Virus-Like Particle Mediated CRISPR/Cas9 Delivery for Efficient and Safe Genome Editing
Abstract
1. An Introduction to Genome Editing Nucleases
2. Lentiviral Capsid Proteins and Their Functions
3. Using VLPs as Safe Gene Editing Delivery Vehicles
3.1. VLPs for Nuclease mRNA Delivery
3.2. Using VLPs for Protein and RNP Delivery
3.2.1. VLP Mediated Nuclease Delivery Using the Fusion Strategy
3.2.2. VLP Mediated Nuclease Delivery Using the ABP/Aptamer Interaction Strategy
3.3. Advantages and Disadvantages of Using VLPs for Delivering Genome Editing Endonucleases
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| DNA | Deoxyribonucleic Acid |
| cDNA | complementary DNA |
| RNA | Ribonucleic Acid |
| gRNA | guide RNA |
| sgRNA | single guide RNA |
| ZFN | Zinc Finger Endonuclease |
| TALEN | Transcription Activator-Like Effector Nuclease |
| CRISPR/Cas | Clustered Regularly Interspaced Short Palindromic Repeats/CRISPR-associated/CRISPR-associated |
| LV | Lentiviral Vector |
| AAV | Adeno-Associated virus-derived Vector |
| HIV-1 | Human Immunodeficiency Virus type 1 |
| Gag | Group-specific antigen |
| MA | Matrix protein |
| CA | Capsid protein |
| NC | Nucleocapsid protein |
| RNP | Ribonucleoprotein |
| INDEL | Insertion and Deletion |
| VLP | Virus-like particle |
References
- Woolf, T.M. Therapeutic repair of mutated nucleic acid sequences. Nat. Biotechnol. 1998, 16, 341–344. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Foden, J.A.; Khayter, C.; Maeder, M.L.; Reyon, D.; Joung, J.K.; Sander, J.D. High-frequency off-target mutagenesis induced by crispr-cas nucleases in human cells. Nat. Biotechnol. 2013, 31, 822–826. [Google Scholar] [CrossRef] [PubMed]
- Khalil, A.M. The genome editing revolution: Review. J. Genet. Eng. Biotechnol. 2020, 18, 68. [Google Scholar] [CrossRef]
- Yang, Z.; Blenner, M. Genome editing systems across yeast species. Curr. Opin. Biotechnol. 2020, 66, 255–266. [Google Scholar] [CrossRef]
- Pramanik, D.; Shelake, R.M.; Kim, M.J.; Kim, J.Y. Crispr-mediated engineering across the central dogma in plant biology for basic research and crop improvement. Mol. Plant 2020. [Google Scholar] [CrossRef]
- Kim, Y.G.; Cha, J.; Chandrasegaran, S. Hybrid restriction enzymes: Zinc finger fusions to fok i cleavage domain. Proc. Natl. Acad. Sci. USA 1996, 93, 1156. [Google Scholar] [CrossRef]
- Tebas, P.; Stein, D.; Tang, W.W.; Frank, I.; Wang, S.Q.; Lee, G.; Spratt, S.K.; Surosky, R.T.; Giedlin, M.A.; Nichol, G.; et al. Gene editing of ccr5 in autologous cd4 t cells of persons infected with hiv. N. Engl. J. Med. 2014, 370, 901–910. [Google Scholar] [CrossRef]
- Li, T.; Huang, S.; Jiang, W.Z.; Wright, D.; Spalding, M.H.; Weeks, D.P.; Yang, B. Tal nucleases (talns): Hybrid proteins composed of tal effectors and foki DNA-cleavage domain. Nucleic Acids Res. 2011, 39, 359–372. [Google Scholar] [CrossRef]
- Moscou, M.J.; Bogdanove, A.J. A simple cipher governs DNA recognition by tal effectors. Science 2009, 326, 1501. [Google Scholar] [CrossRef]
- Qasim, W.; Zhan, H.; Samarasinghe, S.; Adams, S.; Amrolia, P.; Stafford, S.; Butler, K.; Rivat, C.; Wright, G.; Somana, K.; et al. Molecular remission of infant b-all after infusion of universal talen gene-edited car t cells. Sci. Transl. Med. 2017, 9, aam9292. [Google Scholar] [CrossRef]
