RNA Aptamers for a tRNA-Binding Protein from Aeropyrum pernix with Homologous Counterparts Distributed Throughout Evolution
Abstract
1. Introduction
2. Materials and Methods
2.1. Plasmid Construction for Expression of A. pernix Trbp
2.2. Expression and Purification of A. pernix Trbp
2.3. Preparation of RNA Pools for in Vitro Selection of Aptamers
2.4. A. pernix Trbp Binding Assay Using Nitrocellulose Filter
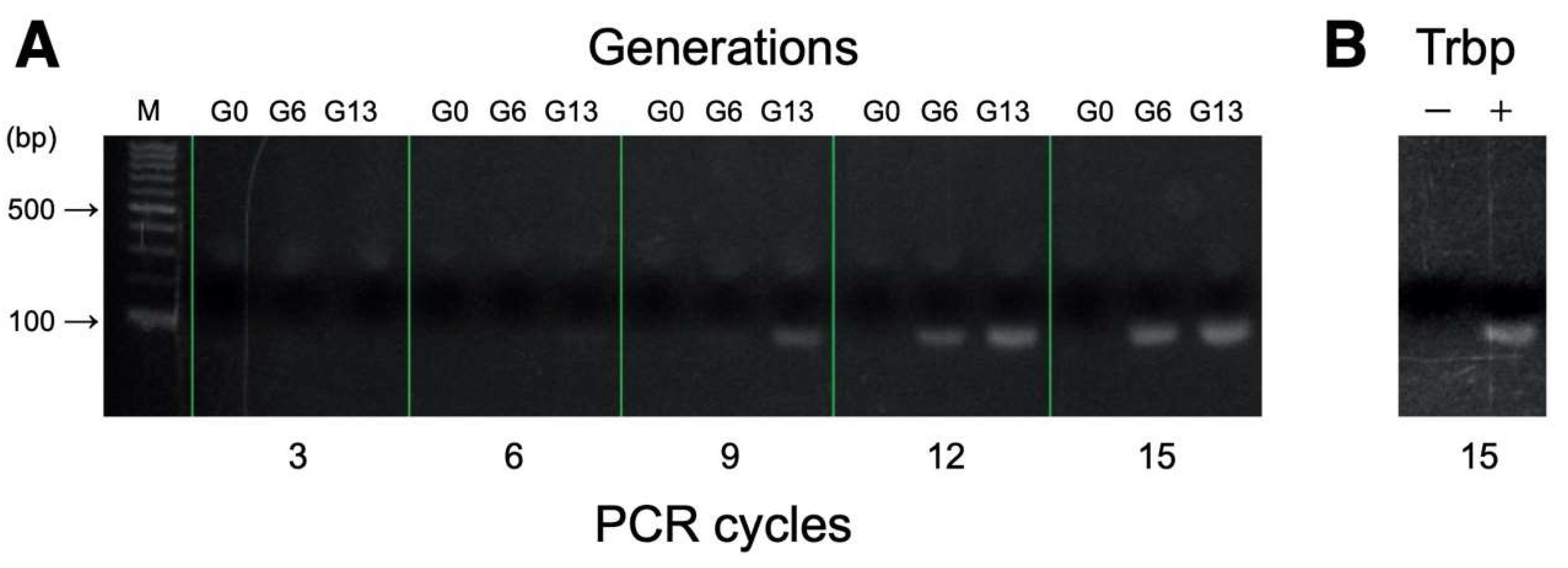
2.5. Binding Assessments of RNA Pools Using PCR
2.6. Electrophoretic Mobility Shift Assay (EMSA)
2.7. Selective 2’-Hydroxyl Acylation Analyzed by Primer Extension (SHAPE)
3. Results
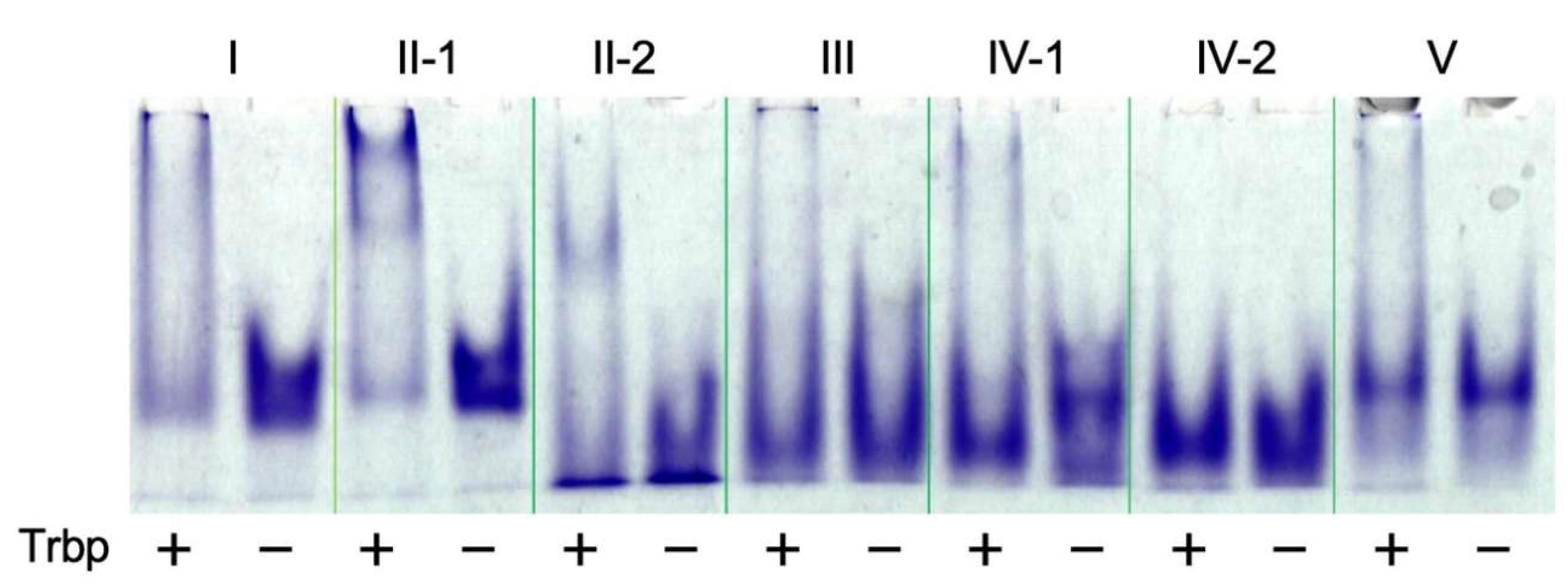
3.1. Selection of A. pernix Trbp Aptamers
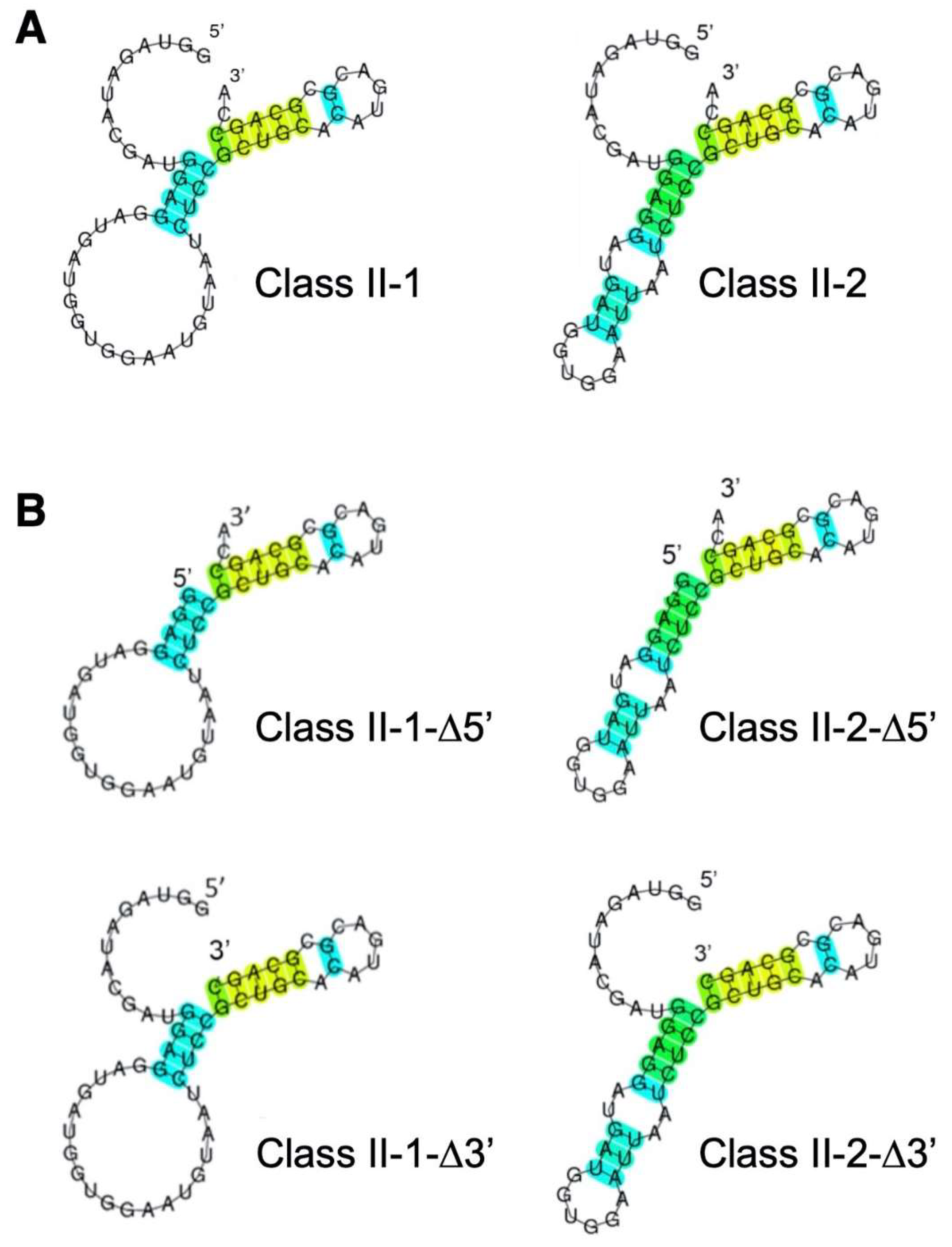
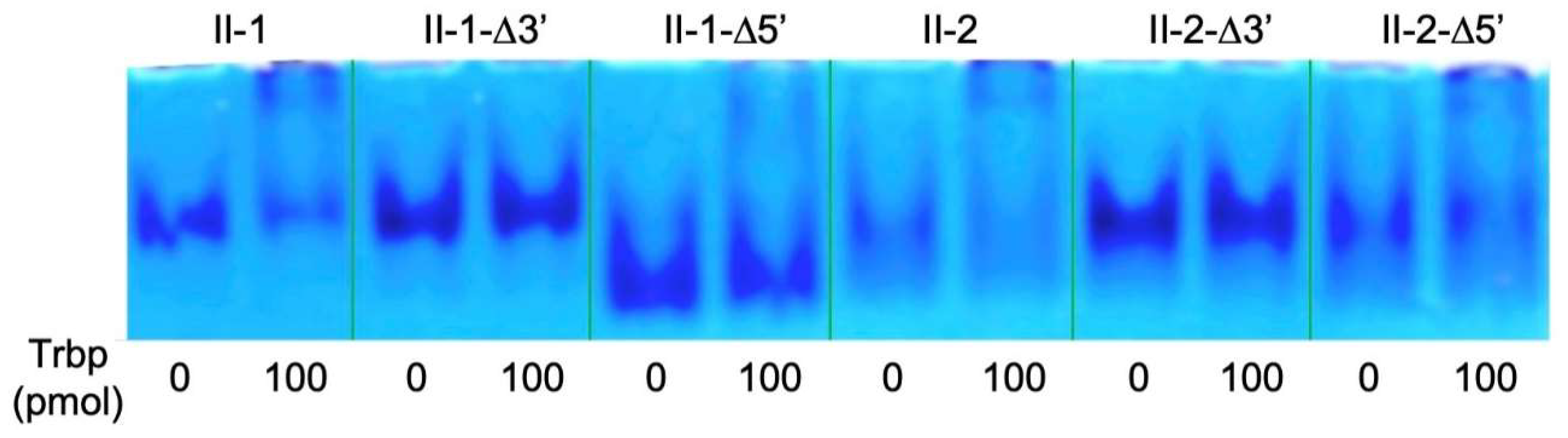
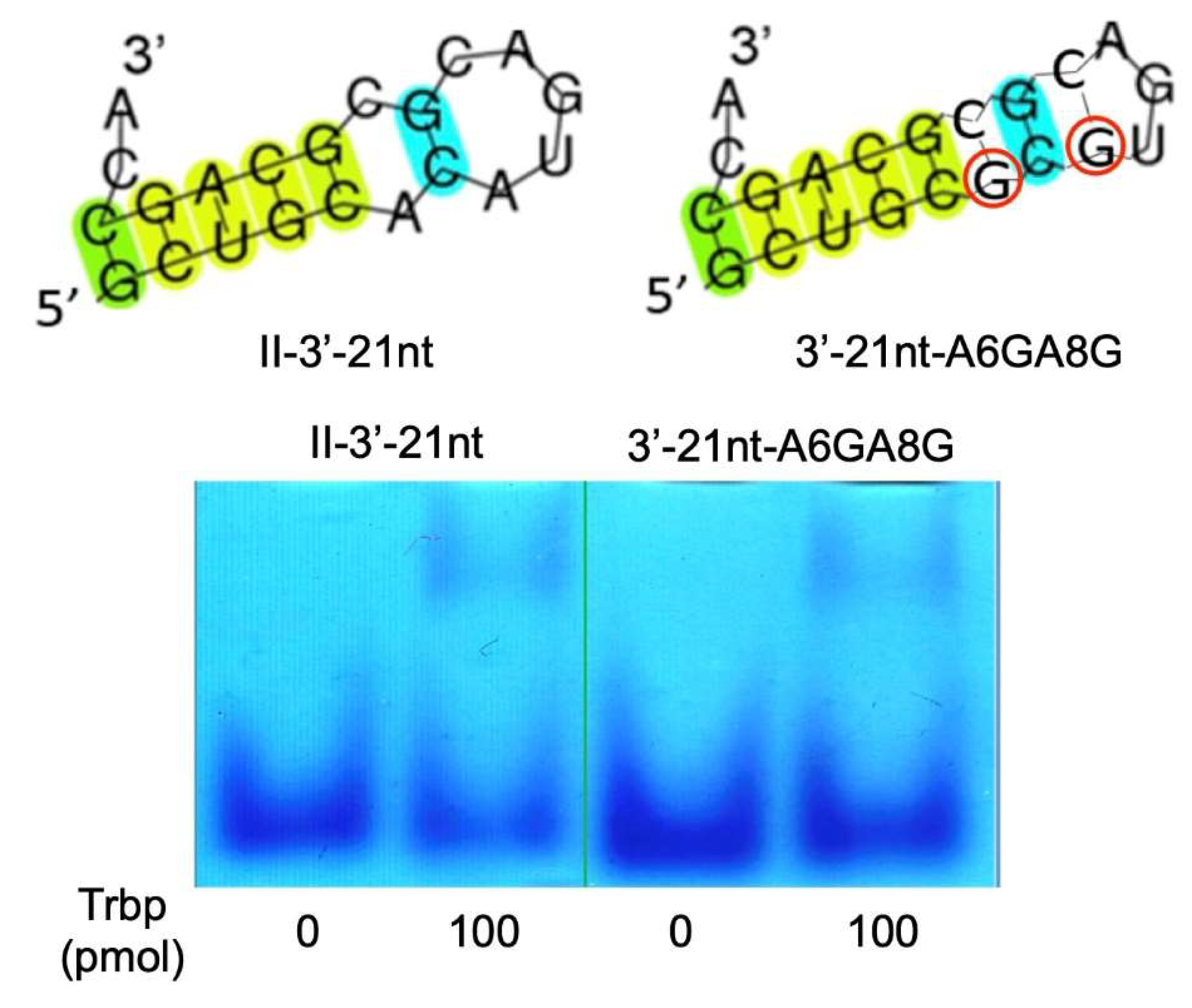
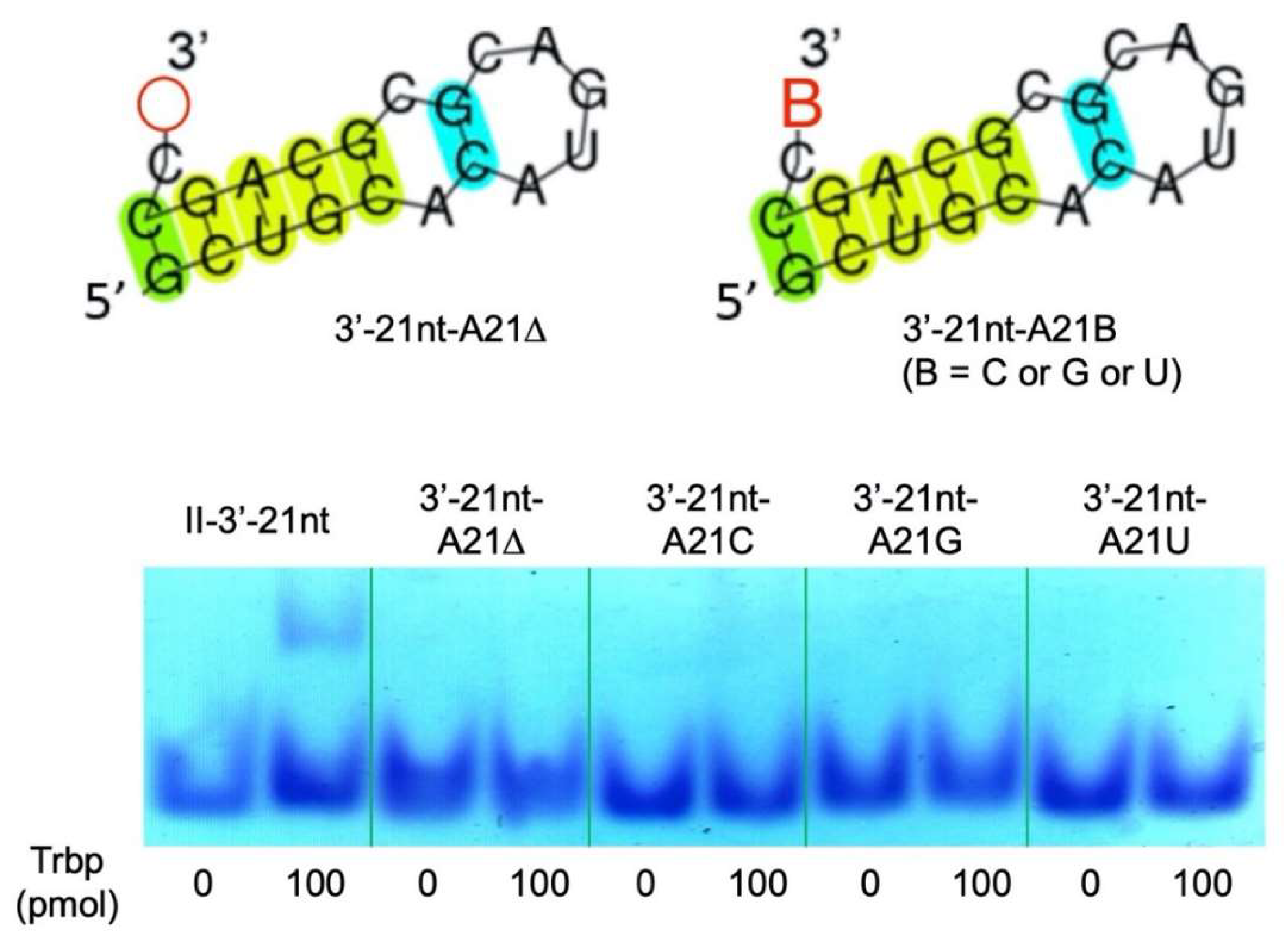
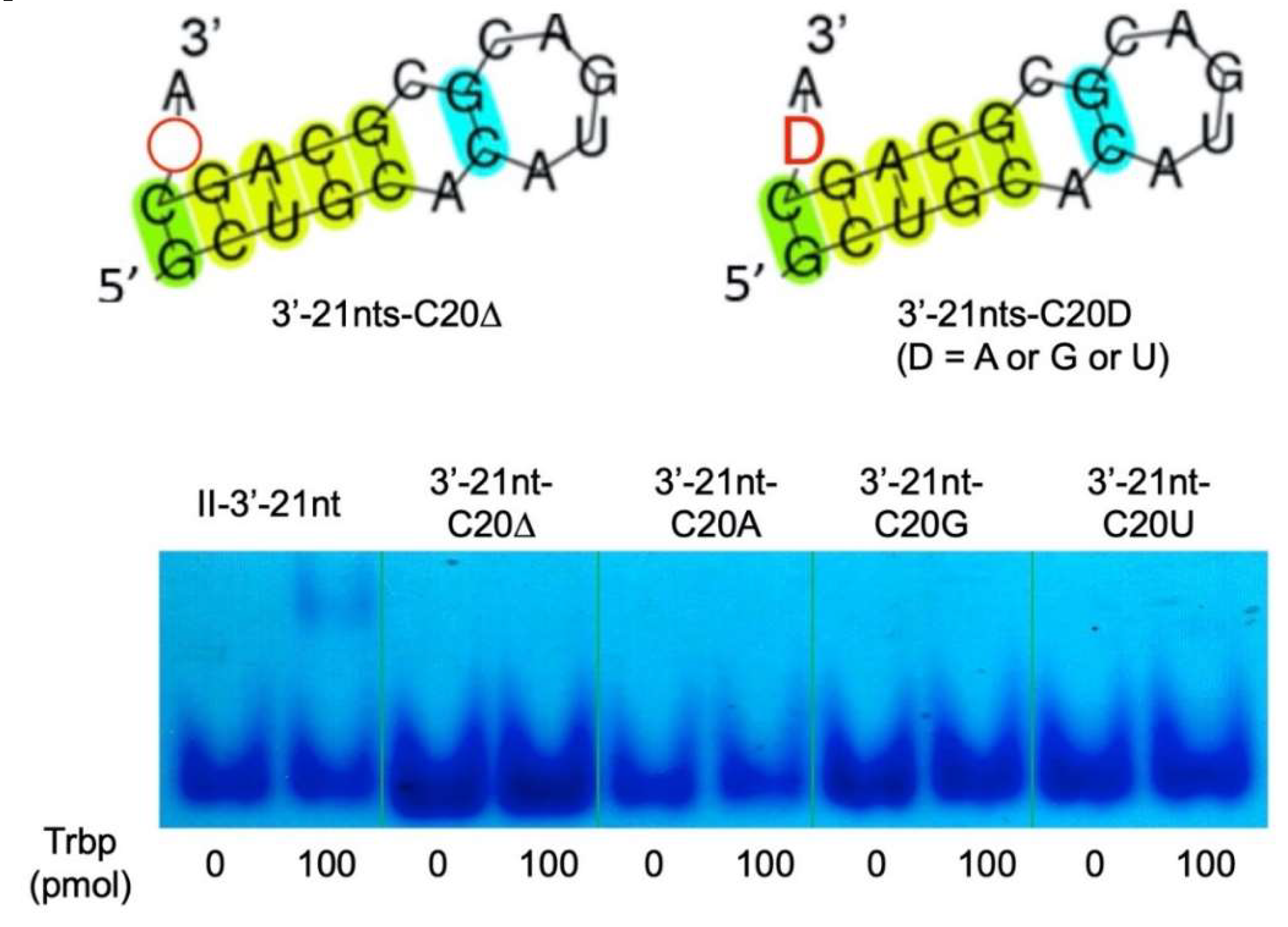
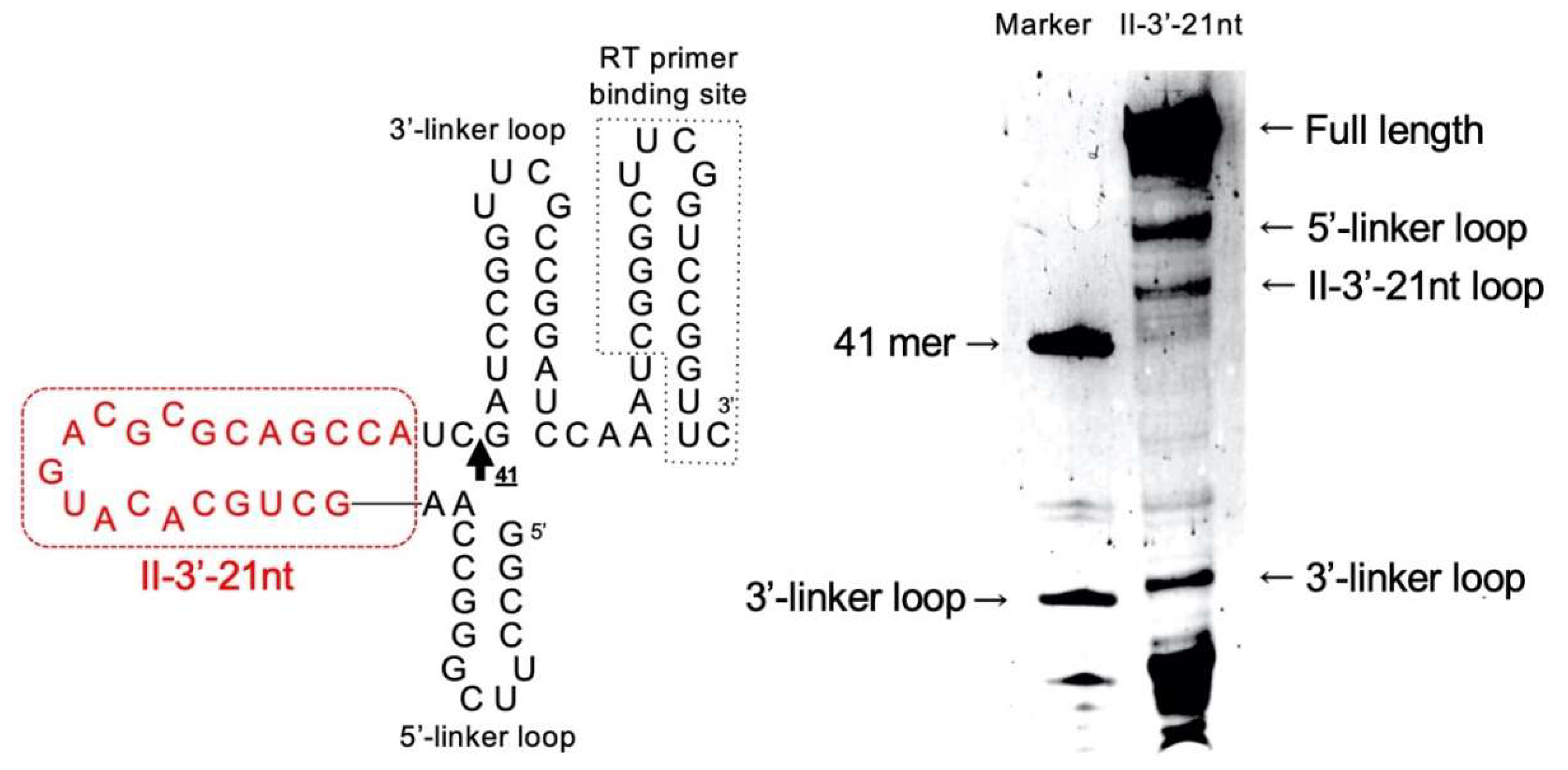
3.2. Reduction of Class II-1 and Class II-2 Aptamers Based on Their Secondary Structure
3.3. Secondary Structure of Class II-3′-21nt Analyzed by SHAPE
4. Discussion
