Genome-Wide HMG Family Investigation and Its Role in Glycoalkaloid Accumulation in Wild Tuber-Bearing Solanum commersonii
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification of Candidate HMG Genes and Phylogenetic Analysis
2.2. Determination of Gene Duplication Patterns
2.3. Plant Material
2.4. Overexpression of ScHMG1 in S. commersonii
2.5. RNA Extraction and Gene Expression Analyses
2.6. Copy Number Determination
2.7. SGA Determination
3. Results and Discussion
3.1. Comparative and Phylogenetic Analysis of the HMG Gene Families in Solanaceae
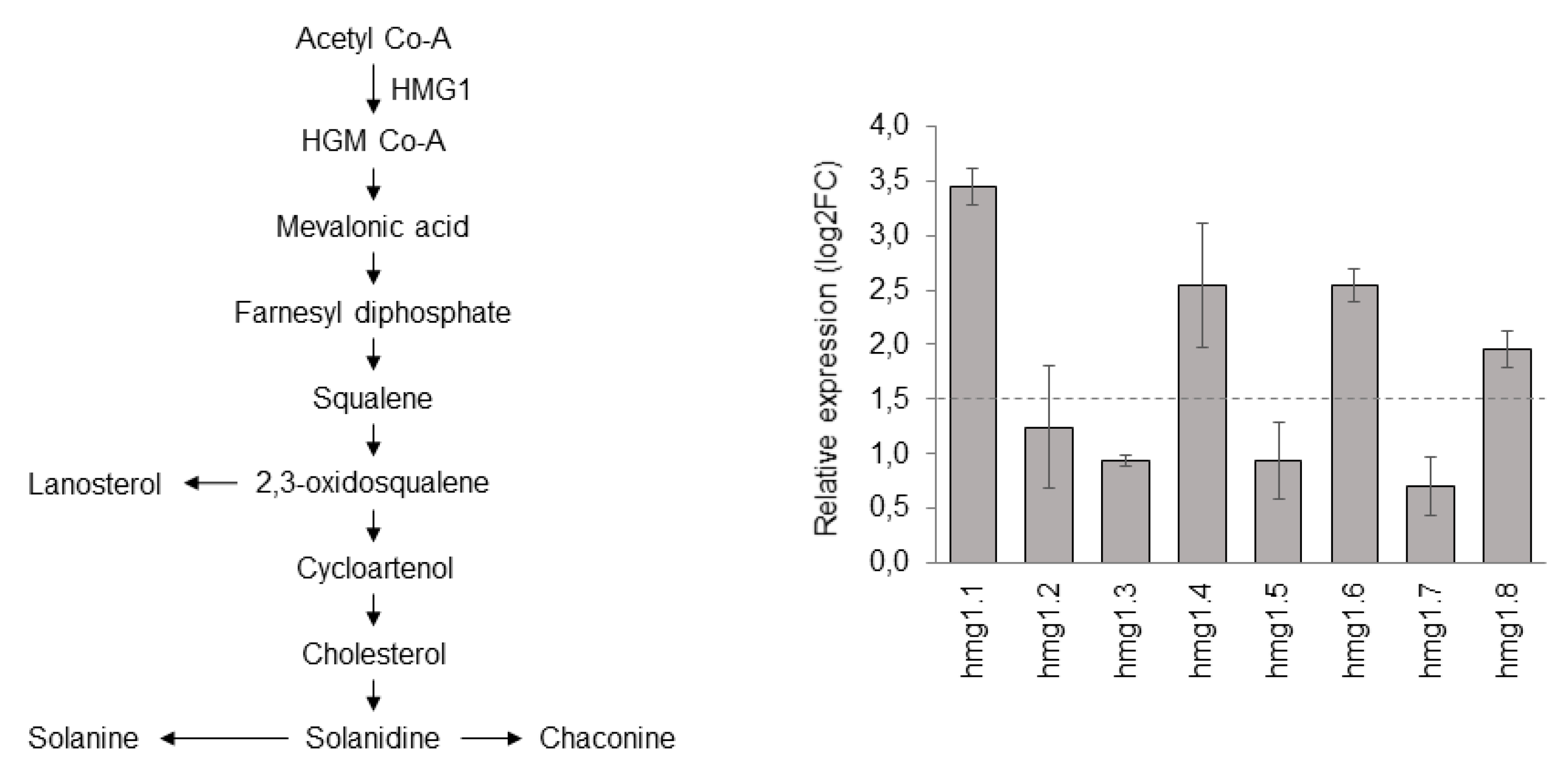
3.2. ScHMG1 Overexpressing Plants and Their Expression Pattern
4. Conclusions
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Uluwaduge, D.I. Glycoalkaloids, bitter tasting toxicants in potatoes: A review. Int. Food Sci. Nutr. 2018, 3, 188–193. [Google Scholar]
- Friedman, M. Chemistry and anticarcinogenic mechanisms of glycoalkaloids produced by eggplants, potatoes, and tomatoes. J. Agric. Food Chem. 2015, 63, 3323–3337. [Google Scholar] [CrossRef] [PubMed]
- Itkin, M.; Heinig, U.; Tzfadia, O.; Bhide, A.J.; Shinde, B.; Cardenas, P.D.; Bocobza, S.E.; Unger, T.; Malitsky, S.; Finkers, R. Biosynthesis of antinutritional alkaloids in solanaceous crops is mediated by clustered genes. Science 2013, 341, 175–179. [Google Scholar] [CrossRef] [PubMed]
- Karlsson, M.F.; Birgersson, G.; Witzgall, P.; Lekfeldt, J.D.S.; Punyasiri, P.N.; Bengtsson, M. Guatemalan potato moth Tecia solanivora distinguish odour profiles from qualitatively different potatoes Solanum tuberosum L. Phytochemistry 2013, 85, 72–81. [Google Scholar] [CrossRef] [PubMed]
- Paudel, J.R.; Davidson, C.; Song, J.; Maxim, I.; Aharoni, A.; Tai, H.H. Pathogen and pest responses are altered due to RNAi-mediated knockdown of GLYCOALKALOID METABOLISM 4 in Solanum tuberosum. Mol. Plant-Microbe Interact. 2017, 30, 876–885. [Google Scholar] [CrossRef]
- Wang, C.C.; Meng, L.H.; Gao, Y.; Grierson, D.; Fu, D.Q. Manipulation of light signal transduction factors as a means of modifying steroidal glycoalkaloids accumulation in tomato leaves. Front. Plant Sci. 2018, 9, 437. [Google Scholar] [CrossRef]
- Zhang, W.; Zuo, C.; Chen, Z.; Kang, Y.; Qin, S. RNA sequencing reveals that both abiotic and biotic stress-responsive genes are induced during expression of steroidal glycoalkaloid in potato tuber subjected to light exposure. Genes 2019, 10, 920. [Google Scholar] [CrossRef]