- Barrangou, R.; Fremaux, C.; Deveau, H.; Richards, M.; Boyaval, P.; Moineau, S.; Romero, D.A.; Horvath, P. Crispr provides acquired resistance against viruses in prokaryotes. Science 2007, 315, 1709–1712. [Google Scholar] [CrossRef] [PubMed]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-rna-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef] [PubMed]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A.; et al. Multiplex genome engineering using crispr/cas systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.W.; Kim, S.; Kim, J.M.; Kim, J.S. Targeted genome engineering in human cells with the cas9 rna-guided endonuclease. Nat. Biotechnol. 2013, 31, 230–232. [Google Scholar] [CrossRef] [PubMed]
- Mali, P.; Yang, L.; Esvelt, K.M.; Aach, J.; Guell, M.; DiCarlo, J.E.; Norville, J.E.; Church, G.M. Rna-guided human genome engineering via cas9. Science 2013, 339, 823–826. [Google Scholar] [CrossRef]
- Jinek, M.; East, A.; Cheng, A.; Lin, S.; Ma, E.; Doudna, J. Rna-programmed genome editing in human cells. Elife 2013, 2, e00471. [Google Scholar] [CrossRef]
- Wang, H.; Yang, H.; Shivalila, C.S.; Dawlaty, M.M.; Cheng, A.W.; Zhang, F.; Jaenisch, R. One-step generation of mice carrying mutations in multiple genes by crispr/cas-mediated genome engineering. Cell 2013, 153, 910–918. [Google Scholar] [CrossRef]
- Bortesi, L.; Fischer, R. The crispr/cas9 system for plant genome editing and beyond. Biotechnol. Adv. 2015, 33, 41–52. [Google Scholar] [CrossRef]
- Zhang, Z.T.; Jiménez-Bonilla, P.; Seo, S.O.; Lu, T.; Jin, Y.S.; Blaschek, H.P.; Wang, Y. Bacterial genome editing with crispr-cas9: Taking clostridium beijerinckii as an example. Methods Mol. Biol. 2018, 1772, 297–325. [Google Scholar]
- Qi, L.S.; Larson, M.H.; Gilbert, L.A.; Doudna, J.A.; Weissman, J.S.; Arkin, A.P.; Lim, W.A. Repurposing crispr as an rna-guided platform for sequence-specific control of gene expression. Cell 2013, 152, 1173–1183. [Google Scholar] [CrossRef]
- Gilbert, L.A.; Larson, M.H.; Morsut, L.; Liu, Z.; Brar, G.A.; Torres, S.E.; Stern-Ginossar, N.; Brandman, O.; Whitehead, E.H.; Doudna, J.A.; et al. Crispr-mediated modular rna-guided regulation of transcription in eukaryotes. Cell 2013, 154, 442–451. [Google Scholar] [CrossRef] [PubMed]
- Bikard, D.; Jiang, W.; Samai, P.; Hochschild, A.; Zhang, F.; Marraffini, L.A. Programmable repression and activation of bacterial gene expression using an engineered crispr-cas system. Nucleic Acids Res. 2013, 41, 7429–7437. [Google Scholar] [CrossRef] [PubMed]
- Nelles, D.A.; Fang, M.Y.; O’Connell, M.R.; Xu, J.L.; Markmiller, S.J.; Doudna, J.A.; Yeo, G.W. Programmable rna tracking in live cells with crispr/cas9. Cell 2016, 165, 488–496. [Google Scholar] [CrossRef]
- Chen, B.; Gilbert, L.A.; Cimini, B.A.; Schnitzbauer, J.; Zhang, W.; Li, G.W.; Park, J.; Blackburn, E.H.; Weissman, J.S.; Qi, L.S.; et al. Dynamic imaging of genomic loci in living human cells by an optimized crispr/cas system. Cell 2013, 155, 1479–1491. [Google Scholar] [CrossRef] [PubMed]
- Nelson, C.E.; Hakim, C.H.; Ousterout, D.G.; Thakore, P.I.; Moreb, E.A.; Castellanos Rivera, R.M.; Madhavan, S.; Pan, X.; Ran, F.A.; Yan, W.X.; et al. In vivo genome editing improves muscle function in a mouse model of duchenne muscular dystrophy. Science 2016, 351, 403–407. [Google Scholar] [CrossRef] [PubMed]