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Morales, A.J.; Swairjo, M.A.; Schimmel, P. Structure-specific tRNA-binding protein from the extreme thermophile Aquifex aeolicus. EMBO J. 1999, 18, 3475–3483. [Google Scholar] [CrossRef]
- Woese, C.R.; Kandler, O.; Wheelis, M.L. Towards a natural system of organisms: Proposal for the domains Archaea, Bacteria, and Eucarya. Proc. Natl. Acad. Sci. USA 1990, 87, 4576–4579. [Google Scholar] [CrossRef]
- Swairjo, M.A.; Morales, A.J.; Wang, C.C.; Ortiz, A.R.; Schimmel, P. Crystal structure of Trbp111: A structure-specific tRNA-binding protein. EMBO J. 2000, 19, 6287–6298. [Google Scholar] [CrossRef]
- Kushiro, T.; Schimmel, P. Trbp111 selectively binds a noncovalently assembled tRNA-like structure. Proc. Natl. Acad. Sci. USA 2002, 99, 16631–16635. [Google Scholar] [CrossRef] [PubMed]
- Kao, J.; Ryan, J.; Brett, G.; Chen, J.; Shen, H.; Fan, Y.-G.; Godman, G.; Familletti, P.C.; Wang, F.; Pan, Y.-C.E.; et al. Endothelial monocyte-activating polypeptide II. A novel tumor-derived polypeptide that activates host-response mechanisms. J. Biol. Chem. 1992, 267, 20239–20247. [Google Scholar] [PubMed]