- Moore, K.B.; Oishi, K.K. 3-Hydroxy-3-methylglutaryl coenzyme A reductase activity in the endosperm of maize vivipary mutants. Plant Physiol. 1994, 105, 119–125. [Google Scholar] [CrossRef]
- Alex, D.; Bach, T.J.; Chye, M.L. Expression of Brassica juncea 3-hydroxy-3-methylglutaryl CoA synthase is developmentally regulated and stress-responsive. Plant J. 2000, 22, 415–426. [Google Scholar] [CrossRef]
- Ha, S.H.; Kim, J.B.; Hwang, Y.S.; Lee, S.W. Molecular characterization of three 3-hydroxy-3-methylglutaryl-CoA reductase genes including pathogen-induced Hmg2 from pepper (Capsicum annuum). Biochim. Biophys. Acta Gene Struct. Expr. 2000, 1625, 253–260. [Google Scholar] [CrossRef]
- Enjuto, M.; Balcells, L.; Campos, N.; Caelles, C.; Arro, M.; Boronat, A. Arabidopsis thaliana contains two differentially expressed 3-hydroxy-3-methylglutaryl-CoA reductase genes, which encode microsomal forms of the enzyme. Proc. Natl. Acad. Sci. USA 1994, 91, 927–931. [Google Scholar] [CrossRef] [PubMed]
- Vollack, K.U.; Dittrich, B.; Ferrer, A.; Boronat, A.; Bach, T.J. Two radish genes for 3-hydroxy-3-methylglutaryl-CoA reductase isozymes complement mevalonate auxotrophy in a yeast mutant and yield membrane-bound active enzyme. J. Plant Physiol. 1994, 143, 479–487. [Google Scholar] [CrossRef]
- Chye, M.-L.; Tan, C.-T.; Chua, N.-H. Three genes encode 3-hydroxy-3-methylglutaryl-coenzyme A reductase in Hevea brasiliensir: Hmgl and hmg3 are differentially expressed. Plant Mol. Biol. 1992, 19, 473–484. [Google Scholar] [CrossRef]
- Bach, T.J.; Wettstein, A.; Boronat, A.; Enjuto, M.; Gruissem, W.; Narita, J.O. Properties and molecular cloning of plant HMG-CoA reductase. In Physiology and Biochemistry of Sterols; Patterson, G.W., Nes, W.D., Eds.; American Oil Chemists Society: Washington, DC, USA, 1991; pp. 29–49. [Google Scholar]
- Yang, Z.; Park, H.; Lacy, G.H.; Cramer, C.L. Differential activation of potato 3-hydroxy-3-methylglutaryl coenzyme: A reductase gene by wounding and pathogen challenge. Plant Cell 1991, 3, 397–405. [Google Scholar] [PubMed]
- Krits, P.; Fogelman, E.; Ginzberg, I. Potato steroidal glycoalkaloid levels and the expression of key isoprenoid metabolic genes. Planta 2007, 227, 143–150. [Google Scholar] [CrossRef]
- Deahl, K.L.; Cantelo, W.W.; Sinden, S.L.; Sanford, L.L. The effect of light intensity on Colorado potato beetle resistance and foliar glycoalkaloid concentration of four Solanum chacoense clones. Am. J. Potato 1991, 68, 659–666. [Google Scholar] [CrossRef]
