Low Long Terminal Repeat (LTR)-Retrotransposon Expression in Leaves of the Marine Phanerogam Posidonia Oceanica L.
Abstract
:1. Introduction
2. Materials and Methods
2.1. P. oceanica DNA and cDNA Illumina Reads Collection
2.2. Retrotransposons Sequence Set of P. oceanica
2.3. Retrotransposons Expression Analysis
3. Results
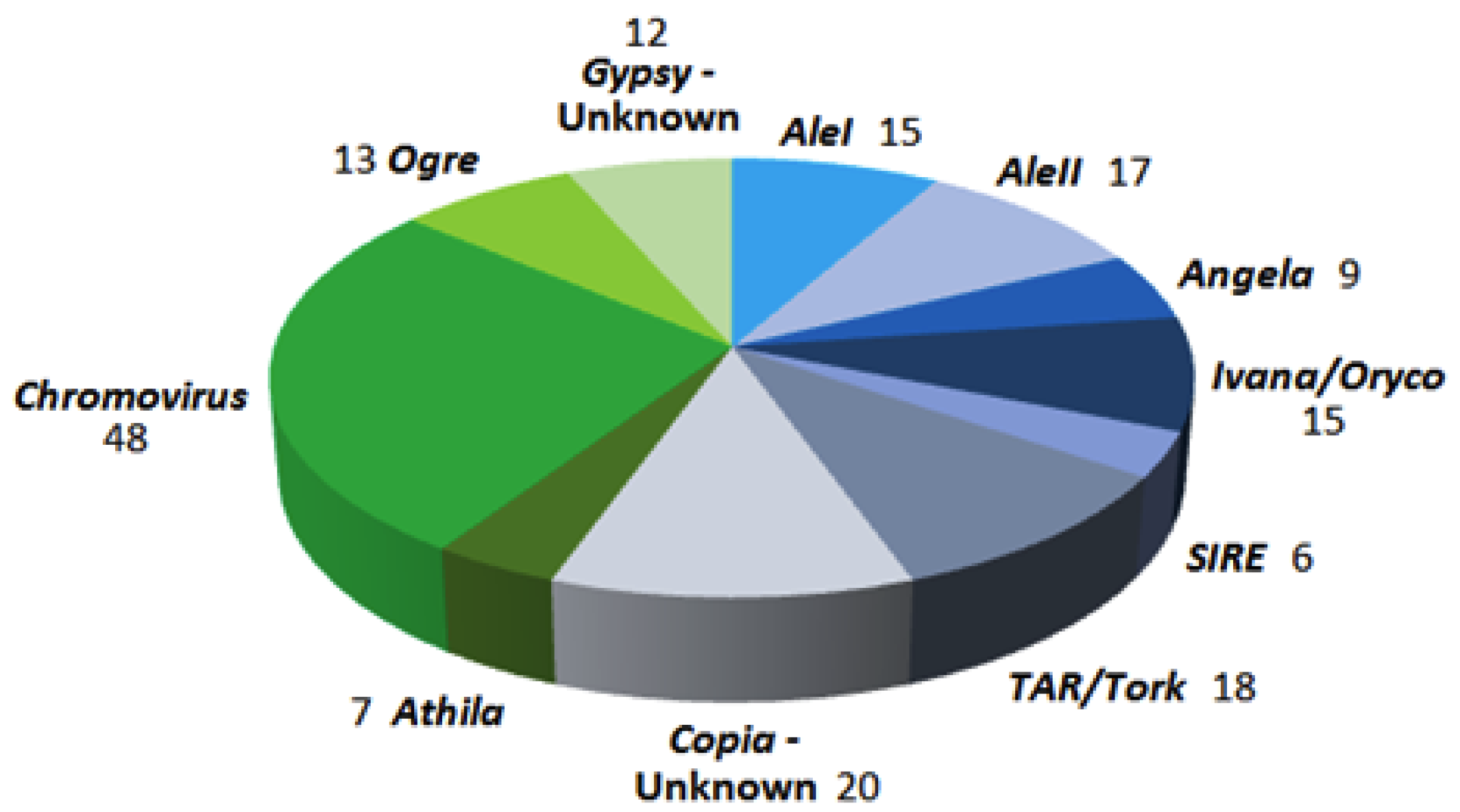
3.1. Preparation of a Set of LTR-Retrotransposon Sequences of P. oceanica
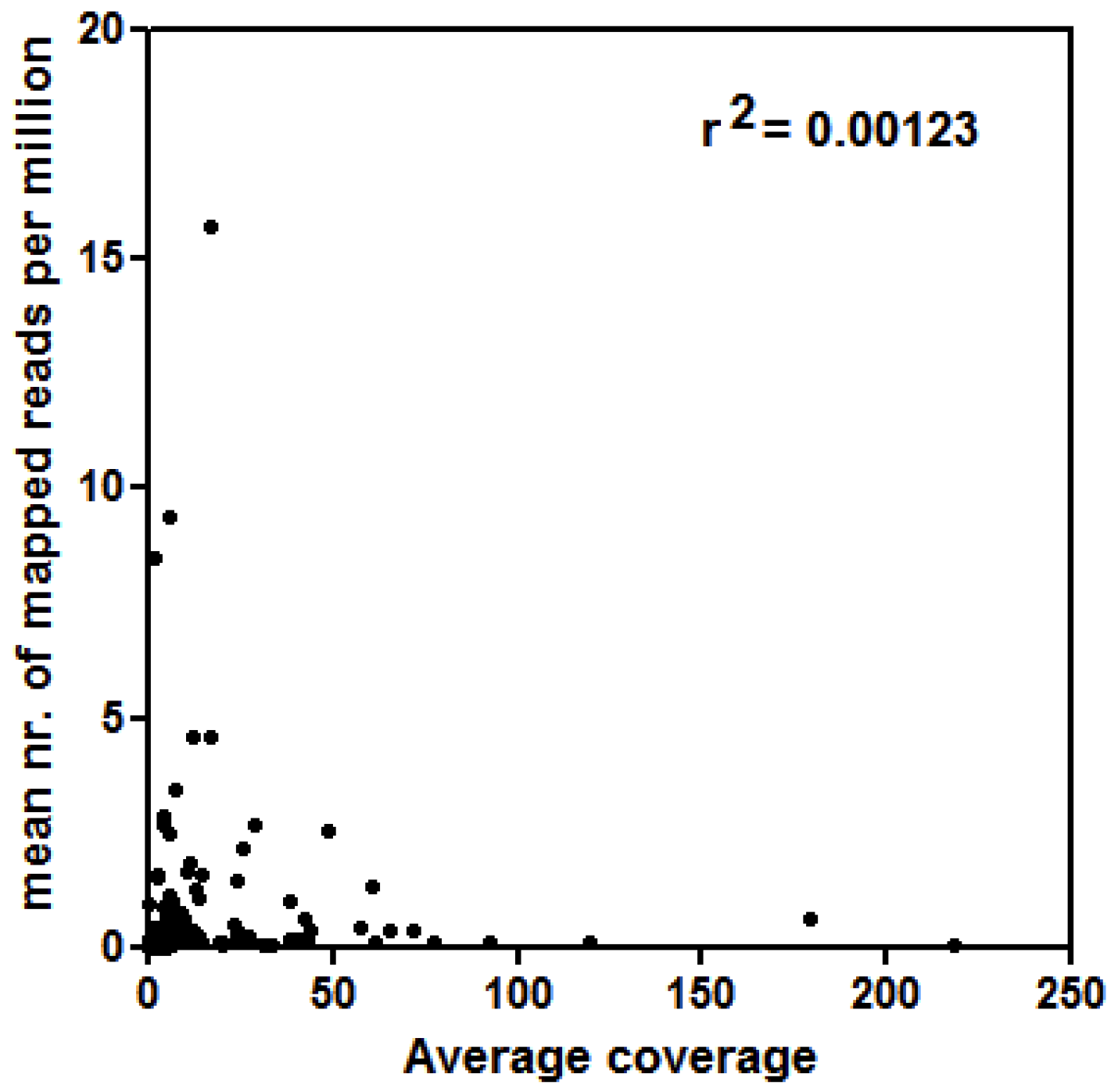
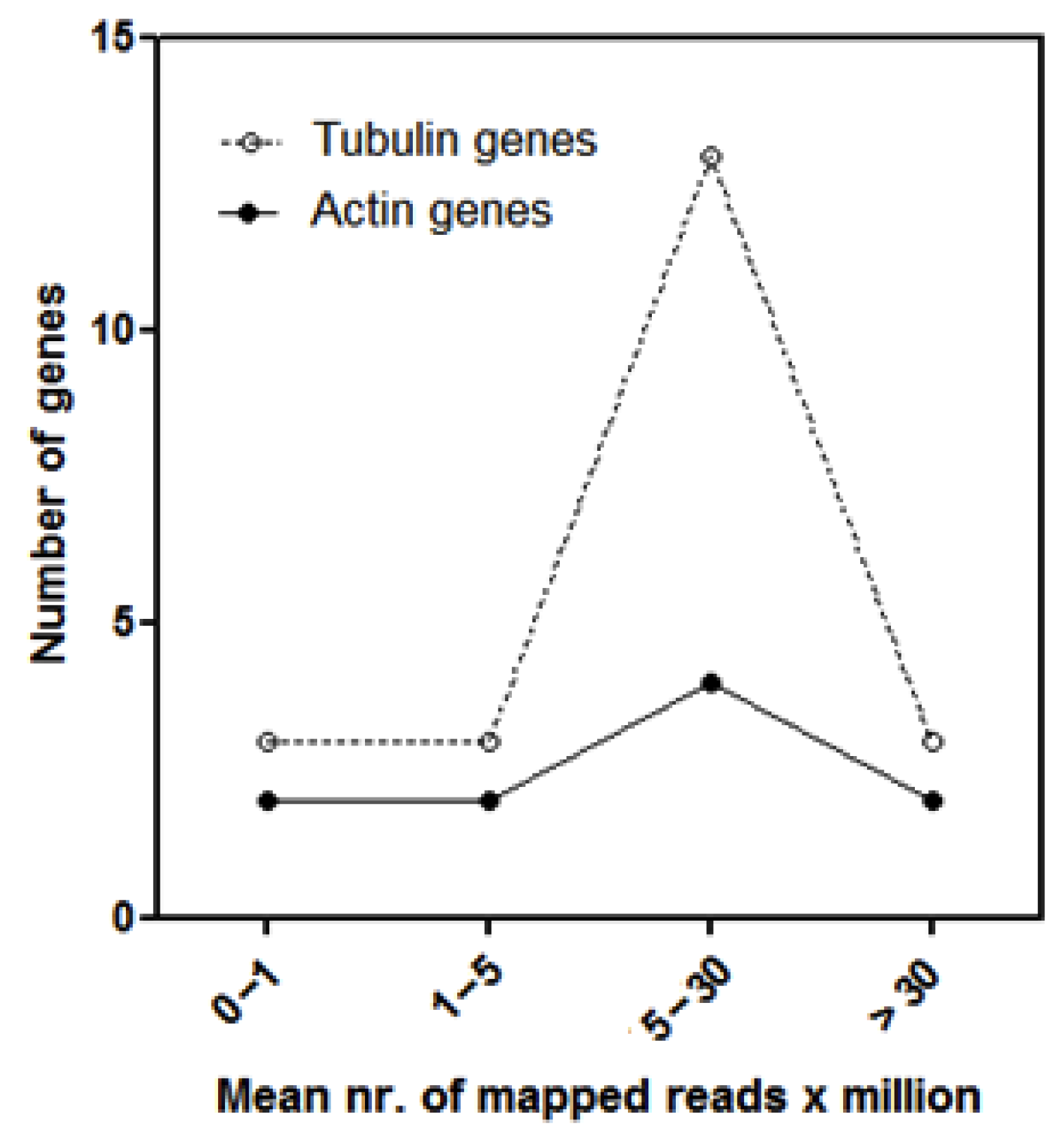
3.2. Analyses of LTR-RE Expression in Deep and Shallow Plants In Vivo
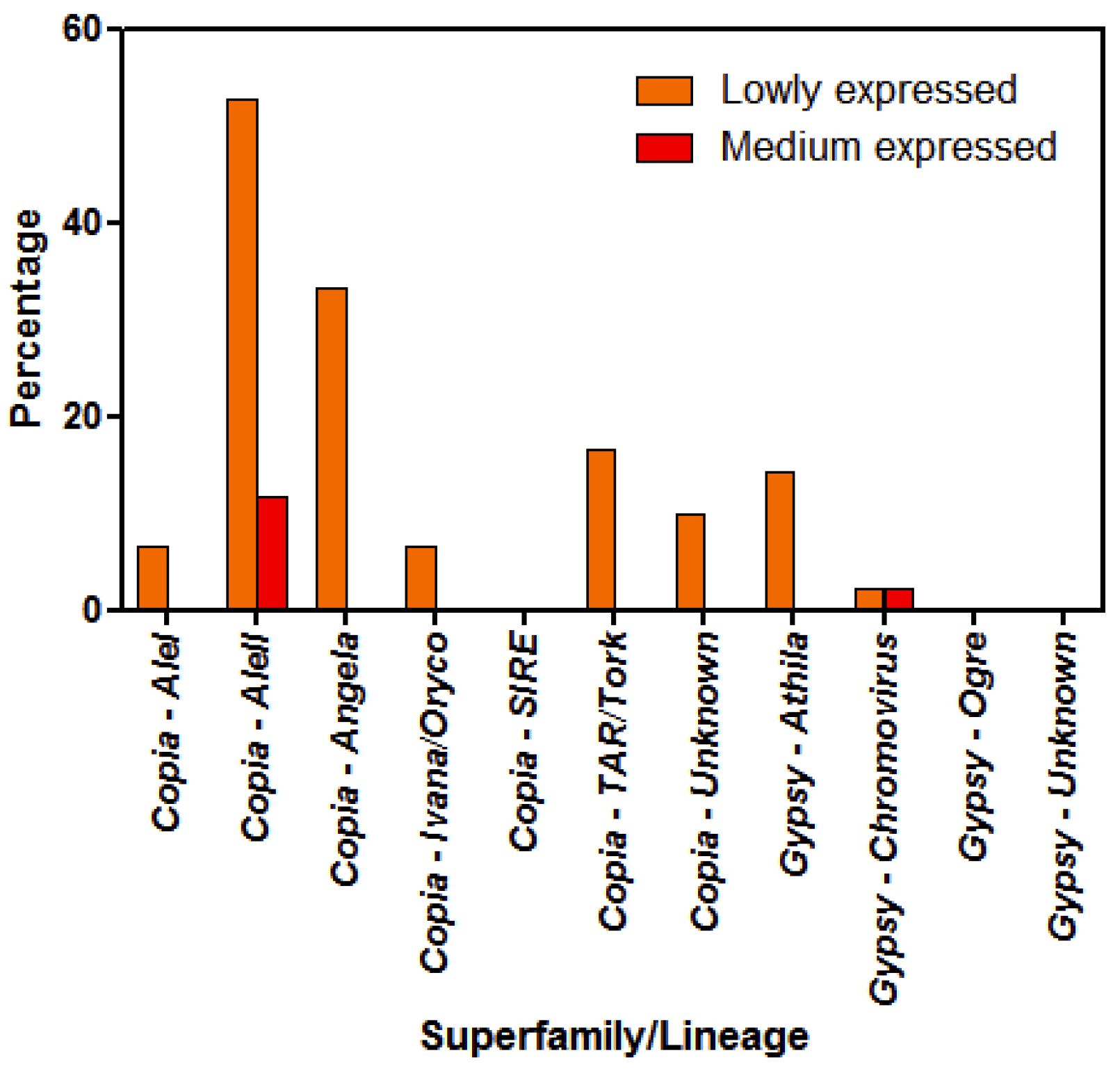
3.3. Differential LTR-Retrotransposon Expression Analyses
4. Discussion
Supplementary Material
Author Contributions
Funding
Conflicts of Interest
Appendix A
References
- Costanza, R. The value of world’s ecosystem services and natural capital. Nature 1997, 287, 253–260. [Google Scholar] [CrossRef]
- McArthur, L.C.; Boland, J.W. The economic contribution of seagrass to secondary production in South Australia. Ecol. Model. 2006, 196, 163–172. [Google Scholar] [CrossRef]
- Duarte, C.M. The future of seagrass meadows. Environ. Conserv. 2002, 29, 192–206. [Google Scholar] [CrossRef] [Green Version]
- Short, F.T.; Polidoro, B.; Livingstone, S.R.; Carpenter, K.E.; Bandeira, S.; Bujang, J.S.; Calumpong, H.P.; Carruthers, T.J.B.; Coles, R.G.; Dennison, W.C.; et al. Extinction risk assessment of the world’s seagrass species. Biol. Conserv. 2011, 144, 1961–1971. [Google Scholar] [CrossRef]
- Peres, J.M. La regression des herbiers a Posidonia oceanica. In Proceedings International Workshop Posidonia oceanica Beds; Boudouresque, C.F., Jeudy de Grissac, A., Olivier, J., Eds.; GIS Posidonie: Marseille, France, 1984; pp. 445–454. [Google Scholar]
- Marbà, N.; Duarte, C.M. Mediterranean warming triggers seagrass (Posidonia oceanica) shoot mortality. Glob. Chang. Biol. 2010, 16, 2366–2375. [Google Scholar] [CrossRef]
- Langar, H.; Djellouli, A.S.; Sellem, F.; El Abed, A. Extension of two Caulerpa species along the Tunisian coast. J. Coast. Conserv. 2002, 8, 163–167. [Google Scholar] [CrossRef]
- Orth, R.J.; Carruthers, T.J.B.; Dennison, W.C.; Duarte, C.M.; Fourqurean, J.W.; Heck, K.L.; Hughes, A.R.; Kendrick, G.A.; Kenworthy, W.J.; Olyarnik, S.; et al. A global crisis for seagrass ecosystems. Bioscience 2006, 56, 987–996. [Google Scholar] [CrossRef] [Green Version]