- Simos, G.; Segref, A.; Fasiolo, F.; Hellmuth, K.; Shevchenko, A.; Mann, M.; Hurt, E.C. The yeast protein Arc1p binds to tRNA and functions as a cofactor for the methionyl- and glutamyl-tRNA synthetases. EMBO J. 1996, 15, 5437–5448. [Google Scholar] [CrossRef] [PubMed]
- Simos, G.; Sauer, A.; Fasiolo, F.; Hurt, E.C. A conserved domain within Arc1p delivers tRNA to aminoacyl-tRNA synthetases. Mol. Cell 1998, 1, 235–242. [Google Scholar] [CrossRef]
- Quevillon, S.; Agou, F.; Robinson, J.C.; Mirande, M. The p43 component of the mammalian multi-synthetase complex is likely to be the precursor of the endothelial monocyte-activating polypeptide II cytokine. J. Biol. Chem. 1997, 272, 32573–32579. [Google Scholar] [CrossRef]
- Schimmel, P. Aminoacyl tRNA synthetases: General scheme of structure-function relationships in the polypeptides and recognition of transfer RNAs. Annu. Rev. Biochem. 1987, 56, 125–158. [Google Scholar] [CrossRef]
- Crepin, T.; Schmitt, E.; Blanquet, S.; Mechulam, Y. Structure and function of the C-terminal domain of methionyl-tRNA synthetase. Biochemistry 2002, 41, 13003–13011. [Google Scholar] [CrossRef]
- Suzuki, H.; Kaneko, A.; Yamamoto, T.; Nambo, M.; Hirasawa, I.; Umehara, T.; Yoshida, H.; Park, S.Y.; Tamura, K. Binding properties of split tRNA to the C-terminal domain of methionyl-tRNA synthetase of Nanoarchaeum equitans. J. Mol. Evol. 2017, 84, 267–278. [Google Scholar] [CrossRef] [PubMed]
