Coupling of Cell Division and Differentiation in Arabidopsis thaliana Cultured Cells with Interaction of Ethylene and ABA Signaling Pathways
Abstract
:1. Introduction
2. Materials and Methods
3. Results and Discussion
3.1. Assessing the Effects of ABA on Growth and Differentiation in Cell Suspension Cultures
3.2. Ethylene and ABA Production during the Sub-Cultivation Cycle and Effect of ABA on DNA Synthesis
3.3. ABA Affects Soluble Proteins Phosphorylaion in Col-0 and Ethylene-Insensitive Mutant Cell Cultures
3.4. The Effect of ABA on Transcription of Individual MPK Genes
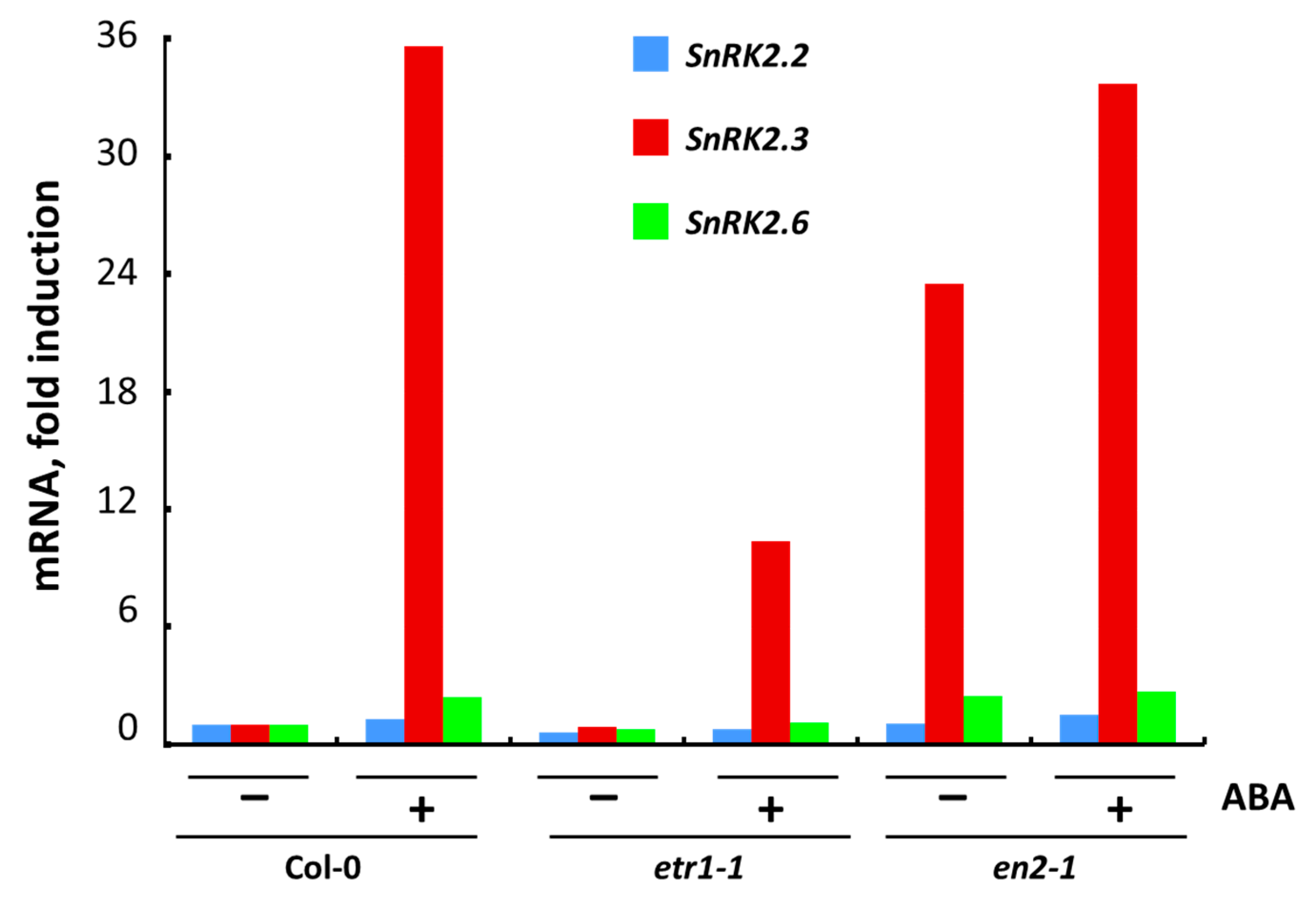
3.5. The Effect of ABA on Transcription of Individual SnRK2 Genes
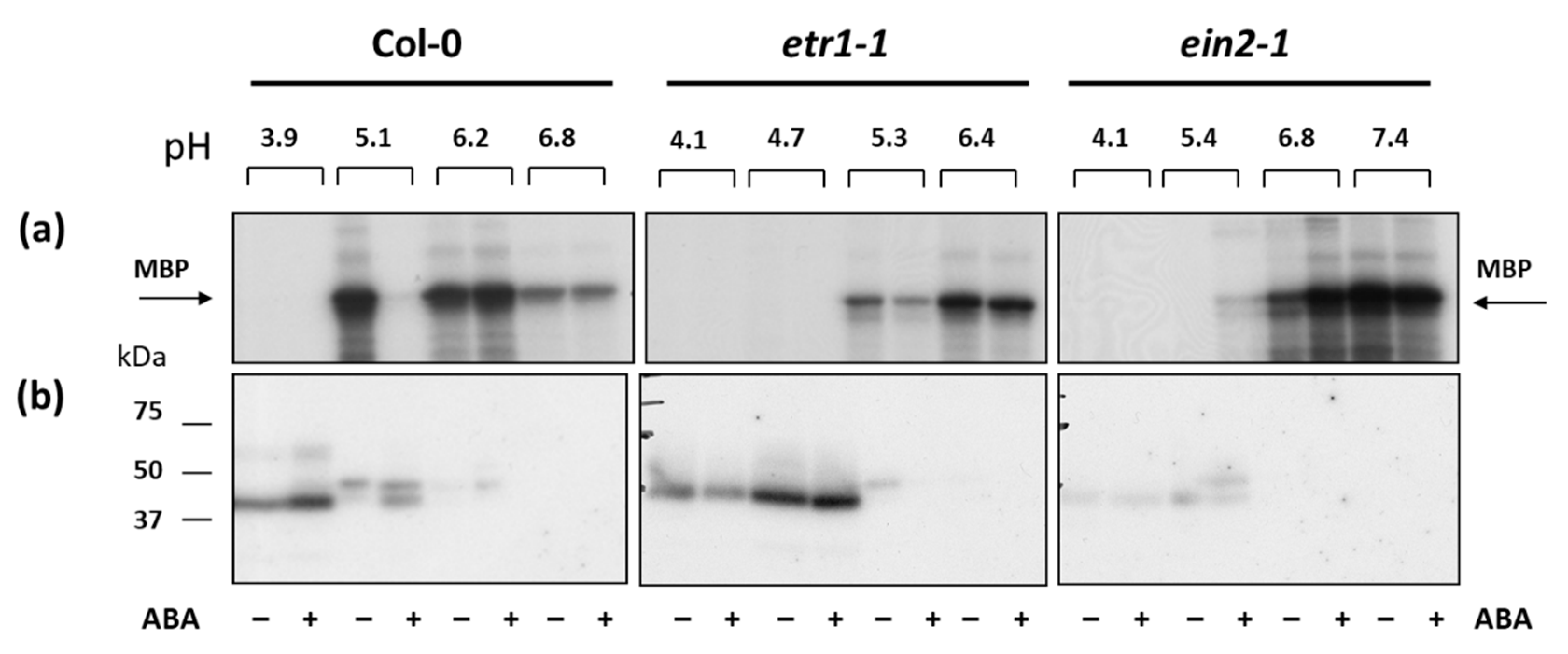
3.6. The effect of ABA on MAPK Activity in Col-0 and Ethylene-Insensitive Mutant Cell Cultures
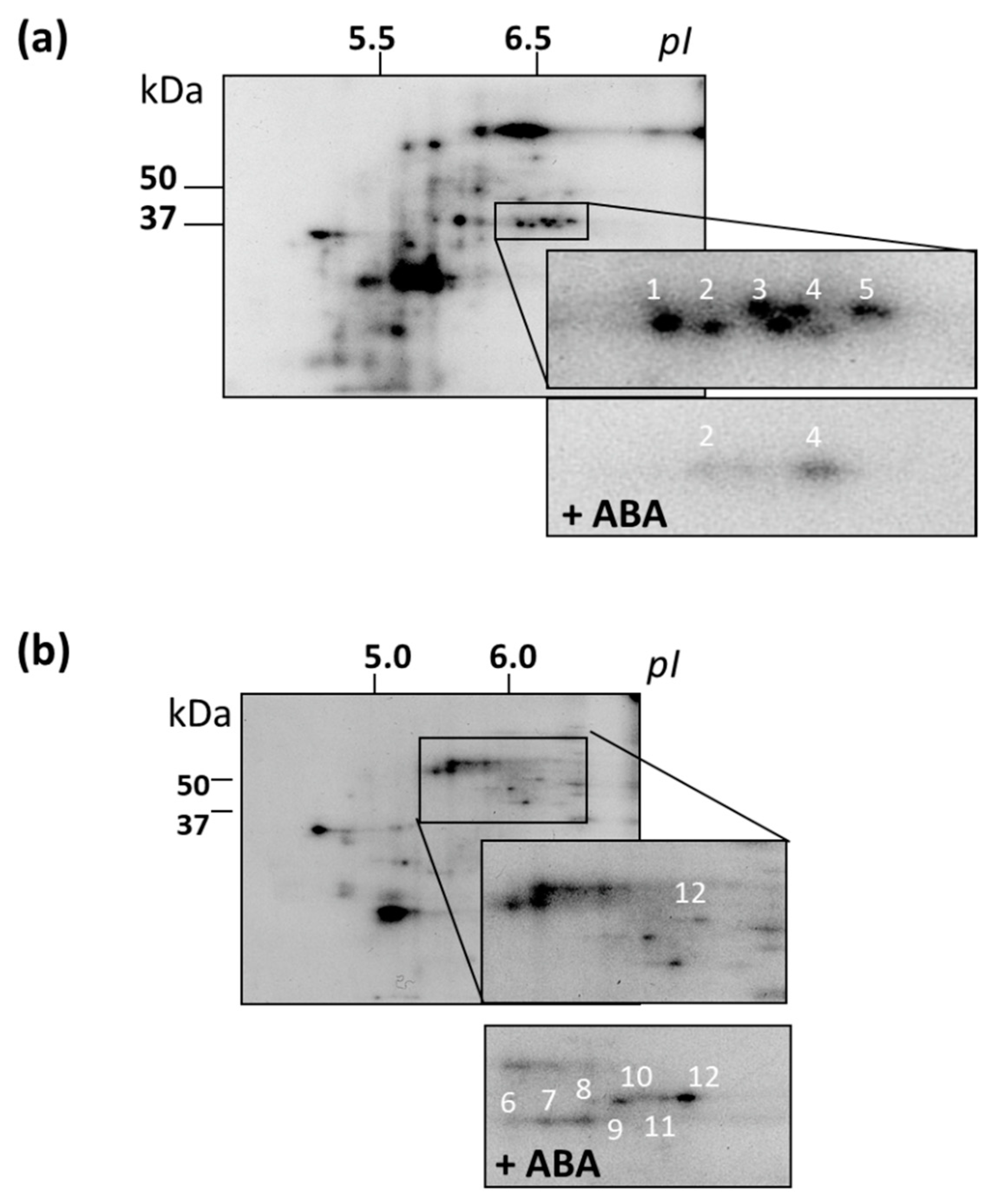
3.7. Identification of MAPKs and Their Putative Substrates with MALDI-TOF MS
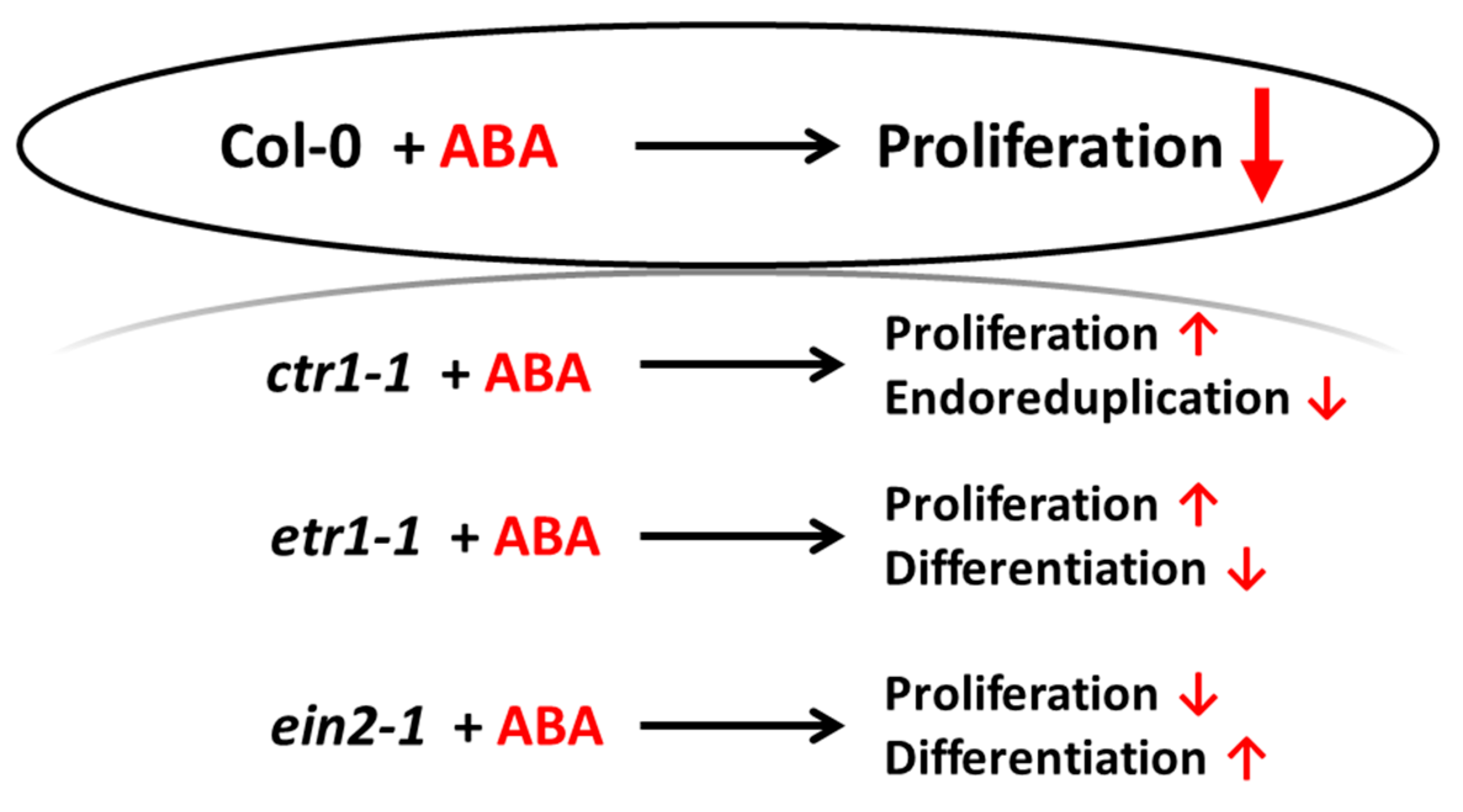
4. Conclusion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Dudits, D.; Abraham, E.; Miskolczi, P.; Ayaydin, F.; Bilgin, M.; Gabor, V.; Horvath, G.V. Cell-cycle control as a target for calcium, hormonal and developmental signals: the role of phosphorylation in the retinoblastoma-centered pathway. Ann. Bot. 2005, 107, 193–1202. [Google Scholar] [CrossRef] [Green Version]
- Ruggiero, B.; Koiwa, H.; Manabe, Y.; Quist, T.M.; Inan, G.; Saccardo, F.; Joly, R.J.; Hasegawa, P.M.; Bressan, R.A.; Maggio, A. Uncoupling the effects of abscisic acid on plant growth and water relations. Analysis of sto1/nced3, an abscisic acid-deficient but salt stress-tolerant mutant in Arabidopsis. Plant Physiol. 2004, 136, 3134–3147. [Google Scholar] [CrossRef] [Green Version]
- Beaudoin, N.; Serizet, C.; Gosti, F.; Giraudat, J. Interactions between abscisic acid and ethylene signaling cascades. Plant Cell 2000, 12, 1103–1116. [Google Scholar] [CrossRef] [Green Version]
- Ghassemian, M.; Nambara, E.; Cutler, S.; Kawaide, H.; Kamiya, Y.; McCourt, P. Regulation of abscisic acid signaling by the ethylene response pathway in Arabidopsis. Plant Cell 2000, 12, 1117–1126. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Ngwenyama, N.; Liu, Y.; Walker, J.C.; Zhang, S. Stomatal development and patterning are regulated by environmentally responsive mitogen-activated protein kinases in Arabidopsis. Plant Cell 2007, 19, 63–73. [Google Scholar] [CrossRef] [Green Version]
- Stepanchenko, N.S.; Fomenkov, A.A.; Moshkov, I.E.; Rakitin, V.Y.; Novikova, G.V.; Nosov, A.V. Phytohormone interplay controls proliferation of in vitro cultivated cells of Arabidopsis thaliana ethylene–insensitive mutants. Dokl. Biol. Sci. 2012, 442, 46–49. [Google Scholar] [CrossRef]
- Ju, C.; Chang, C. Mechanistic insights in ethylene perception and signal transduction. Plant Physiol. 2015, 169, 85–95. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, Y.; Szostkiewicz, I.; Korte, A.; Moes, D.; Yang, Y.; Christmann, A.; Grill, E. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 2009, 324, 1064–1068. [Google Scholar] [CrossRef]