- Waycott, M.; Duarte, C.M.; Carruthers, T.J.B.; Orth, R.J.; Dennison, W.C.; Olyarnik, S.; Calladine, A.; Fourqurean, J.W.; Heck, K.L., Jr.; Hughes, A.R.; et al. Accelerating loss of seagrasses across the globe threatens coastal ecosystems. Proc. Natl. Acad. Sci. USA 2009, 106, 12377–12381. [Google Scholar] [CrossRef] [Green Version]
- Evans, S.M.; Vergés, A.; Poore, A.G.B. Genotypic diversity and short-term response to shading stress in a threatened seagrass: Does low diversity mean low resilience? Front. Plant Sci. 2017, 8, 1417. [Google Scholar] [CrossRef] [Green Version]
- Dodd, R.S.; Douhovnikoff, V. Adjusting to global change through clonal growth and epigenetic variation. Front. Ecol. Evol. 2016, 4, 86. [Google Scholar] [CrossRef] [Green Version]
- Hughes, A.R.; Stachowicz, J.J. Genetic diversity enhances the resistance of a seagrass ecosystem to disturbance. Proc. Natl. Acad. Sci. USA 2004, 101, 8998–9002. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reusch, T.B.H.; Ehlers, A.; Hammerli, A.; Worm, B. Ecosystem recovery after climatic extremes enhanced by genotypic diversity. Proc. Natl. Acad. Sci. USA 2005, 102, 2826–2831. [Google Scholar] [CrossRef] [Green Version]
- De Meester, L.; Louette, G.; Duvivier, C.; Van Damme, C.; Michels, E. Genetic composition of resident populations influences establishment success of immigrant species. Oecologia 2007, 153, 431–440. [Google Scholar] [CrossRef] [PubMed]
- Evans, S.M.; Sinclair, E.A.; Poore, A.G.B.; Steinberg, P.D.; Kendrick, G.A.; Vergés, A. Genetic diversity in threatened Posidonia australis seagrass meadows. Conserv. Genet. 2014, 15, 717–728. [Google Scholar] [CrossRef]
- Loques, F.; Caye, G.; Meinesz, A. Axenic culture of selected tissue of Posidonia oceanica. Aquat. Bot. 1990, 37, 171–188. [Google Scholar] [CrossRef]
- Procaccini, G.; Olsen, J.L.; Reusch, T.B.H. Contribution of genetics and genomics to seagrass biology and conservation. J. Exp. Marine Biol. Ecol. 2007, 350, 234–259. [Google Scholar] [CrossRef]
- Jahnke, M.; Olsen, J.L.; Procaccini, G. A meta-analysis reveals a positive correlation between genetic diversity metrics and environmental status in the long-lived seagrass Posidonia oceanica. Mol. Ecol. 2015, 24, 2336–2348. [Google Scholar] [CrossRef]
- Wicker, T.; Sabot, F.; Hua-Van, A.; Bennetzen, J.L.; Capy, P.; Chalhoub, B.; Flavell, A.; Leroy, P.; Morgante, M.; Panaud, O.; et al. A unified classification system for eukaryotic transposable elements. Nat. Rev. Genet. 2007, 8, 973–982. [Google Scholar] [CrossRef]
- SanMiguel, P.; Gaut, B.S.; Tikhonov, A.; Nakajima, Y.; Bennetzen, J.L. The paleontology of intergene retrotransposons of maize. Nat. Genet. 1998, 20, 43–45. [Google Scholar] [CrossRef]