- Tabebordbar, M.; Zhu, K.; Cheng, J.K.W.; Chew, W.L.; Widrick, J.J.; Yan, W.X.; Maesner, C.; Wu, E.Y.; Xiao, R.; Ran, F.A.; et al. In vivo gene editing in dystrophic mouse muscle and muscle stem cells. Science 2016, 351, 407–411. [Google Scholar] [CrossRef] [PubMed]
- Long, C.; Amoasii, L.; Mireault, A.A.; McAnally, J.R.; Li, H.; Sanchez-Ortiz, E.; Bhattacharyya, S.; Shelton, J.M.; Bassel-Duby, R.; Olson, E.N. Postnatal genome editing partially restores dystrophin expression in a mouse model of muscular dystrophy. Science 2016, 351, 400–403. [Google Scholar] [CrossRef] [PubMed]
- Kleinstiver, B.P.; Prew, M.S.; Tsai, S.Q.; Topkar, V.V.; Nguyen, N.T.; Zheng, Z.; Gonzales, A.P.; Li, Z.; Peterson, R.T.; Yeh, J.R.; et al. Engineered crispr-cas9 nucleases with altered pam specificities. Nature 2015, 523, 481–485. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.H.; Miller, S.M.; Geurts, M.H.; Tang, W.; Chen, L.; Sun, N.; Zeina, C.M.; Gao, X.; Rees, H.A.; Lin, Z.; et al. Evolved cas9 variants with broad pam compatibility and high DNA specificity. Nature 2018, 556, 57–63. [Google Scholar] [CrossRef]
- Walton, R.T.; Christie, K.A.; Whittaker, M.N.; Kleinstiver, B.P. Unconstrained genome targeting with near-pamless engineered crispr-cas9 variants. Science 2020, 368, 290–296. [Google Scholar] [CrossRef]
- Cradick, T.J.; Fine, E.J.; Antico, C.J.; Bao, G. Crispr/cas9 systems targeting beta-globin and ccr5 genes have substantial off-target activity. Nucleic Acids Res. 2013, 41, 9584–9592. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Kim, D.; Cho, S.W.; Kim, J.; Kim, J.S. Highly efficient rna-guided genome editing in human cells via delivery of purified cas9 ribonucleoproteins. Genome Res. 2014, 24, 1012–1019. [Google Scholar] [CrossRef] [PubMed]
- Rees, H.A.; Komor, A.C.; Yeh, W.H.; Caetano-Lopes, J.; Warman, M.; Edge, A.S.B.; Liu, D.R. Improving the DNA specificity and applicability of base editing through protein engineering and protein delivery. Nat. Commun. 2017, 8, 15790. [Google Scholar] [CrossRef] [PubMed]
- Hsu, P.D.; Scott, D.A.; Weinstein, J.A.; Ran, F.A.; Konermann, S.; Agarwala, V.; Li, Y.; Fine, E.J.; Wu, X.; Shalem, O.; et al. DNA targeting specificity of rna-guided cas9 nucleases. Nat. Biotechnol. 2013, 31, 827–832. [Google Scholar] [CrossRef] [PubMed]
- Shalem, O.; Sanjana, N.E.; Hartenian, E.; Shi, X.; Scott, D.A.; Mikkelson, T.; Heckl, D.; Ebert, B.L.; Root, D.E.; Doench, J.G.; et al. Genome-scale crispr-cas9 knockout screening in human cells. Science 2014, 343, 84–87. [Google Scholar] [CrossRef] [PubMed]
- Nelson, C.E.; Wu, Y.; Gemberling, M.P.; Oliver, M.L.; Waller, M.A.; Bohning, J.D.; Robinson-Hamm, J.N.; Bulaklak, K.; Castellanos Rivera, R.M.; Collier, J.H.; et al. Long-term evaluation of aav-crispr genome editing for duchenne muscular dystrophy. Nat. Med. 2019, 25, 427–432. [Google Scholar] [CrossRef]
- Ramakrishna, S.; Kwaku Dad, A.B.; Beloor, J.; Gopalappa, R.; Lee, S.K.; Kim, H. Gene disruption by cell-penetrating peptide-mediated delivery of cas9 protein and guide rna. Genome Res. 2014, 24, 1020–1027. [Google Scholar] [CrossRef]
- Zuris, J.A.; Thompson, D.B.; Shu, Y.; Guilinger, J.P.; Bessen, J.L.; Hu, J.H.; Maeder, M.L.; Joung, J.K.; Chen, Z.Y.; Liu, D.R. Cationic lipid-mediated delivery of proteins enables efficient protein-based genome editing in vitro and in vivo. Nat. Biotechnol. 2015, 33, 73–80. [Google Scholar] [CrossRef]