- Park, S.Y.; Fung, P.; Nishimura, N.; Jensen, D.R.; Fujii, H.; Zhao, Y.; Lumba, S.; Santiago, J.; Rodrigues, A.; Chow, T.F.; et al. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 2009, 324, 1068–1071. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soon, F.F.; Ng, L.-M.; Zhou, X.E.; West, G.M.; Kovach, A.; Tan, M.H.; Suino-Powell, K.M.; He, Y.; Xu, Y.; Chalmers, M.J.; et al. Molecular mimicry regulates ABA signaling by SnRK2 kinases and PP2C phosphatases. Science 2012, 335, 85–88. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fujii, H.; Zhu, J.K. Arabidopsis mutant deficient in 3 abscisic acid-activated protein kinases reveals critical roles in growth, reproduction, and stress. Proc. Natl. Acad. Sci. USA 2009, 106, 8380–8385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fujita, Y.; Nakashima, K.; Yoshida, T.; Katagiri, T.; Kidokoro, S.; Kanamori, N. Three SnRK2 protein kinases are the main positive regulators of abscisic acid signaling in response to water stress in Arabidopsis. Plant Cell Physiol. 2009, 50, 2123–2132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakashima, K.; Nakashima, K.; Fujita, Y.; Kanamori, N.; Katagiri, T.; Umezawa, T.; Kidokoro, S.; Maruyama, K.; Yoshida, T.; Ishiyama, K.; et al. Three Arabidopsis SnRK2 protein kinases, SRK2D/SnRK2.2, SRK2E/SnRK2.6/OST1 and SRK2I/SnRK2.3, involved in ABA signaling are essential for the control of seed development and dormancy. Plant Cell Physiol 2009, 50, 1345–1363. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cutler, S.R.; Rodriguez, P.L.; Finkelstein, R.R.; Abrams, S.R. Abscisic acid: emergence of a core signaling network. Annu. Rev. Plant Biol. 2010, 61, 651–679. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoshida, T.; Fujita, Y.; Sayama, H.; Kidokoro, S.; Maruyama, K.; Mizoi, J.; Yamaguchi-Shinozaki, K. AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant J. 2010, 61, 672–685. [Google Scholar] [CrossRef] [PubMed]
- Harashima, H.; Dissmeyer, N.; Schnittger, A. Cell cycle control across the eukaryotic kingdom. Trends Cell Biol. 2013, 23, 345–356. [Google Scholar] [CrossRef]
- Apelbaum, A.; Burg, S. Effect of ethylene on cell division and deoxyribonucleic acid synthesis in Pisum sativum. Plant Physiol. 1972, 50, 117–124. [Google Scholar] [CrossRef] [Green Version]
- Herbert, R.J.; Vilhar, B.; Evett, C.; Orchard, C.B.; Rogers, H.J.; Davies, M.S.; Francis, D. Ethylene induces cell death at particular phases of the cell cycle in the tobacco TBY-2 cell line. J. Exp. Bot. 2001, 52, 1615–1623. [Google Scholar]
- Dan, H.; Imaseki, H.; Wasteneys, G.O.; Kazama, H. Ethylene stimulates endoreduplication but inhibits cytokinesis in cucumber hypocotyl epidermis. Plant Physiol. 2003, 133, 1726–1731. [Google Scholar] [CrossRef] [Green Version]
- Ortega-Martenez, O.; Pernas, M.; Carol, R.J.; Dolan, L. Ethylene modulates stem cell division in the Arabidopsis thaliana root. Science 2007, 317, 507–510. [Google Scholar] [CrossRef]