- Vicient, C.M.; Suoniemi, A.; Anamthawat-Jonsson, K.; Tanskanen, J.; Beharav, A.; Nevo, E.; Schulman, A.H. Retrotransposon BARE-1 and its role in genome evolution in the genus Hordeum. Plant Cell 1999, 11, 1769–1784. [Google Scholar] [CrossRef] [Green Version]
- Bennetzen, J.L. Transposable elements contributions to plant gene and genome evolution. Plant Mol. Biol. 2000, 42, 251–269. [Google Scholar] [CrossRef] [PubMed]
- Wicker, T.; Keller, B. Genome-wide comparative analysis of copia retrotransposons in Triticeae, rice, and Arabidopsis reveals conserved ancient evolutionary lineages and distinct dynamics of individual copia families. Genome Res. 2007, 17, 1072–1081. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Usai, G.; Mascagni, F.; Natali, L.; Giordani, T.; Cavallini, A. Comparative genome-wide analysis of repetitive DNA in the genus Populus L. Tree Genet. Genomes 2017, 13, 96. [Google Scholar] [CrossRef] [Green Version]
- Buti, M.; Moretto, M.; Barghini, E.; Mascagni, F.; Natali, L.; Brilli, M.; Lomsadze, A.; Sonego, P.; Giongo, L.; Alonge, M.; et al. The genome sequence and transcriptome of Potentilla micrantha and their comparison to Fragaria vesca (the woodland strawberry). GigaScience 2018, 7, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Mascagni, F.; Cavallini, A.; Giordani, T.; Natali, L. Different histories of two highly variable LTR retrotransposons in sunflower species. Gene 2017, 634, 5–14. [Google Scholar] [CrossRef] [Green Version]
- Mascagni, F.; Giordani, T.; Ceccarelli, M.; Cavallini, A.; Natali, L. Genome-wide analysis of LTR-retrotransposon diversity and its impact on the evolution of the genus Helianthus (L.). BMC Genomics 2017, 18, 634. [Google Scholar] [CrossRef] [Green Version]
- Mascagni, F.; Vangelisti, A.; Giordani, T.; Cavallini, A.; Natali, L. Specific LTR-retrotransposons show copy number variations between wild and cultivated sunflowers. Genes 2018, 9, 433. [Google Scholar] [CrossRef] [Green Version]
- Neumann, P.; Novák, P.; Hoštáková, N.; Macas, J. Systematic survey of plant LTR retrotransposons elucidates phylogenetic relationships of their polyprotein domains and provides a reference for element classification. Mobile DNA 2019, 10, 1. [Google Scholar] [CrossRef]
- Springer, N.M.; Ying, K.; Fu, Y.; Ji, T.; Yeh, C.T.; Jia, Y.; Wu, W.; Richmond, T.; Kitzman, J.; Rosenbaum, H.; et al. Maize inbreds exhibit high levels of copy number variation (CNV) and presence/absence variation (PAV) in genome content. PLoS Genet. 2009, 5, e1000734. [Google Scholar] [CrossRef] [Green Version]