- Mout, R.; Ray, M.; Yesilbag Tonga, G.; Lee, Y.W.; Tay, T.; Sasaki, K.; Rotello, V.M. Direct cytosolic delivery of crispr/cas9-ribonucleoprotein for efficient gene editing. Acs Nano 2017, 11, 2452–2458. [Google Scholar] [CrossRef]
- Mellott, A.J.; Forrest, M.L.; Detamore, M.S. Physical non-viral gene delivery methods for tissue engineering. Ann. Biomed. Eng. 2013, 41, 446–468. [Google Scholar] [CrossRef]
- Roos, W.H.; Ivanovska, I.L.; Evilevitch, A.; Wuite, G.J.L. Viral capsids: Mechanical characteristics, genome packaging and delivery mechanisms. Cell Mol. Life Sci. 2007, 64, 1484–1497. [Google Scholar] [CrossRef] [PubMed]
- Lidmar, J.; Mirny, L.; Nelson, D.R. Virus shapes and buckling transitions in spherical shells. Phys. Rev. E Stat. Nonlinear Soft Matter Phys. 2003, 68, 051910. [Google Scholar] [CrossRef] [PubMed]
- Vernizzi, G.; Olvera de la Cruz, M. Faceting ionic shells into icosahedra via electrostatics. Proc. Natl. Acad. Sci. USA 2007, 104, 18382–18386. [Google Scholar] [CrossRef] [PubMed]
- Durand, S.; Cimarelli, A. The inside out of lentiviral vectors. Viruses 2011, 3, 132–159. [Google Scholar] [CrossRef]
- Mock, U.; Riecken, K.; Berdien, B.; Qasim, W.; Chan, E.; Cathomen, T.; Fehse, B. Novel lentiviral vectors with mutated reverse transcriptase for mrna delivery of tale nucleases. Sci. Rep. 2014, 4, 6409. [Google Scholar] [CrossRef]
- Prel, A.; Caval, V.; Gayon, R.; Ravassard, P.; Duthoit, C.; Payen, E.; Maouche-Chretien, L.; Creneguy, A.; Nguyen, T.H.; Martin, N.; et al. Highly efficient in vitro and in vivo delivery of functional rnas using new versatile ms2-chimeric retrovirus-like particles. Mol. Methods Clin. Dev. 2015, 2, 15039. [Google Scholar] [CrossRef]
- Knopp, Y.; Geis, F.K.; Heckl, D.; Horn, S.; Neumann, T.; Kuehle, J.; Meyer, J.; Fehse, B.; Baum, C.; Morgan, M.; et al. Transient retrovirus-based crispr/cas9 all-in-one particles for efficient, targeted gene knockout. Mol. Ther. Nucleic Acids 2018, 13, 256–274. [Google Scholar] [CrossRef]
- Lu, B.; Javidi-Parsijani, P.; Makani, V.; Mehraein-Ghomi, F.; Sarhan, W.M.; Sun, D.; Yoo, K.W.; Atala, Z.P.; Lyu, P.; Atala, A. Delivering sacas9 mrna by lentivirus-like bionanoparticles for transient expression and efficient genome editing. Nucleic Acids Res. 2019, 47, e44. [Google Scholar] [CrossRef]
- Lindel, F.; Dodt, C.R.; Weidner, N.; Noll, M.; Bergemann, F.; Behrendt, R.; Fischer, S.; Dietrich, J.; Cartellieri, M.; Hamann, M.V.; et al. Trafo-crispr: Enhanced genome engineering by transient foamy virus vector-mediated delivery of crispr/cas9 components. Mol. Ther. Nucleic Acids 2019, 18, 708–726. [Google Scholar] [CrossRef]
- Cai, Y.; Bak, R.O.; Mikkelsen, J.G. Targeted genome editing by lentiviral protein transduction of zinc-finger and tal-effector nucleases. Elife 2014, 3, e01911. [Google Scholar] [CrossRef]
- Choi, J.G.; Dang, Y.; Abraham, S.; Ma, H.; Zhang, J.; Guo, H.; Cai, Y.; Mikkelsen, J.G.; Wu, H.; Shankar, P.; et al. Lentivirus pre-packed with cas9 protein for safer gene editing. Gene Ther. 2016, 23, 627–633. [Google Scholar] [CrossRef] [PubMed]