- Kawarabayasi, Y.; Hino, Y.; Horikawa, H.; Yamazaki, S.; Haikawa, Y.; Jin-no, K.; Takahashi, M.; Sekine, M.; Baba, S.; Ankai, A.; et al. Complete genome sequence of an aerobic hyper-thermophilic crenarchaeon, Aeropyrum pernix K1. DNA Res. 1999, 6, 83–101. [Google Scholar] [CrossRef] [PubMed]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef]
- Kikuchi, K.; Umehara, T.; Fukuda, K.; Hwang, J.; Kuno, A.; Hasegawa, T.; Nishikawa, S. RNA aptamers targeted to domain II of hepatitis C virus IRES that bind to its apical loop region. J. Biochem. 2003, 133, 263–270. [Google Scholar] [CrossRef] [PubMed]
- Sampson, J.R.; Uhlenbeck, O.C. Biochemical and physical characterization of an unmodified yeast phenylalanine transfer RNA transcribed in vitro. Proc. Natl. Acad. Sci. USA 1988, 85, 1033–1037. [Google Scholar] [CrossRef] [PubMed]
- Hamachi, K.; Hayashi, H.; Shimamura, M.; Yamaji, Y.; Kaneko, A.; Fujisawa, A.; Umehara, T.; Tamura, K. Glycols modulate terminator stem stability and ligand-dependency of a glycine riboswitch. BioSystems 