- Vitte, C.; Fustier, M.A.; Alix, K.; Tenaillon, M.I. The bright side of transposons in crop evolution. Briefings Funct. Genom. 2014, 13, 276–295. [Google Scholar] [CrossRef] [Green Version]
- Slotkin, R.K.; Martienssen, R. Transposable elements and the epigenetic regulation of the genome. Nat. Rev. Genet. 2007, 8, 272–285. [Google Scholar] [CrossRef] [PubMed]
- Butelli, E.; Licciardello, C.; Zhang, Y.; Liu, J.; Mackay, S.; Bailey, P. Retrotransposons control fruit-specific, cold-dependent accumulation of anthocyanins in blood oranges. Plant Cell 2012, 24, 1242–1255. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Falchi, R.; Vendramin, E.; Zanon, L.; Scalabrin, S.; Cipriani, G.; Verde, I.; Vizzotto, G.; Morgante, M. Three distinct mutational mechanisms acting on a single gene underpin the origin of yellow flesh in peach. Plant J. 2013, 76, 175–187. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lisch, D. How important are transposons for plant evolution? Nat. Rev. Genet. 2013, 14, 49–61. [Google Scholar] [CrossRef]
- Rey, O.; Danchin, E.; Mirouze, M.; Loot, C.; Blanchet, S. Adaptation to global change: A transposable element–epigenetics perspective. Trends Ecol. Evol. 2016, 31, 514–526. [Google Scholar] [CrossRef] [PubMed]
- Cavallini, A.; Natali, L.; Giordani, T.; Polizzi, E.; Balestri, E.; Cinelli, F.; Maserti, B.E.; Ferrara, R. Cytophotometric and biochemical characterization of Posidonia oceanica L. (Potamogetonaceae) genome. Caryologia 1995, 48, 201–209. [Google Scholar] [CrossRef]
- Barghini, E.; Mascagni, F.; Natali, L.; Giordani, T.; Cavallini, A. Analysis of the repetitive component and retrotransposons population in the genome of a marine angiosperm, Posidonia oceanica (L.) Delile. Mar. Genomics 2015, 24, 397–404. [Google Scholar] [CrossRef]
- Marín-Guirao, L.; Entrambasaguas, L.; Dattolo, E.; Ruiz, J.M.; Procaccini, G. Molecular mechanisms behind the physiological resistance to intense transient warming in an iconic marine plant. Front. Plant Sci. 2017, 8, 1142. [Google Scholar] [CrossRef] [Green Version]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed on 18 February 2020).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucl. Acid Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Olsen, J.L.; Rouzé, P.; Verhelst, B.; Lin, Y.C.; Bayer, T.; Collen, J.; Dattolo, E.; De Paoli, E.; Dittami, S.; Maumus, F.; et al. The genome of the seagrass Zostera marina reveals angiosperm adaptation to the sea. Naure 2016, 530, 331–334. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ellinghaus, D.; Kurtz, S.; Willhoeft, U. LTRharvest, an efficient and flexible software for de novo detection of LTR retrotransposons. BMC Bioinform. 2008, 9, 18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Copetti, D.; Zhang, J.; El Baidouri, M.; Gao, D.; Wang, J.; Barghini, E.; Cossu, R.M.; Angelova, A.; Maldonado, C.E.; Roffler, S.; et al. RiTE database: A resource database for genus-wide rice genomics and evolutionary biology. BMC Genom. 2015, 16, 538. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, P.; Binns, D.; Chang, H.; Fraser, M.; Li, W.; McAnulla, C.; McWilliam, H.; Maslen, J.; Mitchell, A.; Nuka, G.; et al. InterProScan 5: Genome-scale protein function classification. Bioinformatics 2014, 30, 1236–1240. [Google Scholar] [CrossRef] [Green Version]
- Sonnhammer, E.L.; Eddy, S.R.; Durbin, R. Pfam: A comprehensive database of protein domain families based on seed alignments. Proteins 1997, 28, 405–420. [Google Scholar] [CrossRef]
- D’Esposito, D.; Orrù, L.; Dattolo, E.; Bernardo, L.; Lamontanara, A.; Orsini, L.; Serra, I.A.; Mazzuca, S.; Procaccini, G. Transcriptome characterisation and simple sequence repeat marker discovery in the seagrass Posidonia oceanica. Sci. Data 2016, 3, 160115. [Google Scholar] [CrossRef]
- Baggerley, K.; Deng, L.; Morris, J.; Aldaz, C. Differential expression in SAGE: Accounting for normal between-library variation. Bioinformatics 2003, 19, 1477–1483. [Google Scholar] [CrossRef]
- Lu, X.; Chen, D.; Shu, D.; Zhang, Z.; Wang, W.; Klukas, C.; Chen, L.L.; Fan, Y.; Chen, M.; Zhang, C. The differential transcription network between embryo and endosperm in the early developing maize seed. Plant Physiol. 2013, 162, 440–455. [Google Scholar] [CrossRef] [Green Version]
- Giordani, T.; Cossu, R.M.; Mascagni, F.; Marroni, F.; Morgante, M.; Cavallini, A.; Natali, L. Genome-wide analysis of LTR-retrotransposons expression in leaves of Populus x canadensis water-deprived plants. Tree Genet. Genomes 2016, 12, 75. [Google Scholar] [CrossRef] [Green Version]
- Cavallini, A.; Mascagni, F.; Giordani, T.; Natali, L. Genome skimming for plant retrotransposon 442 identification and expression analysis. Agrochimica 2019, 63, 367–378. [Google Scholar]