- Mangeot, P.E.; Risson, V.; Fusil, F.; Marnef, A.; Laurent, E.; Blin, J.; Mournetas, V.; Massourides, E.; Sohier, T.J.M.; Corbin, A.; et al. Genome editing in primary cells and in vivo using viral-derived nanoblades loaded with cas9-sgrna ribonucleoproteins. Nat. Commun. 2019, 10, 45. [Google Scholar] [CrossRef] [PubMed]
- Gee, P.; Lung, M.S.Y.; Okuzaki, Y.; Sasakawa, N.; Iguchi, T.; Makita, Y.; Hozumi, H.; Miura, Y.; Yang, L.F.; Iwasaki, M.; et al. Extracellular nanovesicles for packaging of crispr-cas9 protein and sgrna to induce therapeutic exon skipping. Nat. Commun. 2020, 11, 1334. [Google Scholar] [CrossRef] [PubMed]
- Indikova, I.; Indik, S. Highly efficient ’hit-and-run’ genome editing with unconcentrated lentivectors carrying vpr.Prot.Cas9 protein produced from rre-containing transcripts. Nucleic Acids Res. 2020, 48, 8178–8187. [Google Scholar] [CrossRef]
- Lyu, P.; Javidi-Parsijani, P.; Atala, A.; Lu, B. Delivering cas9/sgrna ribonucleoprotein (rnp) by lentiviral capsid-based bionanoparticles for efficient ’hit-and-run’ genome editing. Nucleic Acids Res. 2019, 47, e99. [Google Scholar] [CrossRef]
- Lu, Z.; Yao, X.; Lyu, P.; Yadav, M.; Yoo, K.; Atala, A.; Lu, B. Lentiviral capsid-mediated spcas9 ribonucleoprotein delivery for efficient and safe multiplex genome editing. Cris. J. 2021, in press. [Google Scholar]
- Lyu, P.; Lu, Z.; Cho, S.I.; Yadav, M.; Yoo, K.; Atala, A.; Kim, J.S.; Lu, B. Adenine base editor ribonucleoproteins delivered by lentivirus-like particles show high on-target base editing and undetectable rna off-target activities. Cris. J. 2021, in press. [Google Scholar]
- Payne, S. Chapter 36-family retroviridae. In Viruses; Payne, S., Ed.; Academic Press: Cambridge, MA, USA, 2017; pp. 287–301. [Google Scholar]
- Lingappa, J.R.; Reed, J.C.; Tanaka, M.; Chutiraka, K.; Robinson, B.A. How hiv-1 gag assembles in cells: Putting together pieces of the puzzle. Virus Res. 2014, 193, 89–107. [Google Scholar] [CrossRef]
- Briggs, J.A.; Simon, M.N.; Gross, I.; Krausslich, H.G.; Fuller, S.D.; Vogt, V.M.; Johnson, M.C. The stoichiometry of gag protein in hiv-1. Nat. Struct. Mol. Biol. 2004, 11, 672–675. [Google Scholar] [CrossRef]
- Cockrell, A.S.; Kafri, T. Gene delivery by lentivirus vectors. Mol. Biotechnol. 2007, 36, 184–204. [Google Scholar] [CrossRef]
- Parolin, C.; Sodroski, J. A defective hiv-1 vector for gene transfer to human lymphocytes. J. Mol. Med. (Berl.) 1995, 73, 279–288. [Google Scholar] [CrossRef] [PubMed]
- Kartikeyan, S.; Bharmal, R.N.; Tiwari, R.P.; Bisen, P.S. Hiv and Aids: Basic Elements and Priorities; Springer: Dordrecht, The Netherlands, 2007; p. XIV, 418. [Google Scholar]
- Spearman, P.; Wang, J.J.; Vander Heyden, N.; Ratner, L. Identification of human immunodeficiency virus type 1 gag protein domains essential to membrane binding and particle assembly. J. Virol. 1994, 68, 3232–3242. [Google Scholar] [CrossRef] [PubMed]
- Ganser-Pornillos, B.K.; von Schwedler, U.K.; Stray, K.M.; Aiken, C.; Sundquist, W.I. Assembly properties of the human immunodeficiency virus type 1 ca protein. J. Virol. 2004, 78, 2545–2552. [Google Scholar] [CrossRef] [PubMed]
- De Guzman, R.N.; Wu, Z.R.; Stalling, C.C.; Pappalardo, L.; Borer, P.N.; Summers, M.F. Structure of the hiv-1 nucleocapsid protein bound to the sl3 psi-rna recognition element. Science 1998, 279, 384–388. [Google Scholar] [CrossRef]