2013, 113, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Hamada, M.; Kiryu, H.; Sato, K.; Mituyama, T.; Asai, K. Prediction of RNA secondary structure using generalized centroid estimators. Bioinformatics 2009, 25, 465–473. [Google Scholar] [CrossRef]
- Sato, K.; Hamada, M.; Asai, K.; Mituyama, T. CENTROIDFOLD: A web server for RNA secondary structure prediction. Nucleic Acids Res. 2009, 37, W277–W280. [Google Scholar] [CrossRef]
- Takahashi, K.I.; Baba, S.; Chattopadhyay, P.; Koyanagi, Y.; Yamamoto, N.; Takaku, H.; Kawai, G. Structural requirement for the two-step dimerization of human immunodeficiency virus type 1 genome. RNA 2000, 6, 96–102. [Google Scholar] [CrossRef]
- Merino, E.J.; Wilkinson, K.A.; Coughlan, J.L.; Weeks, K.M. RNA structure analysis at single nucleotide resolution by selective 2′-hydroxyl acylation and primer extension (SHAPE). J. Am. Chem. Soc. 2005, 127, 4223–4231. [Google Scholar] [CrossRef]
- Kurihara, E.; Uchida, S.; Umehara, T.; Tamura, K. Development of a functionally minimized mutant of the R3C ligase ribozyme offers insight into the plausibility of the RNA world hypothesis. Biology 2014, 3, 452–465. [Google Scholar] [CrossRef] [PubMed]