- Vicient, C.M.; Jaaskelainen, M.J.; Kalendar, R.; Schulman, A.H. Active retrotransposons are a common feature of grass genomes. Plant Physiol. 2001, 125, 1283–1292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ishiguro, S.; Ogasawara, K.; Fujino, K.; Sato, Y.; Kishima, Y. Low temperature-responsive changes in the anther transcriptome’s repeat sequences are indicative of stress sensitivity and pollen sterility in rice strains. Plant Physiol. 2014, 164, 671–682. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, X.; Zhou, J.; Li, J.; Zou, X.; Zhao, J.; Li, Q.; Xia, R.; Yang, R.; Wang, D.; Zuo, Z.; et al. Efficient generation of marker-free transgenic rice plants using an improved transposon-mediated transgene reintegration strategy. Plant Physiol. 2015, 167, 11–24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Usai, G.; Mascagni, F.; Vangelisti, A.; Giordani, T.; Ceccarelli, M.; Cavallini, A.; Natali, L. Interspecific hybridisation and LTR-retrotransposon mobilisation-related structural variation in plants: A case study. Genomics 2020, 112, 1611–1621. [Google Scholar] [CrossRef] [PubMed]
- Kashkush, K.; Feldman, M.; Levy, A.A. Transcriptional activation of retrotransposons alters the expression of adjacent genes in wheat. Nat. Genet. 2003, 33, 102–106. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Han, P.F.; Tan, M.; Shan, X.H.; Dong, Y.Z.; Wang, X.Z.; Fedak, G.; Hao, S.; Liu, B. Activation of a rice endogenous retrotransposon Tos17 in tissue culture is accompanied by cytosine demethylation and causes heritable alteration in methylation pattern of flanking genomic regions. Theor. Appl. Genet. 2004, 109, 200–209. [Google Scholar] [CrossRef]
- He, P.; Ma, Y.; Dai, H.Y.; Li, L.; Liu, Y.; Li, H.; Zhao, G.; Zhang, Z. Characterization of the hormone and stress-induced expression of FaRE1 retrotransposon promoter in strawberry. J. Plant Biol. 2012, 55, 1–7. [Google Scholar] [CrossRef]
- Ito, H.; Yoshida, T.; Tsukahara, S.; Kawabe, A. Evolution of the ONSEN retrotransposon family activated upon heat stress in Brassicaceae. Gene 2013, 518, 256–261. [Google Scholar] [CrossRef] [Green Version]
- Voronova, A.; Belevich, V.; Rungis, D.; Jansons, A. Stress-induced transcriptional activation of retrotransposon-like sequences in the Scots pine (Pinus sylvestris L.) genome. Tree Genet. Genomes 2014, 10, 937–951. [Google Scholar] [CrossRef]
- Mascagni, F.; Vangelisti, A.; Usai, G.; Giordani, T.; Cavallini, A.; Natali, L. A computational genome-wide analysis of long terminal repeats retrotransposon expression in sunflower roots (Helianthus annuus L.). Genetica 2020, 148, 13–23. [Google Scholar] [CrossRef]