- Kozukue, N.; Yoon, K.S.; Byun, G.I.; Misoo, S.; Levin, C.E.; Friedman, M. Distribution of glycoalkaloids in potato tubers of 59 accessions of two wild and five cultivated Solanum species. J. Agric. Food Chem. 2008, 56, 11920–11928. [Google Scholar] [CrossRef]
- Hanneman, R.E. The potato germplasm resource. Am. Potato J. 1989, 66, 655–667. [Google Scholar] [CrossRef]
- Hawkes, J.G. The Potato: Evolution, Biodiversity and Genetic Resources; Belhaven Press: Birmingham, UK, 1990. [Google Scholar]
- Micheletto, S.; Boland, R.; Huarte, M. Argentinian wild diploid Solanum species as sources of quantitative late blight resistance. Theor. Appl. Genet. 2000, 101, 902–906. [Google Scholar] [CrossRef]
- D’Amelia, V.; Villano, C.; Aversano, R. Emerging genetic technologies to improve crop productivity. Encycl. Food Secur. Sustain. 2019, 3, 152–158. [Google Scholar]
- Aversano, R.; Contaldi, F.; Ercolano, M.R.; Grosso, V.; Iorizzo, M.; Tatino, F.; Xumerle, L.; Dal Molin, A.; Avanzato, C.; Ferrarini, A. The Solanum commersonii genome sequence provides insights into adaptation to stress conditions and genome evolution of wild potato relatives. Plant Cell 2015, 27, 954–968. [Google Scholar] [CrossRef] [PubMed]
- Esposito, S.; Aversano, R.; D’Amelia, V.; Villano, C.; Alioto, D.; Mirouze, M.; Carputo, D. Dicer-like and RNA-dependent RNA polymerase gene family identification and annotation in the cultivated Solanum tuberosum and its wild relative S. commersonii. Planta 2018, 248, 729–743. [Google Scholar] [CrossRef] [PubMed]
- Villano, C.; Miraglia, V.; Iorizzo, M.; Aversano, R.; Carputo, D. Combined use of molecular markers and high-resolution melting (HRM) to assess chromosome dosage in potato hybrids. J. Hered. 2016, 107, 187–192. [Google Scholar] [CrossRef] [PubMed]
- Manrique-Carpintero, N.C.; Tokuhisa, J.G.; Ginzberg, I.; Holliday, J.A.; Veilleux, R.E. Sequence diversity in coding regions of candidate genes in the glycoalkaloid biosynthetic pathway of wild potato species. G3 Genes Genomes Genet. 2013, 3, 1467–1479. [Google Scholar] [CrossRef] [PubMed]
- Vázquez, A.; González, G.; Ferreira, F.; Moyna, P.; Kenne, L. Glycoalkaloids of Solanum commersonii Dun. Ex Poir. Euphytica 1997, 95, 195–201. [Google Scholar] [CrossRef]
- NCBI Search Domain. Available online: https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi. (accessed on 4 March 2018).