- Tinoco Jr, I.; Borer, P.N.; Dengler, B.; Levin, M.D.; Uhlenbeck, O.C.; Crothers, D.M.; Bralla, J. Improved estimation of secondary structure in ribonucleic acids. Nat. New Biol. 1973, 246, 40–41. [Google Scholar] [CrossRef] [PubMed]
- Caetano-Anollés, G.; Kim, H.S.; Mittenthal, J.E. The origin of modern metabolic networks inferred from phylogenomic analysis of protein architecture. Proc. Natl. Acad. Sci. USA 2007, 104, 9358–9363. [Google Scholar] [CrossRef] [PubMed]
- Nomanbhoy, T.; Morales, A.J.; Abraham, A.T.; Vörtler, C.S.; Giegé, R.; Schimmel, P. Simultaneous binding of two proteins to opposite sides of a single transfer RNA. Nat. Struct. Biol. 2001, 8, 344–348. [Google Scholar] [CrossRef]
- Tamura, K.; Schimmel, P. Chiral-selective aminoacylation of an RNA minihelix. Science 2004, 305, 1253. [Google Scholar] [CrossRef]
- Tamura, K.; Schimmel, P.R. Chiral-selective aminoacylation of an RNA minihelix: Mechanistic features and chiral suppression. Proc. Natl. Acad. Sci. USA 2006, 103, 13750–13752. [Google Scholar] [CrossRef]
- Ando, T.; Takahashi, S.; Tamura, K. Principles of chemical geometry underlying chiral selectivity in RNA minihelix aminoacylation. Nucleic Acids Res. 2018, 46, 11144–11152. [Google Scholar] [CrossRef]