- Ma, Y.; Sun, H.Y.; Zhao, G.L.; Dai, H.; Gao, X.; Li, H.; Zhang, Z. Isolation and characterization of genomic retrotransposon sequences from octoploid strawberry (Fragaria × ananassa Duch.). Plant Cell Rep. 2008, 27, 499–507. [Google Scholar] [CrossRef] [PubMed]
- Hawkins, J.S.; Kim, H.R.; Nason, J.D.; Wing, R.A.; Wendel, J.F. Differential lineage-specific amplification of transposable elements is responsible for genome size variation in Gossypium. Genome Res. 2006, 16, 1252–1261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vangelisti, A.; Mascagni, F.; Giordani, T.; Sbrana, C.; Turrini, A.; Cavallini, A.; Giovannetti, M.; Natali, L. Arbuscular mycorrhizal fungi induce the expression of specific retrotransposons in roots of sunflower (Helianthus annuus L.). PLoS ONE 2019, 14, e0212371. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Legrand, S.; Caron, T.; Maumus, F.; Schvartzman, S.; Quadrana, L.; Durand, E.; Gallina, S.; Pauwels, M.; Mazoyer, C.; Huyghe, L.; et al. Differential retention of transposable element-derived sequences in outcrossing Arabidopsis genomes. Mob. DNA 2019, 10, 30. [Google Scholar] [CrossRef] [Green Version]
- Tenaillon, M.I.; Hollister, J.D.; Gaut, B.S. A triptych of the evolution of plant transposable elements. Trends Plant Sci. 2010, 15, 471–478. [Google Scholar] [CrossRef]
- Grandbastien, M.A. Activation of plant retrotransposons under stress conditions. Trends Plant Sci. 1998, 3, 181–189. [Google Scholar] [CrossRef]
- Vukich, M.; Giordani, T.; Natali, L.; Cavallini, A. Copia and Gypsy retrotransposons activity in sunflower (Helianthus annuus L.). BMC Plant Biol. 2009, 9, 150. [Google Scholar] [CrossRef] [Green Version]
- Meyers, B.C.; Tingey, S.V.; Morgante, M. Abundance, distribution, and transcriptional activity of repetitive elements in the maize genome. Genome Res. 2001, 11, 1660–1676. [Google Scholar] [CrossRef] [Green Version]
- Yamazaki, M.; Tsugawa, H.; Miyao, A.; Yano, M.; Wu, J.; Yamamoto, S.; Matsumoto, T.; Sasaki, T.; Hirochika, H. The rice retrotransposon Tos17 prefers low-copy-number sequences as integration targets. Mol. Genet. Genom. 2001, 265, 336–344. [Google Scholar]
- Ruocco, M.; Musacchia, F.; Olivé, I.; Costa, M.M.; Barrote, I.; Santos, R.; Sanges, R.; Procaccini, G.; Silva, J. Genome wide transcriptional reprogramming in the seagrass Cymodocea nodosa under experimental ocean acidification. Mol. Ecol. 2017, 26, 4241–4259. [Google Scholar] [CrossRef]
| ID | Superfamily | Lineage | Length (nt) | Mean Number of Mapped Reads Per Million | |
|---|---|---|---|---|---|
| Deep | Shallow | ||||