- Tamura, K.; Dudley, J.; Nei, M.; Kumar, S. MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Boil. Evol. 2007, 24, 1596–1599. [Google Scholar] [CrossRef]
- Wheelan, S.J.; Church, D.M.; Ostell, J.M. Spidey: A tool for mRNA-to-genomic alignments. Genome Res. 2001, 11, 1952–1957. [Google Scholar] [CrossRef]
- Esposito, S.; Aversano, R.; Bradeen, J.M.; Di Matteo, A.; Villano, C.; Carputo, D. Deep-sequencing of Solanum commersonii small RNA libraries reveals riboregulators involved in cold stress response. Plant Biol. 2020, 22, 133–142. [Google Scholar] [CrossRef]
- Esposito, S.; D’Amelia, V.; Carputo, D.; Aversano, R. Genes involved in stress signals: The CBLs-CIPKs network in cold tolerant Solanum commersonii. Biol. Plant. 2019, 63, 699–709. [Google Scholar] [CrossRef]
- Zhao, P.; Zhu, K.Y.; Jiang, H. Heterologous expression, purification, and biochemical characterization of a greenbug (Schizaphis graminum) acetylcholinesterase encoded by a paralogous gene (ace-1). J. Biochem. Mol. Toxicol. 2010, 24, 51–59. [Google Scholar] [CrossRef]
- Xia, K.; Liu, T.; Ouyang, J.; Wang, R.; Fan, T.; Zhang, M. Genome-wide identification, classification, and expression analysis of autophagy-associated gene homologues in rice (Oryza sativa L.). DNA Res. 2011, 18, 363–377. [Google Scholar] [CrossRef] [PubMed]
- Plant Genome Duplication Database. Available online: http://chibba.agtec.uga.edu/duplication/ (accessed on 4 April 2020).
- Zhang, Z.; Yu, J. Evaluation of six methods for estimating synonymous and nonsynonymous substitution rates. Genom. Proteom. Bioinf. 2006, 4, 173–181. [Google Scholar] [CrossRef]
- Blanc, G.; Wolfe, K.H. Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes. Plant Cell 2004, 16, 1667–1678. [Google Scholar] [CrossRef] [PubMed]
- D’Amelia, V.; Aversano, R.; Batelli, G.; Caruso, I.; Castellano Moreno, M.; Castro-Sanz, A.B.; Chiaiese, P.; Fasano, C.; Palomba, F.; Carputo, D. High AN1 variability and interaction with basic helix-loop-helix co-factors related to anthocyanin biosynthesis in potato leaves. Plant J. 2014, 80, 527–540. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, T.; Kurose, T.; Hino, T.; Tanaka, K.; Kawamukai, M.; Niwa, Y.; Toyooka, K.; Matsuoka, K.; Jinbo, T.; Kimura, T. Development of series of Gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J. Biosci. Bioeng. 2007, 104, 34–41. [Google Scholar] [CrossRef] [PubMed]
- Cardi, T.; Carputo, D.; Frusciante, L. In vitro shoot regeneration and chromosome doubling in 2x and 3x potato clones. Am. Potato J. 1992, 69, 1–12. [Google Scholar] [CrossRef]
- Brulé, D.; Villano, C.; Davies, L.J.; Trdá, L.; Claverie, J.; Héloir, M.C.; Chiltz, A.; Adrian, M.; Darblade, B.; Tornero, P.; et al. The grapevine (Vitis vinifera) LysM receptor kinases VvLYK1-1 and VvLYK1-2 mediate chitooligosaccharide-triggered immunity. Plant Biotechnol. J. 2019, 17, 812–825. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Villano, C.; Rinaldi, A.; Lanzillo, C.; Moio, L.; Tamburrino, A.J.; Carputo, D.; Aversano, R. Polyphenol content and differential expression of flavonoid biosynthetic pathway genes in berries of Aglianico. Acta Hortic. 2017, 1188, 141–148. [Google Scholar] [CrossRef]
- Di Meo, F.; Aversano, R.; Diretto, G.; Demurtas, O.C.; Villano, C.; Cozzolino, S.; Carputo, D.; Crispi, S. Anti-cancer activity of grape seed semi-polar extracts in human mesothelioma cell lines. J. Funct. Foods 2019, 61, 103515. [Google Scholar] [CrossRef]
- Bradeen, J.M.; Iorizzo, M.; Mollov, D.S.; Raasch, J.; Kramer, L.C.; Millett, B.P.; Carputo, D. Higher copy numbers of the potato RB transgene correspond to enhanced transcript and late blight resistance levels. Mol. Plant-Microbe Interact. 2009, 22, 437–446. [Google Scholar] [CrossRef] [PubMed]