- Love, J.; Bjorklund, S.; Vahala, J.; Hertzberg, M.; Kangasjarvi, J.; Sundberg, B. Ethylene is an endogenous stimulator of cell division in the cambial meristem of Populus. Proc. Natl. Acad. Sci. USA 2009, 106, 5984–5989. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bystrova, E.I.; Zhukovskaya, N.V.; Rakitin, V.Y.; Ivanov, V.B. Role of ethylene in activation of cell division in quiescent center of excised maize roots. Rus. J. Dev. Biol. 2015, 46, 60–64. [Google Scholar] [CrossRef]
- Swiatek, A.; Lenjou, M.; van Bockstaele, D.; Inze, D.; van Onckelen, H. Differential effect of jasmonic acid and abscisic acid on cell cycle progression in tobacco BY-2 cells. Plant Physiol. 2002, 128, 201–211. [Google Scholar] [CrossRef] [PubMed]
- Skirycz, A.; Claeys, H.; De Bodt, S.; Oikawa, A.; Shinoda, S.; Andriankaja, M.; Maleux, K.; Eloy, N.B.; Coppens, F.; Yoo, S.-D.; et al. Pause-and-stop: The effects of osmotic stress on cell proliferation during early leaf development in Arabidopsis and a role for ethylene signaling in cell cycle arrest. Plant Cell 2011, 23, 1876–1888. [Google Scholar] [CrossRef] [Green Version]
- Meszaros, T.; Miskolczi, P.; Ayaydin, F.; Pettko-Szandtner, A.; Peres, A.; Magyar, Z.; Horvath, G.V.; Bako, L.; Feher, A.; Dudits, D. Multiple cyclin-dependent kinase complexes and phosphatases control G2/M progression in alfalfa cells. Plant Mol. Biol. 2000, 43, 595–605. [Google Scholar] [CrossRef]
- West, G.; Inze, D.; Beemster, G.T. Cell cycle modulation in the response of the primary root of Arabidopsis to salt stress. Plant Physiol. 2004, 135, 1050–1058. [Google Scholar] [CrossRef]
- Cheng, W.H.; Endo, A.; Zhou, L.; Penney, J.; Chen, H.-C.; Arroyo, A.; Leon, P.; Nambara, E.; Asami, T.; Seo, M.; et al. A unique short-chain dehydrogenase/reductase in Arabidopsis glucose signaling and abscisic acid biosynthesis and functions. Plant Cell 2002, 14, 2723–2743. [Google Scholar] [CrossRef]
- Sharp, R.E.; LeNoble, M.E.; Else, M.A.; Thorne, E.T.; Gherardi, F. Endogenous ABA maintains shoot growth in tomato independently of effects on plant water balance: evidence for an interaction with ethylene. J. Exp. Bot. 2000, 51, 1575–1584. [Google Scholar] [CrossRef] [Green Version]
- Shenk, R.U.; Hildebrandt, A. Medium and techniques for induction and growth of monocotyledons and dicotyledons plant cell cultures. Can. J. Bot. 1972, 50, 199–204. [Google Scholar] [CrossRef]
- Sato, Y.; Yajima, Y.; Tokunaga, N.; Whetten, R. Comparison between tracheary element lignin formation and extracellular lignin-like substance formation during the culture of isolated Zinnia elegans mesophyll cells. Biologia 2011, 66, 88–95. [Google Scholar] [CrossRef]
- Ueda, J.; Saito, H.; Watanabe, H.; Evers, B.M. Novel and quantitative DNA dot-blotting methods for assessment of in vivo proliferation. Am. J. Physiol. 2005, 288, 842–847. [Google Scholar] [CrossRef] [PubMed]
- Fomenkov, A.A.; Nosov, A.V.; Rakitin, V.Y.; Sukhanova, E.S.; Mamaeva, A.S.; Sobol’kova, G.I.; Nosov, A.M.; Novikova, G.V. Ethylene in the proliferation of cultured plant cells: Regulating or just going along? Rus. J. Plant Physiol. 2015, 62, 815–822. [Google Scholar] [CrossRef]