- Freed, E.O. Hiv-1 gag proteins: Diverse functions in the virus life cycle. Virology 1998, 251, 1–15. [Google Scholar] [CrossRef]
- Lu, K.; Heng, X.; Summers, M.F. Structural determinants and mechanism of hiv-1 genome packaging. J. Mol. Biol. 2011, 410, 609–633. [Google Scholar] [CrossRef]
- Grigorov, B.; Decimo, D.; Smagulova, F.; Pechoux, C.; Mougel, M.; Muriaux, D.; Darlix, J.L. Intracellular hiv-1 gag localization is impaired by mutations in the nucleocapsid zinc fingers. Retrovirology 2007, 4, 54. [Google Scholar] [CrossRef]
- Hoshikawa, N.; Kojima, A.; Yasuda, A.; Takayashiki, E.; Masuko, S.; Chiba, J.; Sata, T.; Kurata, T. Role of the gag and pol genes of human immunodeficiency virus in the morphogenesis and maturation of retrovirus-like particles expressed by recombinant vaccinia virus: An ultrastructural study. J. Gen. Virol. 1991, 72 Pt 10, 2509–2517. [Google Scholar] [CrossRef]
- Akkina, R.K.; Walton, R.M.; Chen, M.L.; Li, Q.X.; Planelles, V.; Chen, I.S. High-efficiency gene transfer into cd34+ cells with a human immunodeficiency virus type 1-based retroviral vector pseudotyped with vesicular stomatitis virus envelope glycoprotein g. J. Virol. 1996, 70, 2581–2585. [Google Scholar] [CrossRef]
- Naldini, L.; Blomer, U.; Gallay, P.; Ory, D.; Mulligan, R.; Gage, F.H.; Verma, I.M.; Trono, D. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science 1996, 272, 263–267. [Google Scholar] [CrossRef]
- Reiser, J.; Harmison, G.; Kluepfel-Stahl, S.; Brady, R.O.; Karlsson, S.; Schubert, M. Transduction of nondividing cells using pseudotyped defective high-titer hiv type 1 particles. Proc. Natl. Acad. Sci. USA 1996, 93, 15266–15271. [Google Scholar] [CrossRef] [PubMed]
- Mattei, S.; Flemming, A.; Anders-Osswein, M.; Krausslich, H.G.; Briggs, J.A.; Muller, B. Rna and nucleocapsid are dispensable for mature hiv-1 capsid assembly. J. Virol. 2015, 89, 9739–9747. [Google Scholar] [CrossRef] [PubMed]
- Ortinski, P.I.; O’Donovan, B.; Dong, X.; Kantor, B. Integrase-deficient lentiviral vector as an all-in-one platform for highly efficient crispr/cas9-mediated gene editing. Mol. Ther. Methods Clin. Dev. 2017, 5, 153–164. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Schokrpur, S.; Archang, M.; Hermann, K.; Sharrow, A.C.; Khanna, P.; Novak, J.; Signoretti, S.; Bhatt, R.S.; Knudsen, B.S.; et al. A non-integrating lentiviral approach overcomes cas9-induced immune rejection to establish an immunocompetent metastatic renal cancer model. Mol. Ther. Methods Clin. Dev. 2018, 9, 203–210. [Google Scholar] [CrossRef]
- Luis, A. The old and the new: Prospects for non-integrating lentiviral vector technology. Viruses 2020, 12, 1103. [Google Scholar]
- Fouts, D.E.; True, H.L.; Celander, D.W. Functional recognition of fragmented operator sites by r17/ms2 coat protein, a translational repressor. Nucleic Acids Res. 1997, 25, 4464–4473. [Google Scholar] [CrossRef][Green Version]
- Bertrand, E.; Chartrand, P.; Schaefer, M.; Shenoy, S.M.; Singer, R.H.; Long, R.M. Localization of ash1 mrna particles in living yeast. Mol. Cell 1998, 2, 437–445. [Google Scholar] [CrossRef]
- Wu, B.; Chao, J.A.; Singer, R.H. Fluorescence fluctuation spectroscopy enables quantitative imaging of single mrnas in living cells. Biophys. J. 2012, 102, 2936–2944. [Google Scholar] [CrossRef]