- Lumbreras, V.; Campos, N.; Boronat, A. The use of an alternative promoter in the Arabidopsis thaliana HMG1 gene generates an mRNA that encodes a novel 3-hydroxy-3-methylglutaryl coenzyme A reductase isoform with an extended N-terminal region. Plant J. 1995, 8, 541–549. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, M.; Kamide, Y.; Nagata, N.; Seki, H.; Ohyama, K.; Kato, H.; Yoshida, S. Loss of function of 3-hydroxy-3-methylglutaryl coenzyme A reductase 1 (HMG1) in Arabidopsis leads to dwarfing, early senescence and male sterility, and reduced sterol levels. Plant J. 2004, 37, 750–761. [Google Scholar] [CrossRef] [PubMed]
- Cui, T.; Bai, J.; Zhang, J.; Zhang, J.; Wang, D. Transcriptional expression of seven key genes involved in steroidal glycoalkaloid biosynthesis in potato microtubers. N. Z. J. Crop Hortic. 2014, 42, 118–126. [Google Scholar] [CrossRef][Green Version]
- Suzuki, M.; Muranaka, T. Molecular genetics of plant sterol backbone synthesis. Lipids 2007, 42, 47–54. [Google Scholar] [CrossRef]
- Devarenne, T.P.; Ghosh, A.; Chappell, J. Regulation of squalene synthase, a key enzyme of sterol biosynthesis, in tobacco. Plant Physiol. 2002, 129, 1095–1106. [Google Scholar] [CrossRef] [PubMed]
- Ginzberg, I.; Thippeswamy, M.; Fogelman, E.; Demirel, U.; Mweetwa, A.M.; Tokuhisa, J.; Veilleux, R.E. Induction of potato steroidal glycoalkaloid biosynthetic pathway by overexpression of cDNA encoding primary metabolism HMG-CoA reductase and squalene synthase. Planta 2012, 235, 1341–1353. [Google Scholar] [CrossRef]
- Mlcek, J.; Rop, O. Fresh edible flowers of ornamental plants–A new source of nutraceutical foods. Trends Food Sci. Technol. 2011, 22, 561–569. [Google Scholar] [CrossRef]
- Chappell, J. Biochemistry and molecular biology of the isoprenoid biosynthetic pathway in plants. Annu. Rev. Plant Biol. 1995, 46, 521–547. [Google Scholar] [CrossRef]
- Schaller, H.; Grausem, B.; Benveniste, P.; Chye, M.L.; Tan, C.T.; Song, Y.H.; Chua, N.H. Expression of the Hevea brasiliensis (HBK) Mull. Arg. 3-hydroxy-3-methylglutaryl-coenzyme A reductase 1 in tobacco results in sterol overproduction. Plant Physiol. 1995, 109, 761–770. [Google Scholar] [CrossRef]
- Ohyama, K.; Suzuki, M.; Masuda, K.; Yoshida, S.; Muranaka, T. Chemical phenotypes of the hmg1 and hmg2 mutants of Arabidopsis demonstrate the in-planta role of HMG-CoA reductase in triterpene biosynthesis. Chem. Pharm. Bull. 2007, 55, 1518–1521. [Google Scholar] [CrossRef] [PubMed]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Villano, C.; D’Amelia, V.; Esposito, S.; Adelfi, M.G.; Contaldi, F.; Ferracane, R.; Vitaglione, P.; Aversano, R.; Carputo, D. Genome-Wide HMG Family Investigation and Its Role in Glycoalkaloid Accumulation in Wild Tuber-Bearing Solanum commersonii. Life 2020, 10, 37. https://doi.org/10.3390/life10040037
Villano C, D’Amelia V, Esposito S, Adelfi MG, Contaldi F, Ferracane R, Vitaglione P, Aversano R, Carputo D. Genome-Wide HMG Family Investigation and Its Role in Glycoalkaloid Accumulation in Wild Tuber-Bearing Solanum commersonii. Life. 2020; 10(4):37. https://doi.org/10.3390/life10040037
Chicago/Turabian StyleVillano, Clizia, Vincenzo D’Amelia, Salvatore Esposito, Maria Grazia Adelfi, Felice Contaldi, Rosalia Ferracane, Paola Vitaglione, Riccardo Aversano, and Domenico Carputo. 2020. "Genome-Wide HMG Family Investigation and Its Role in Glycoalkaloid Accumulation in Wild Tuber-Bearing Solanum commersonii" Life 10, no. 4: 37. https://doi.org/10.3390/life10040037
APA StyleVillano, C., D’Amelia, V., Esposito, S., Adelfi, M. G., Contaldi, F., Ferracane, R., Vitaglione, P., Aversano, R., & Carputo, D. (2020). Genome-Wide HMG Family Investigation and Its Role in Glycoalkaloid Accumulation in Wild Tuber-Bearing Solanum commersonii. Life, 10(4), 37. https://doi.org/10.3390/life10040037