- Novikova, G.V.; Mur, L.A.J.; Nosov, A.V.; Fomenkov, A.A.; Mironov, K.S.; Mamaeva, A.S.; Shilov, E.S.; Rakitin, V.Y.; Hall, M.A. Nitric oxide has a concentration-dependent effect on the cell cycle acting via EIN2 in Arabidopsis thaliana cultured cells. Front. Physiol. 2017, 8, 142. [Google Scholar] [CrossRef] [Green Version]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Shevchenko, A.; Wilm, M.; Vorm, O.; Mann, M. Mass spectrometric sequencing of proteins from silver-stained polyacrylamide gels. Anal. Chem. 1996, 68, 850–858. [Google Scholar] [CrossRef]
- Hey, J.; Posch, A.; Cohen, A.; Liu, N.; Harbers, A. Fractionation of complex protein mixtures by liquid-phase isoelectric focusing. Methods Mol. Biol. 2008, 424, 225–239. [Google Scholar] [CrossRef]
- Neuhoff, V.; Stamm, R.; Eibl, H. Clear background and highly sensitive protein staining with Coomassie Blue dyes in polyacrylamide gels: a systematic analysis. Electrophoresis 1985, 6, 427–448. [Google Scholar] [CrossRef]
- Fomenkov, A.A.; Nosov, A.V.; Rakitin, V.Y.; Mamaeva, A.S.; Novikova, G.V. Cytophysiological characteristics of Arabidopsis thaliana cultivated cells with disable perception of ethylene signal by the ETR1 receptor. Rus. J. Plant Physiol. 2014, 61, 598–607. [Google Scholar] [CrossRef]
- Oda, Y.; Fukuda, H. Secondary cell wall patterning during xylem differentiation. Curr. Opin. Plant Biol. 2012, 15, 38–44. [Google Scholar] [CrossRef]
- Oda, Y.; Mimura, T.; Hasezawa, S. Regulation of secondary cell wall development by cortical microtubules during tracheary element differentiation in Arabidopsis cell suspensions. Plant Physiol. 2005, 137, 1027–1036. [Google Scholar] [CrossRef] [Green Version]
- Novikova, G.V.; Moshkov, I.E.; Smith, A.R.; Hall, M.A. The effect of ethylene on MAPKinase-like activity in Arabidopsis thaliana. FEBS Lett. 2000, 474, 29–32. [Google Scholar] [CrossRef] [Green Version]
- Ouaked, F.; Rozhon, W.; Lecourieux, D.; Hirt, H. A MAPK pathway mediates ethylene signaling in plants. EMBO J. 2003, 22, 1282–1288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoo, S.-D.; Cho, Y.-H.; Tena, G.; Xiong, Y.; Sheen, J. Dual control of nuclear EIN3 by bifurcate MAPK cascades in C2H4 signalling. Nature 2008, 451, 789–797. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y. Roles of mitogen-activated protein kinase cascades in ABA signaling. Plant Cell Rep. 2012, 31, 1–12. [Google Scholar] [CrossRef] [PubMed]
- de Zelicourt, A.; Colcombet, J.; Hirt, H. The role of MAPK modules and ABA during abiotic stress signaling. Trends Plant Sci. 2016, 21, 677–685. [Google Scholar] [CrossRef] [PubMed]
- Jagodzik, P.; Tajdel-Zielinska, M.; Ciesla, A.; Marczak, M.; Ludwikow, A. Mitogen-activated protein kinase cascades in plant hormone signaling. Front. Plant Sci. 2018, 9, 1387. [Google Scholar] [CrossRef] [PubMed]
- Sasabe, M.; Boudolf, V.; De Veylder, L.; Inze, D.; Genschik, P.; Machida, Y. Phosphorylation of a mitotic kinesin-like protein and a MAPKKK by cyclin-dependent kinases (CDKs) is involved in the transition to cytokinesis in plants. Proc. Natl. Acad. Sci. USA 2011, 108, 17844–17849. [Google Scholar] [CrossRef] [Green Version]
