, T.; Zhang, M. Genome-wide identification, classification, and expression analysis of autophagy-associated gene homologues in rice (Oryza sativa L.). DNA Res. 2011, 18, 363–377. [Google Scholar] [CrossRef] [PubMed]
- Plant Genome Duplication Database. Available online: http://chibba.agtec.uga.edu/duplication/ (accessed on 4 April 2020).
- Zhang, Z.; Yu, J. Evaluation of six methods for estimating synonymous and nonsynonymous substitution rates. Genom. Proteom. Bioinf. 2006, 4, 173–181. [Google Scholar] [CrossRef]
- Blanc, G.; Wolfe, K.H. Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes. Plant Cell 2004, 16, 1667–1678. [Google Scholar] [CrossRef] [PubMed]
- D’Amelia, V.; Aversano, R.; Batelli, G.; Caruso, I.; Castellano Moreno, M.; Castro-Sanz, A.B.; Chiaiese, P.; Fasano, C.; Palomba, F.; Carputo, D. High AN1 variability and interaction with basic helix-loop-helix co-factors related to anthocyanin biosynthesis in potato leaves. Plant J. 2014, 80, 527–540. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, T.; Kurose, T.; Hino, T.; Tanaka, K.; Kawamukai, M.; Niwa, Y.; Toyooka, K.; Matsuoka, K.; Jinbo, T.; Kimura, T. Development of series of Gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J. Biosci. Bioeng. 2007, 104, 34–41. [Google Scholar] [CrossRef] [PubMed]
- Cardi, T.; Carputo, D.; Frusciante, L. In vitro shoot regeneration and chromosome doubling in 2x and 3x potato clones. Am. Potato J. 1992, 69, 1–12. [Google Scholar] [CrossRef]
- Brulé, D.; Villano, C.; Davies, L.J.; Trdá, L.; Claverie, J.; Héloir, M.C.; Chiltz, A.; Adrian, M.; Darblade, B.; Tornero, P.; et al. The grapevine (Vitis vinifera) LysM receptor kinases VvLYK1-1 and VvLYK1-2 mediate chitooligosaccharide-triggered immunity. Plant Biotechnol. J. 2019, 17, 812–825. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Villano, C.; Rinaldi, A.; Lanzillo, C.; Moio, L.; Tamburrino, A.J.; Carputo, D.; Aversano, R. Polyphenol content and differential expression of flavonoid biosynthetic pathway genes in berries of Aglianico. Acta Hortic. 2017, 1188, 141–148. [Google Scholar] [CrossRef]
- Di Meo, F.; Aversano, R.; Diretto, G.; Demurtas, O.C.; Villano, C.; Cozzolino, S.; Carputo, D.; Crispi, S. Anti-cancer activity of grape seed semi-polar extracts in human mesothelioma cell lines. J. Funct. Foods 2019, 61, 103515. [Google Scholar] [CrossRef]
- Bradeen, J.M.; Iorizzo, M.; Mollov, D.S.; Raasch, J.; Kramer, L.C.; Millett, B.P.; Carputo, D. Higher copy numbers of the potato RB transgene correspond to enhanced transcript and late blight resistance levels. Mol. Plant-Microbe Interact. 2009, 22, 437–446. [Google Scholar] [CrossRef] [PubMed]
- Lumbreras, V.; Campos, N.; Boronat, A. The use of an alternative promoter in the Arabidopsis thaliana HMG1 gene generates an mRNA that encodes a novel 3-hydroxy-3-methylglutaryl coenzyme A reductase isoform with an extended N-terminal region. Plant J. 1995, 8, 541–549. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, M.; Kamide, Y.; Nagata, N.; Seki, H.; Ohyama, K.; Kato, H.; Yoshida, S. Loss of function of 3-hydroxy-3-methylglutaryl coenzyme A reductase 1 (HMG1) in Arabidopsis leads to dwarfing, early senescence and male sterility, and reduced sterol levels. Plant J. 2004, 37, 750–761. [Google Scholar] [CrossRef] [PubMed]
- Cui, T.; Bai, J.; Zhang, J.; Zhang, J.; Wang, D. Transcriptional expression of seven key genes involved in steroidal glycoalkaloid biosynthesis in potato microtubers. N. Z. J. Crop Hortic. 2014, 42, 118–126. [Google Scholar] [CrossRef][Green Version]
- Suzuki, M.; Muranaka, T. Molecular genetics of plant sterol backbone synthesis. Lipids 2007, 42, 47–54. [Google Scholar] [CrossRef]
- Devarenne, T.P.; Ghosh, A.; Chappell, J. Regulation of squalene synthase, a key enzyme of sterol biosynthesis, in tobacco. Plant Physiol. 2002, 129, 1095–1106. [Google Scholar] [CrossRef] [PubMed]
- Ginzberg, I.; Thippeswamy, M.; Fogelman, E.; Demirel, U.; Mweetwa, A.M.; Tokuhisa, J.; Veilleux, R.E. Induction of potato steroidal glycoalkaloid biosynthetic pathway by overexpression of cDNA encoding primary metabolism HMG-CoA reductase and squalene synthase. Planta 2012, 235, 1341–1353. [Google Scholar] [CrossRef]
- Mlcek, J.; Rop, O. Fresh edible flowers of ornamental plants–A new source of nutraceutical foods. Trends Food Sci. Technol. 2011, 22, 561–569. [Google Scholar] [CrossRef]
- Chappell, J. Biochemistry and molecular biology of the isoprenoid biosynthetic pathway in plants. Annu. Rev. Plant Biol. 1995, 46, 521–547. [Google Scholar] [CrossRef]
- Schaller, H.; Grausem, B.; Benveniste, P.; Chye, M.L.; Tan, C.T.; Song, Y.H.; Chua, N.H. Expression of the Hevea brasiliensis (HBK) Mull. Arg. 3-hydroxy-3-methylglutaryl-coenzyme A reductase 1 in tobacco results in sterol overproduction. Plant Physiol. 1995, 109, 761–770. [Google Scholar] [CrossRef]
- Ohyama, K.; Suzuki, M.; Masuda, K.; Yoshida, S.; Muranaka, T. Chemical phenotypes of the hmg1 and hmg2 mutants of Arabidopsis demonstrate the in-planta role of HMG-CoA reductase in triterpene biosynthesis. Chem. Pharm. Bull. 2007, 55, 1518–1521. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).