- Tamura, K. Perspectives on the origin of biological homochirality on Earth. J. Mol. Evol. 2019, 87, 143–146. [Google Scholar] [CrossRef]
- Di Giulio, M. On the origin of the transfer RNA molecule. J. Theor. Biol. 1992, 159, 199–214. [Google Scholar] [CrossRef]
- Schimmel, P.; Giegé, R.; Moras, D.; Yokoyama, S. An operational RNA code for amino acids and possible relationship to genetic code. Proc. Natl. Acad. Sci. USA 1993, 90, 8763–8768. [Google Scholar] [CrossRef]
- Schimmel, P.; Ribas de Pouplana, L. Transfer RNA: From minihelix to genetic code. Cell 1995, 81, 983–986. [Google Scholar] [CrossRef]
- Tamura, K. Origins and early evolution of the tRNA molecule. Life 2015, 5, 1687–1699. [Google Scholar] [CrossRef]


| Generation | A. pernix Trbp (pmol) | RNA (pmol) | Wash (mL × times) |
|---|---|---|---|
| 1 | 40 | 1000 | 0.5 × 3 |
| 2 | 10 | 100 | 0.5 × 3 |
| 3 | 1 | 10 | 0.5 × 3 |
| 4 | 1 | 10 | 0.5 × 3 |
| 5 | 0.5 | 5 | 0.5 × 3 |
| 6 | 0.3 | 3 | 0.5 × 3 |
| 7 | 0.3 | 3 | 0.5 × 3 |
| 8 | 1 | 1 | 5 × 1 |
| 9 | 1 | 1 | 5 × 1 |
| 10 | 1 | 1 | 5 × 1 |
| 11 | 1 | 1 | 5 × 1 |
| 12 | 1 | 1 | 5 × 1 |