- Fujita, M.; Fujita, Y.; Noutoshi, Y.; Takahashi, F.; Narusaka, Y.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Crosstalk between abiotic and biotic stress responses, a current view from the points of convergence in the stress signaling networks. Curr. Opin. Plant Biol. 2006, 9, 436–442. [Google Scholar] [CrossRef]
- Xing, Y.; Jia, W.; Zhang, J. AtMKK1 mediates ABA-induced CAT1 expression and H2O2 production via AtMPK6-coupled signaling in Arabidopsis. Plant J. 2008, 54, 440–451. [Google Scholar] [CrossRef]
- Zhang, H.; Zhu, H.; Pan, Y.; Yu, Y.; Luan, S.; Li, L. A DTX/MATE-type transporter facilitates abscisic acid efflux and modulates ABA sensitivity and drought tolerance in Arabidopsis. Mol. Plant 2014, 7, 1522–1532. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Zou, D.; Li, Y.; Sun, X.; Wang, N.N.; Gong, S.Y.; Zheng, Y.; Li, X.-B. GhMPK17, a cotton mitogen-activated protein kinase, is involved in plant response to high salinity and osmotic stresses and ABA signaling. PLoS ONE 2014, 9, e95642. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Cai, H.; Liu, P.; Wang, C.; Gao, H.; Wu, C.; Yan, K.; Zhang, S.; Huang, J.; Zheng, C. Arabidopsis MAPKKK18 positively regulates drought stress resistance via downstream MAPKK3. Biochem. Biophys. Res. Commun. 2017, 484, 292–297. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Yang, F.; Zhang, G.; Song, S.; Li, Y.; Ren, D.; Miao, Y.; Song, C.-P. AIK1, a mitogen-activated protein kinase, modulates abscisic acid responses through the MKK5-MPK6 kinase cascade. Plant Physiol. 2017, 173, 1391–1408. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Q.; Wang, Y.; Liu, C.; Pei, Z.M.; Shi, W.-L. The crosstalk between ABA, nitric oxide, hydrogen peroxide, and calcium in stomatal closing of Arabidopsis thaliana. Biologia 2017, 72, 1140–1146. [Google Scholar] [CrossRef]
- MAPK Group. Mitogen-activated protein kinase cascades in plants: A new nomenclature. Trends Plant Sci. 2002, 7, 301–308. [Google Scholar] [CrossRef]
- Ortiz-Masia, D.; Perez-Amador, M.A.; Carbonell, J.; Marcote, M.J. Diverse stress signals activate the C1 subgroup MAP kinases of Arabidopsis. FEBS Lett. 2007, 581, 1834–1840. [Google Scholar] [CrossRef] [Green Version]
- Kobayashi, Y.; Yamamoto, S.; Minami, H.; Kagaya, Y.; Hattori, T. Differential activation of the rice sucrose nonfermenting 1- related protein kinase 2 family by hyperosmotic stress and abscisic acid. Plant Cell 2004, 16, 1163–1177. [Google Scholar] [CrossRef] [Green Version]
- Mustilli, A.C.; Merlot, S.; Vavasseurb, A.; Fenzia, F.; Giraudat, J. Arabidopsis OST1 protein kinase mediates the regulation of stomatal aperture by abscisic acid and acts upstream of reactive oxygen species production. Plant Cell 2002, 14, 3089–3099. [Google Scholar] [CrossRef] [Green Version]
- Yoshida, R.; Hobo, T.; Ichimura, K.; Mizoguchi, T.; Takahashi, F.; Aronso, J.; Ecker, J.R.; Shinozaki, K. ABA-activated SnRK2 protein kinase is required for dehydration stress signaling in Arabidopsis. Plant Cell Physiol. 2002, 43, 1473–1483. [Google Scholar] [CrossRef]