| Poc_contig_4694 | Copia | AleI | 237 | 1.68 | 2.04 |
| Poc_contig_11919 | Copia | AleII | 303 | 1.04 | 1.16 |
| Poc_contig_13155 | Copia | AleII | 350 | 0.96 | 1.31 |
| Poc_contig_16252 | Copia | AleII | 292 | 1.49 | 1.50 |
| Poc_contig_2925 | Copia | AleII | 265 | 1.47 | 1.74 |
| Poc_contig_11953 | Copia | AleII | 264 | 2.39 | 2.53 |
| Poc_contig_11947 | Copia | AleII | 233 | 2.29 | 2.73 |
| Poc_contig_7641 | Copia | AleII | 258 | 2.46 | 2.91 |
| Poc_contig_5573 | Copia | AleII | 235 | 2.70 | 3.06 |
| Poc_contig_10207 | Copia | AleII | 364 | 3.25 | 3.56 |
| Poc_contig_14966 | Copia | AleII | 330 | 8.64 | 10.07 |
| Poc_contig_10239 | Copia | AleII | 434 | 14.39 | 17.02 |
| Poc_contig_13500 | Copia | Angela | 326 | 0.90 | 1.24 |
| Poc_contig_3616 | Copia | Angela | 223 | 1.57 | 1.62 |
| Poc_contig_6783 | Copia | Angela | 279 | 4.39 | 4.74 |
| Poc_contig_2856 | Copia | Ivana/Oryco | 429 | 1.77 | 2.58 |
| Poc_contig_13693 | Copia | TAR/Tork | 152 | 1.29 | 1.20 |
| Poc_contig_16069 | Copia | TAR/Tork | 312 | 1.47 | 1.75 |
| Poc_contig_3029 | Copia | TAR/Tork | 395 | 3.77 | 5.34 |
| Poc_contig_12140 | Copia | Unknown | 207 | 1.02 | 1.04 |
| Poc_contig_14451 | Copia | Unknown | 176 | 1.40 | 1.47 |
| Poc_contig_3356 | Gypsy | Athila | 828 | 2.28 | 3.11 |
| Poc_contig_6648 | Gypsy | Chromovirus | 301 | 1.19 | 1.41 |
| Poc_contig_16201 | Gypsy | Chromovirus | 260 | 9.11 | 7.91 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vangelisti, A.; Mascagni, F.; Usai, G.; Natali, L.; Giordani, T.; Cavallini, A. Low Long Terminal Repeat (LTR)-Retrotransposon Expression in Leaves of the Marine Phanerogam Posidonia Oceanica L. Life 2020, 10, 30. https://doi.org/10.3390/life10030030
Vangelisti A, Mascagni F, Usai G, Natali L, Giordani T, Cavallini A. Low Long Terminal Repeat (LTR)-Retrotransposon Expression in Leaves of the Marine Phanerogam Posidonia Oceanica L. Life. 2020; 10(3):30. https://doi.org/10.3390/life10030030
Chicago/Turabian StyleVangelisti, Alberto, Flavia Mascagni, Gabriele Usai, Lucia Natali, Tommaso Giordani, and Andrea Cavallini. 2020. "Low Long Terminal Repeat (LTR)-Retrotransposon Expression in Leaves of the Marine Phanerogam Posidonia Oceanica L." Life 10, no. 3: 30. https://doi.org/10.3390/life10030030
APA StyleVangelisti, A., Mascagni, F., Usai, G., Natali, L., Giordani, T., & Cavallini, A. (2020). Low Long Terminal Repeat (LTR)-Retrotransposon Expression in Leaves of the Marine Phanerogam Posidonia Oceanica L. Life, 10(3), 30. https://doi.org/10.3390/life10030030