- Ma, H.; Tu, L.C.; Naseri, A.; Huisman, M.; Zhang, S.; Grunwald, D.; Pederson, T. Multiplexed labeling of genomic loci with dcas9 and engineered sgrnas using crisprainbow. Nat. Biotechnol. 2016, 34, 528–530. [Google Scholar] [CrossRef]
- Zalatan, J.G.; Lee, M.E.; Almeida, R.; Gilbert, L.A.; Whitehead, E.H.; La Russa, M.; Tsai, J.C.; Weissman, J.S.; Dueber, J.E.; Qi, L.S.; et al. Engineering complex synthetic transcriptional programs with crispr rna scaffolds. Cell 2015, 160, 339–350. [Google Scholar] [CrossRef]
- Muriaux, D.; Costes, S.; Nagashima, K.; Mirro, J.; Cho, E.; Lockett, S.; Rein, A. Role of murine leukemia virus nucleocapsid protein in virus assembly. J. Virol. 2004, 78, 12378–12385. [Google Scholar] [CrossRef] [PubMed]
- Holtkamp, S.; Kreiter, S.; Selmi, A.; Simon, P.; Koslowski, M.; Huber, C.; Tureci, O.; Sahin, U. Modification of antigen-encoding rna increases stability, translational efficacy, and t-cell stimulatory capacity of dendritic cells. Blood 2006, 108, 4009–4017. [Google Scholar] [CrossRef] [PubMed]
- Schrom, E.; Huber, M.; Aneja, M.; Dohmen, C.; Emrich, D.; Geiger, J.; Hasenpusch, G.; Herrmann-Janson, A.; Kretzschmann, V.; Mykhailyk, O.; et al. Translation of angiotensin-converting enzyme 2 upon liver- and lung-targeted delivery of optimized chemically modified mrna. Mol. Ther. Nucleic Acids 2017, 7, 350–365. [Google Scholar] [CrossRef] [PubMed]
- Hamann, M.V.; Stanke, N.; Müllers, E.; Stirnnagel, K.; Hütter, S.; Artegiani, B.; Bragado Alonso, S.; Calegari, F.; Lindemann, D. Efficient transient genetic manipulation in vitro and in vivo by prototype foamy virus-mediated nonviral rna transfer. Mol. Ther. J. Am. Soc. Gene Ther. 2014, 22, 1460–1471. [Google Scholar] [CrossRef] [PubMed]
- Hamann, M.V.; Müllers, E.; Reh, J.; Stanke, N.; Effantin, G.; Weissenhorn, W.; Lindemann, D. The cooperative function of arginine residues in the prototype foamy virus gag c-terminus mediates viral and cellular rna encapsidation. Retrovirology 2014, 11, 87. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Tu, L.C.; Naseri, A.; Huisman, M.; Zhang, S.; Grunwald, D.; Pederson, T. Crispr-cas9 nuclear dynamics and target recognition in living cells. J. Cell Biol. 2016, 214, 529–537. [Google Scholar] [CrossRef] [PubMed]
- Kaczmarczyk, S.J.; Sitaraman, K.; Young, H.A.; Hughes, S.H.; Chatterjee, D.K. Protein delivery using engineered virus-like particles. Proc. Natl. Acad. Sci. USA 2011, 108, 16998–17003. [Google Scholar] [CrossRef]
- Peretti, S.; Schiavoni, I.; Pugliese, K.; Federico, M. Cell death induced by the herpes simplex virus-1 thymidine kinase delivered by human immunodeficiency virus-1-based virus-like particles. Mol. Ther. 2005, 12, 1185–1196. [Google Scholar] [CrossRef]
- Joo, K.I.; Wang, P. Visualization of targeted transduction by engineered lentiviral vectors. Gene Ther. 2008, 15, 1384–1396. [Google Scholar] [CrossRef]
- Vindry, C.; Guillin, O.; Mangeot, P.E.; Ohlmann, T.; Chavatte, L. A versatile strategy to reduce uga-selenocysteine recoding efficiency of the ribosome using crispr-cas9-viral-like-particles targeting selenocysteine-trna([ser]sec) gene. Cells 2019, 8, 574. [Google Scholar] [CrossRef]
- Brown, E.J.; Albers, M.W.; Shin, T.B.; Ichikawa, K.; Keith, C.T.; Lane, W.S.; Schreiber, S.L. A mammalian protein targeted by g1-arresting rapamycin-receptor complex. Nature 1994, 369, 756–758. [Google Scholar] [CrossRef] [PubMed]