- Hrabak, E.M.; Chan, C.W.M.; Gribskov, M.; Harper, J.F.; Choi, J.H.; Halford, N.; Kudla, J.; Luan, S.; Nimmo, H.G.; Sussman, M.R.; et al. The Arabidopsis CDPK-SnRK superfamily of protein kinases. Plant Physiol. 2003, 132, 666–680. [Google Scholar] [CrossRef] [Green Version]
- Boudsocq, M.; Barbier-Brygoo, H.; Laurière, C. Identification of nine sucrose nonfermenting 1-related protein kinases 2 activated by hyperosmotic and saline stresses in Arabidopsis thaliana. J. Biol. Chem. 2004, 279, 41758–41766. [Google Scholar] [CrossRef] [Green Version]
- Yoshida, R.; Umezawa, T.; Mizoguchi, T.; Takahashi, S.; Takahashi, F.; Shinozaki, K. The regulatory domain of SRK2E/OST1/SnRK2.6 interacts with ABI1 and integrates abscisic acid (ABA) and osmotic stress signals controlling stomatal closure in Arabidopsis. J. Biol. Chem. 2006, 281, 5310–5318. [Google Scholar] [CrossRef] [Green Version]
- Righetti, P.G.; Castagna, A.; Herbert, B.; Candiano, G. How to bring the “unseen” proteome to the limelight via electrophoretic pre-fractionation techniques. Biosci. Rep. 2005, 25, 3–17. [Google Scholar] [CrossRef] [PubMed]
- Taj, G.; Agarwal, P.; Kumar, A. In-silico approaches for studying cross-talk of different kinases associated in diverse biological processes with their interacting substrates partners. J. Proteomics Bioinform. 2011, 4, 91–97. [Google Scholar] [CrossRef] [Green Version]
- Kosetsu, K.; Matsunaga, S.; Nakagami, H.; Colcombet, J.; Sasabe, M.; Soyano, T.; Takahashi, Y.; Hirt, H.; Machida, Y. The MAP kinase MPK4 is required for cytokinesis in Arabidopsis thaliana. Plant Cell 2010, 22, 3778–3790. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Di Laurenzio, L.; Wysocka-Diller, J.; Malamy, J.E.; Pysh, L.; Helariutta, Y.; Freshour, G.; Hahn, M.G.; Feldmann, K.A.; Benfey, P.N. The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell 1996, 86, 423–433. [Google Scholar] [CrossRef] [Green Version]
- Wysocka-Diller, J.; Helariutta, Y.; Hukaki, H.; Malamy, J.; Benfey, P.N. Molecular analysis of SCARECROW functions reveals a radial patterning mechanism common to root and shoot. Development 2000, 127, 595–603. [Google Scholar]
- Heidstra, R.; Welch, D.; Scheres, B. Mosaic analysis using marked activation and deletion clones dissect Arabidopsis SCARECROW action in asymmetric cell division. Genes Dev. 2004, 18, 1964–1969. [Google Scholar] [CrossRef] [Green Version]
- Nakai, T.; Kato, K.; Shinmyo, A.; Sekine, M. Arabidopsis KRPs have distinct inhibitory activity toward cyclin D2-associated kinases, including plant-specific B-type cyclin-dependent kinase. FEBS Lett. 2006, 580, 336–340. [Google Scholar] [CrossRef] [Green Version]
- Verkest, A.; Manes, C.L.D.; Vercruysse, S.; Maes, S.; Van der Schueren, E.; Beekman, R.T.; Genschik, P.; Kuiper, M.; Inze, D.; De Veylder, I. The cyclin-dependent kinase inhibitor KRP2 controls the onset of the endoreduplication cycle during Arabidopsis leaf development through inhibition of mitotic CDKA;1 kinase complexes. Plant Cell 2005, 17, 1723–1736. [Google Scholar] [CrossRef] [Green Version]