| 13 | 1 | 1 | 5 × 1 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ohmori, S.; Wani, M.; Kitabatake, S.; Nakatsugawa, Y.; Ando, T.; Umehara, T.; Tamura, K. RNA Aptamers for a tRNA-Binding Protein from Aeropyrum pernix with Homologous Counterparts Distributed Throughout Evolution. Life 2020, 10, 11. https://doi.org/10.3390/life10020011
Ohmori S, Wani M, Kitabatake S, Nakatsugawa Y, Ando T, Umehara T, Tamura K. RNA Aptamers for a tRNA-Binding Protein from Aeropyrum pernix with Homologous Counterparts Distributed Throughout Evolution. Life. 2020; 10(2):11. https://doi.org/10.3390/life10020011
Chicago/Turabian StyleOhmori, Senri, Marina Wani, Saki Kitabatake, Yuka Nakatsugawa, Tadashi Ando, Takuya Umehara, and Koji Tamura. 2020. "RNA Aptamers for a tRNA-Binding Protein from Aeropyrum pernix with Homologous Counterparts Distributed Throughout Evolution" Life 10, no. 2: 11. https://doi.org/10.3390/life10020011
APA StyleOhmori, S., Wani, M., Kitabatake, S., Nakatsugawa, Y., Ando, T., Umehara, T., & Tamura, K. (2020). RNA Aptamers for a tRNA-Binding Protein from Aeropyrum pernix with Homologous Counterparts Distributed Throughout Evolution. Life, 10(2), 11. https://doi.org/10.3390/life10020011