- Schnittger, A.; Weinl, C.; Bouyer, D.; Schöbinger, U.; Hülskamp, M. Misexpression of the cyclin-dependent kinase inhibitor ICK1/KRP1 in single-celled Arabidopsis trichomes reduces endoreduplication and cell size and indices cell death. Plant Cell 2003, 15, 303–315. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Menges, M.; de Jager, S.M.; Gruissem, W.; Murray, J.A.H. Global analysis of the core cell cycle regulators of Arabidopsis identifies novel genes, reveals multiple and highly specific profiles of expression and provides a coherent model for plant cell cycle control. Plant J. 2005, 41, 546–566. [Google Scholar] [CrossRef] [PubMed]
- Ormenese, S.; de Almeida Engler, J.; De Groodt, R.; De Veylder, L.; Inzé, D.; Jacqmard, A. Analysis of the spatial expression pattern of seven Kip related proteins (KRPs) in the shoot apex of Arabidopsis thaliana. Ann. Bot. 2004, 93, 575–580. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neufeld, B.; Grosse-Wilde, A.; Hoffmeyer, A.; Jordan, B.W.; Chen, P.; Dinev, D.; Ludwig, S.; Rapp, U.R. Serine/Threonine kinases 3pK and MAPK-activated protein kinase 2 interact with the basic helix-loop-helix factor E47 and repress its transcriptional activity. J. Biol. Chem. 2000, 275, 20239–20242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manke, I.A.; Nguyen, A.; Lim, D.; Stewart, M.Q.; Elia, A.E.H.; Yaffe, M.B. MAPKAP kinase-2 is a cell cycle checkpoint kinase that regulates the G2/M transition and S phase progression in response to UV irradiation. Mol. Cell 2005, 17, 37–48. [Google Scholar] [CrossRef] [PubMed]
- Menges, M.; Hennig, L.; Gruissem, W.; Murray, J.A.H. Cell cycle-regulated gene expression in Arabidopsis. J. Biol. Chem. 2002, 277, 41987–42002. [Google Scholar] [CrossRef] [Green Version]
- Chiwocha, S.D.S.; Cutler, A.J.; Abrams, S.R.; Ambrose, S.J.; Yang, J.; Ross, A.R.S.; Kermode, A.R. The etr1-2 mutation in Arabidopsis thaliana affects the abscisic acid, auxin, cytokinin and gibberellin metabolic pathways during maintenance of seed dormancy, moist-chilling and germination. Plant J. 2005, 42, 35–48. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, Y.; Sano, T.; Tamaoki, M.; Nakajima, N.; Kondo, N.; Hasezawa, S. Ethylene inhibits abscisic acid-induced stomatal closure in Arabidopsis. Plant Physiol. 2005, 138, 2337–2343. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, Y.; Sano, T.; Tamaoki, M.; Nakajima, N.; Kondo, N.; Hasezawa, S. Cytokinin and auxin inhibit abscisic acid-induced stomatal closure by enhancing ethylene production in Arabidopsis. J. Exp. Bot. 2006, 57, 2259–2266. [Google Scholar] [CrossRef]
- Sharp, R.E.; LeNoble, M.E. ABA, Ethylene and the control of shoot and root growth under water stress. J. Exp. Bot. 2002, 53, 33–37. [Google Scholar] [CrossRef]
- Spollen, W.G.; LeNoble, M.E.; Samuels, T.D.; Bernstein, N.; Sharp, R.E. Abscisic acid accumulation maintains maize primary root elongation at low water potentials by restricting ethylene production. Plant Physiol. 2000, 122, 967–976. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schweighofer, A.; Kazanaviciute, V.; Scheikl, E.; Teige, M.; Doczi, R.; Hirt, H.; Schwanninger, M.; Kant, M.; Schuurink, R.; Mauch, F.; et al. The PP2C-type phosphatase AP2C1, which negatively regulates MPK4 and MPK6, modulates innate immunity, jasmonic acid, and ethylene levels in Arabidopsis. Plant Cell 2007, 19, 2213–2224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
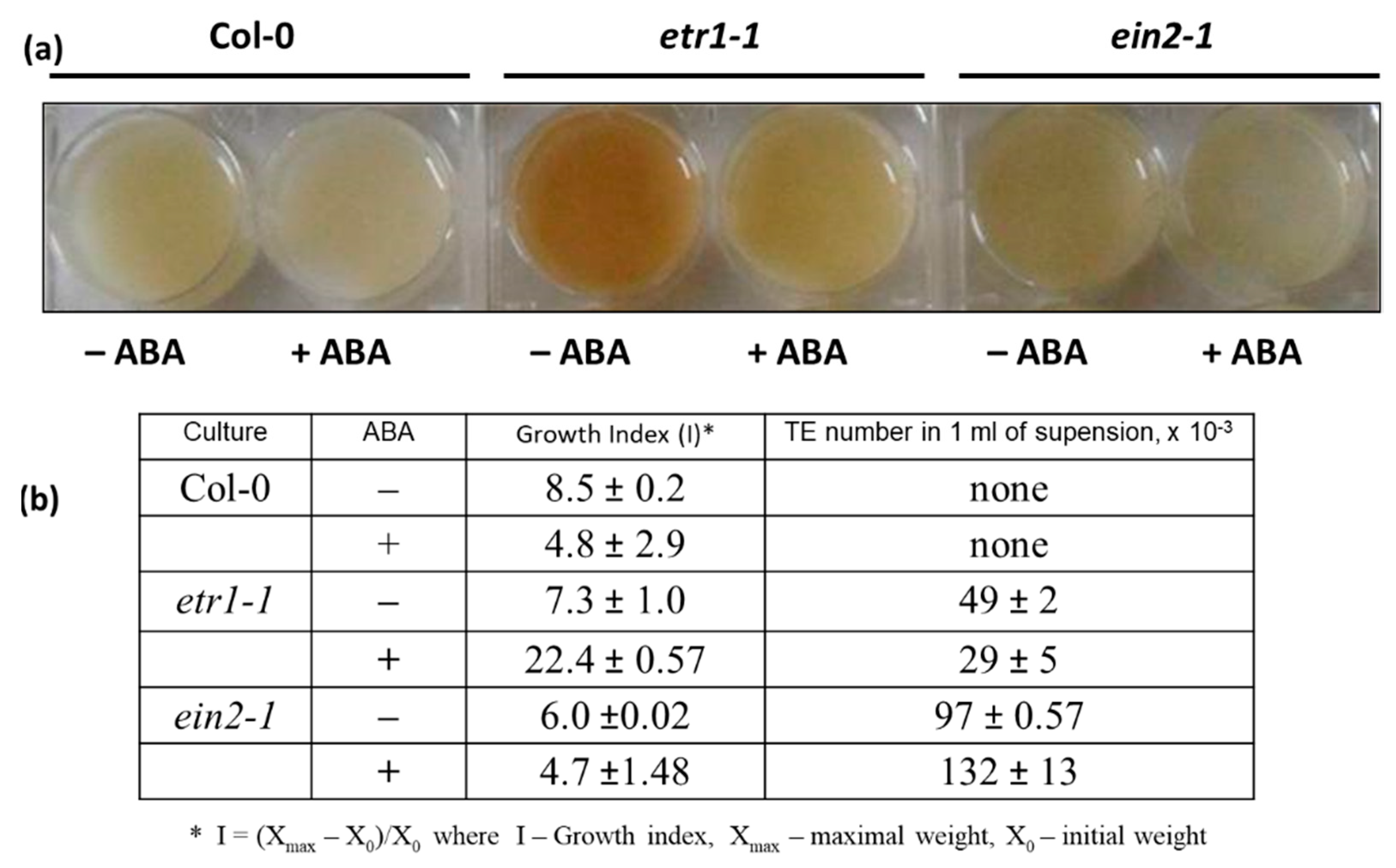
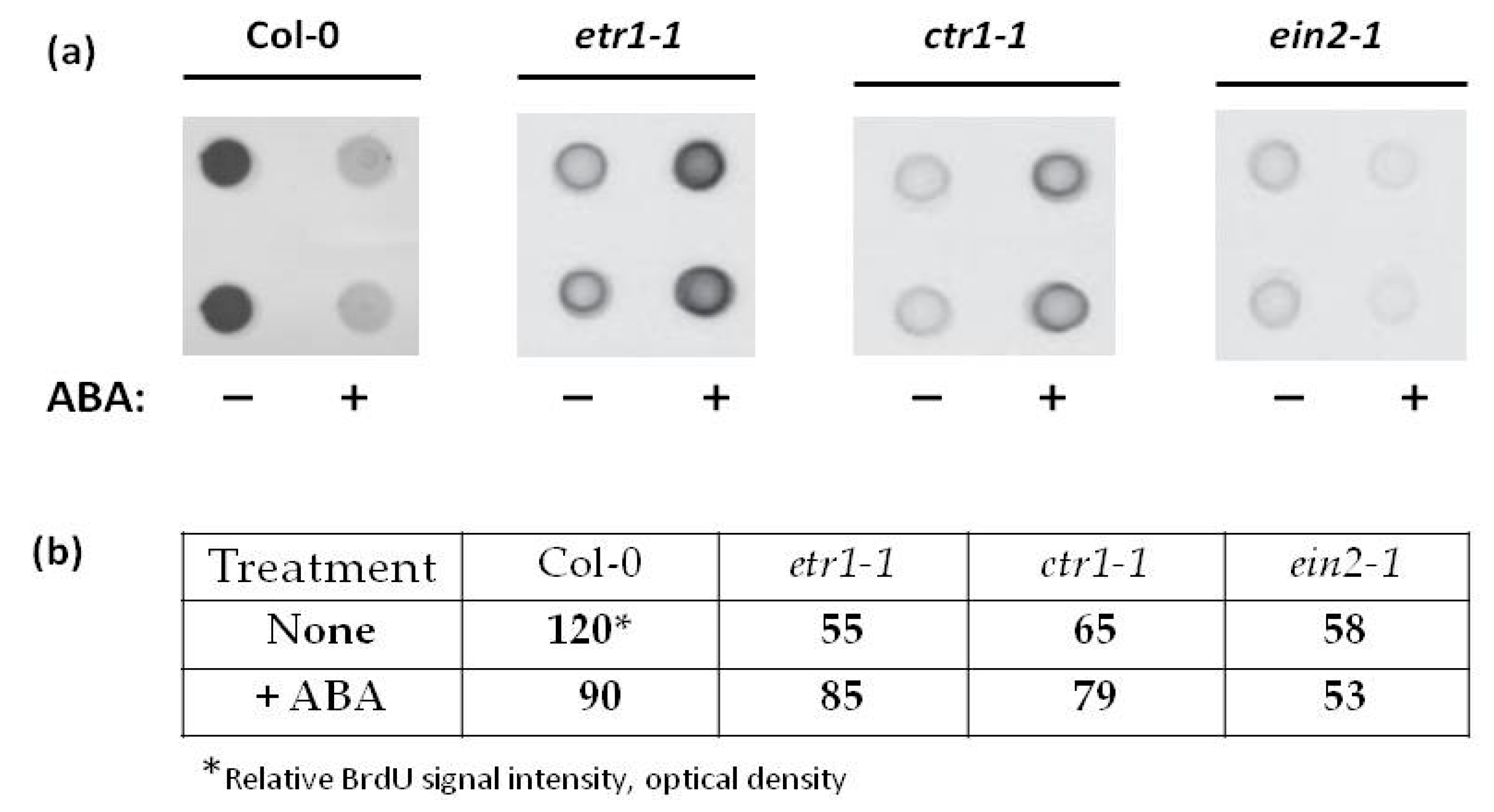
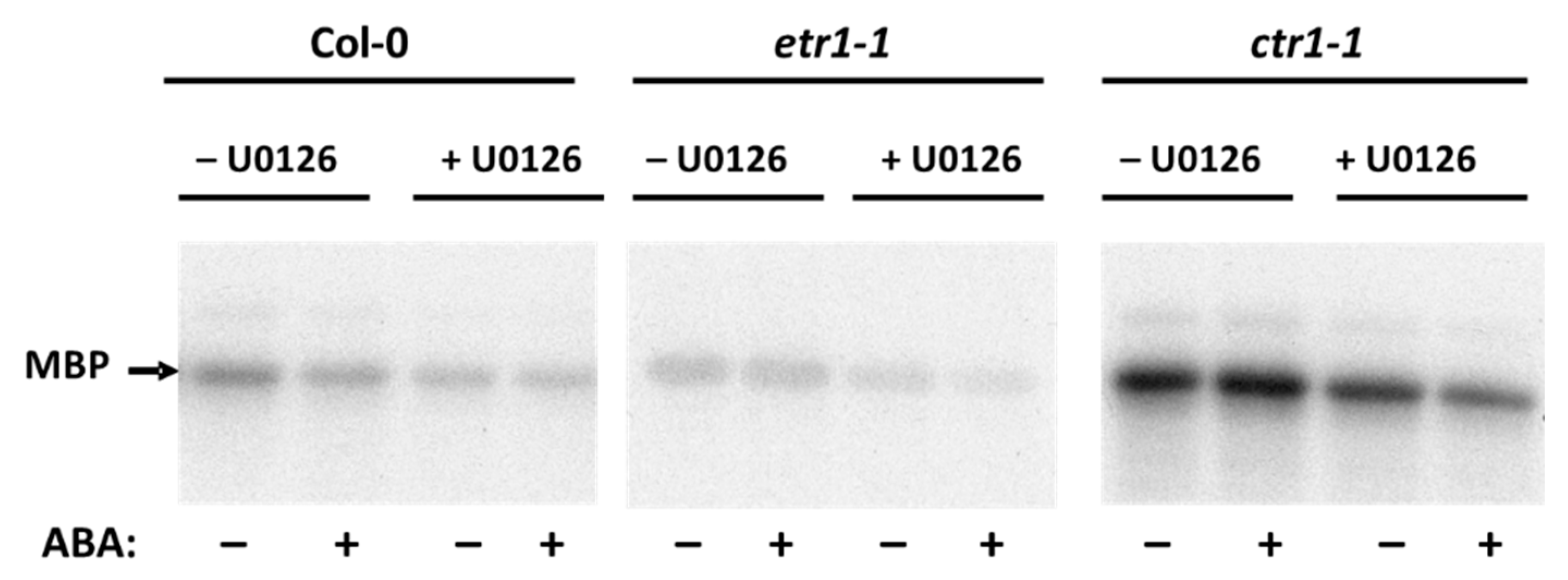
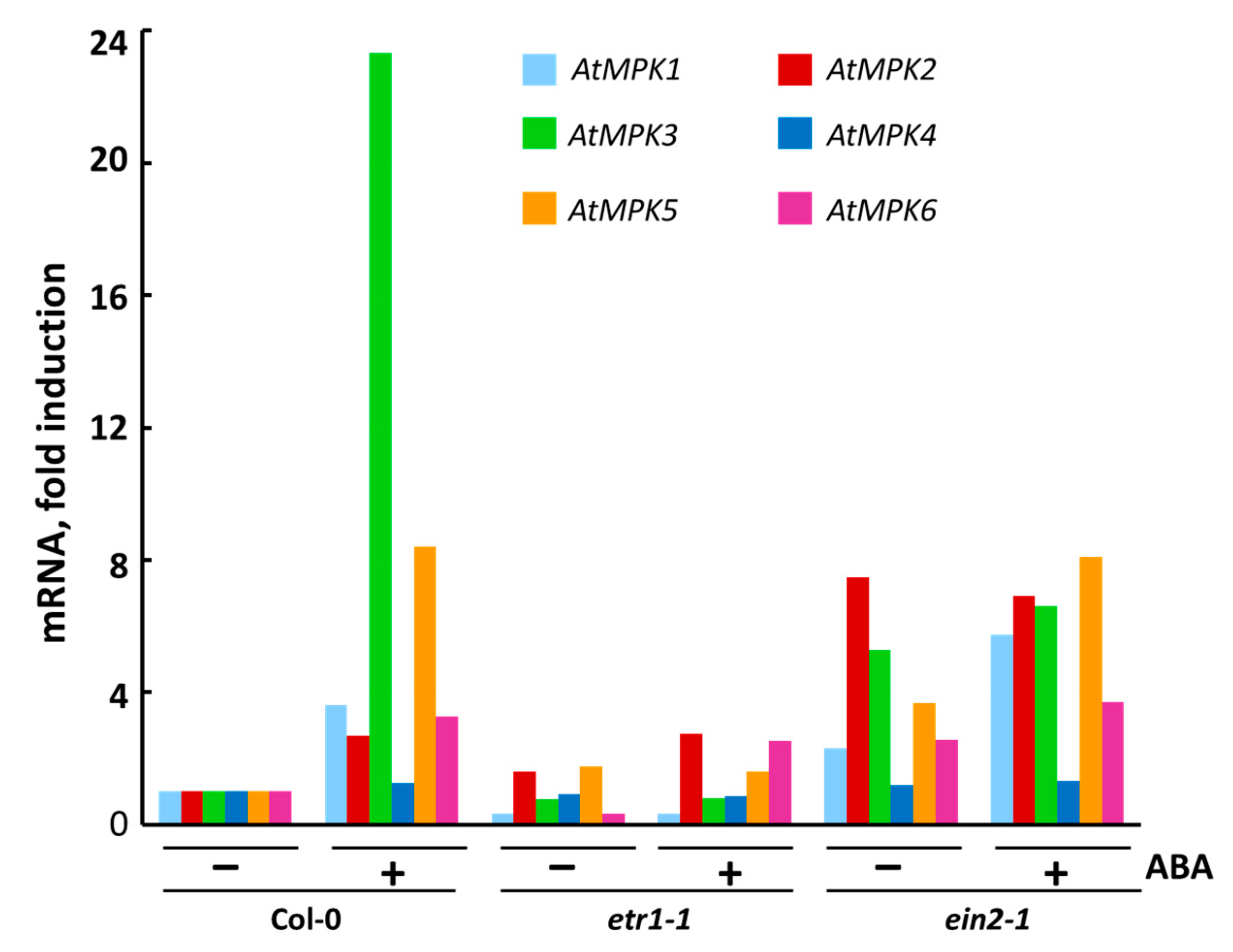
| Days | Col-0 | etr1-1 | ein2-1 |
|---|---|---|---|
| 2 | 4.3 ± 0.21 | 2.9 ± 0.15 | 2.4 ± 0.12 |
| 4 | 4.0 ± 0.20 | 3.5 ± 0.18 | 2.6 ± 0.13 |
| 6 | 3.6 ± 0.18 | 4.4 ± 0.22 | 3.0 ± 0.15 |
| 9 | 3.3 ± 0.16 | 5.6 ± 0.28 | 4.8 ± 0.24 |
| Cultivation, Days | ABA | Ethylene Evolution, nL g−1 FW h−1 | ||
|---|---|---|---|---|
| Col-0 | etr1-1 | ein2-1 | ||
| 4 | − | 595 ± 15 | 281 ± 4 | 18 ± 3 |
| + | 482 ± 27 | 288 ± 18 | 31 ± 1 | |
| 6 | − | 323 ± 7 | 268 ± 1 | 12 ± 2 |
| + | 464 ± 7 | 266 ± 23 | 22 ± 1 | |
| 10 | − | 34 ± 5 | 82 ± 45 | 2.7 ± 0.04 |
| + | 238 ± 22 | 66 ± 27 | 3.5 ± 0.25 | |
| Culture | Untreated | ABA Treated |
|---|---|---|
| Col-0 | 280 ± 6 | 250 ± 12 |
| etr1-1 | 160 ± 6 | 150 ± 8 |
| ctr1-1 | 130 ± 8 | 200 ± 20 |
| Protein | pI | Mr, Da |
|---|---|---|
| NADP-dependent oxidoreductase (spot 1) | 5.8 | 38133 |
| Annexin (ANNAT2) (spot 2) | 5.76 | 36243 |
| Aldo/keto reductase (spot 3) | 5.92 | 37900 |
| Pyrabactin resistance-like 8 (PYL8) (spot 4) | 6.07 | 21397 |
| Glyceraldehyde-3-phophate-dehydrogenase (spot 5) | 6.67 | 36913 |
| Kip-related protein 4 (KRP4) (spot 6) | 9.61 | 31699 |
| Enolase (spot 7) | 5.79 | 51477 |
| ATP synthase β-subunit (spot 8) | 6.18 | 59713 |
| Bifunctional enolase (spot 9) | 5.54 | 47689 |
| Translation initiation factor 4A (eF4A), (spot 10) | 5.47 | 46674 |
| Mitogen-activated protein kinase11 (MPK11) (spot 11) | 5.54 | 47689 |
| Scarecrow-like 9 (SCL9) (spot 12) | 5.66 | 53344 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Novikova, G.V.; Stepanchenko, N.S.; Zorina, A.A.; Nosov, A.V.; Rakitin, V.Y.; Moshkov, I.E.; Los, D.A. Coupling of Cell Division and Differentiation in Arabidopsis thaliana Cultured Cells with Interaction of Ethylene and ABA Signaling Pathways. Life 2020, 10, 15. https://doi.org/10.3390/life10020015
Novikova GV, Stepanchenko NS, Zorina AA, Nosov AV, Rakitin VY, Moshkov IE, Los DA. Coupling of Cell Division and Differentiation in Arabidopsis thaliana Cultured Cells with Interaction of Ethylene and ABA Signaling Pathways. Life. 2020; 10(2):15. https://doi.org/10.3390/life10020015
Chicago/Turabian StyleNovikova, Galina V., Natalia S. Stepanchenko, Anna A. Zorina, Alexander V. Nosov, Victor Y. Rakitin, Igor E. Moshkov, and Dmitry A. Los. 2020. "Coupling of Cell Division and Differentiation in Arabidopsis thaliana Cultured Cells with Interaction of Ethylene and ABA Signaling Pathways" Life 10, no. 2: 15. https://doi.org/10.3390/life10020015
APA StyleNovikova, G. V., Stepanchenko, N. S., Zorina, A. A., Nosov, A. V., Rakitin, V. Y., Moshkov, I. E., & Los, D. A. (2020). Coupling of Cell Division and Differentiation in Arabidopsis thaliana Cultured Cells with Interaction of Ethylene and ABA Signaling Pathways. Life, 10(2), 15. https://doi.org/10.3390/life10020015