- Sabatini, D.M.; Erdjument-Bromage, H.; Lui, M.; Tempst, P.; Snyder, S.H. Raft1: A mammalian protein that binds to fkbp12 in a rapamycin-dependent fashion and is homologous to yeast tors. Cell 1994, 78, 35–43. [Google Scholar] [CrossRef]
- Gonzalez, M.E. The hiv-1 vpr protein: A multifaceted target for therapeutic intervention. Int. J. Mol. Sci. 2017, 18, 126. [Google Scholar] [CrossRef] [PubMed]
- Muller, B.; Tessmer, U.; Schubert, U.; Krausslich, H.G. Human immunodeficiency virus type 1 vpr protein is incorporated into the virion in significantly smaller amounts than gag and is phosphorylated in infected cells. J. Virol. 2000, 74, 9727–9731. [Google Scholar] [CrossRef] [PubMed]
- Selig, L.; Pages, J.C.; Tanchou, V.; Preveral, S.; Berlioz-Torrent, C.; Liu, L.X.; Erdtmann, L.; Darlix, J.; Benarous, R.; Benichou, S. Interaction with the p6 domain of the gag precursor mediates incorporation into virions of vpr and vpx proteins from primate lentiviruses. J. Virol. 1999, 73, 592–600. [Google Scholar] [CrossRef]
- Mukerjee, R.; Chang, J.R.; Del Valle, L.; Bagashev, A.; Gayed, M.M.; Lyde, R.B.; Hawkins, B.J.; Brailoiu, E.; Cohen, E.; Power, C.; et al. Deregulation of micrornas by hiv-1 vpr protein leads to the development of neurocognitive disorders. J. Biol. Chem. 2011, 286, 34976–34985. [Google Scholar] [CrossRef]
- Lyu, P.; Yoo, K.W.; Yadav, M.K.; Atala, A.; Aartsma-Rus, A.; Putten, M.V.; Duan, D.; Lu, B. Sensitive and reliable evaluation of single-cut sgrnas to restore dystrophin by a gfp-reporter assay. PLoS ONE 2020, 15, e0239468. [Google Scholar] [CrossRef]
- Lim, F.; Downey, T.P.; Peabody, D.S. Translational repression and specific rna binding by the coat protein of the pseudomonas phage pp7. J. Biol. Chem. 2001, 276, 22507–22513. [Google Scholar] [CrossRef]
- Austin, R.J.; Xia, T.; Ren, J.; Takahashi, T.T.; Roberts, R.W. Designed arginine-rich rna-binding peptides with picomolar affinity. J. Am. Chem. Soc. 2002, 124, 10966–10967. [Google Scholar] [CrossRef]
- Wulczyn, F.G.; Kahmann, R. Translational stimulation: Rna sequence and structure requirements for binding of com protein. Cell 1991, 65, 259–269. [Google Scholar] [CrossRef]
- Grunewald, J.; Zhou, R.; Garcia, S.P.; Iyer, S.; Lareau, C.A.; Aryee, M.J.; Joung, J.K. Transcriptome-wide off-target rna editing induced by crispr-guided DNA base editors. Nature 2019, 569, 433–437. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Sun, Y.; Yan, R.; Liu, Y.; Zuo, E.; Gu, C.; Han, L.; Wei, Y.; Hu, X.; Zeng, R.; et al. Off-target rna mutation induced by DNA base editing and its elimination by mutagenesis. Nature 2019, 571, 275–278. [Google Scholar] [CrossRef] [PubMed]
- Rees, H.A.; Wilson, C.; Doman, J.L.; Liu, D.R. Analysis and minimization of cellular rna editing by DNA adenine base editors. Sci. Adv. 2019, 5, eaax5717. [Google Scholar] [CrossRef] [PubMed]
- Farboud, B.; Jarvis, E.; Roth, T.L.; Shin, J.; Corn, J.E.; Marson, A.; Meyer, B.J.; Patel, N.H.; Hochstrasser, M.L. Enhanced genome editing with cas9 ribonucleoprotein in diverse cells and organisms. J. Vis. Exp. 2018, 135, 57350. [Google Scholar] [CrossRef] [PubMed]
- Hinz, J.M.; Laughery, M.F.; Wyrick, J.J. Nucleosomes inhibit cas9 endonuclease activity in vitro. Biochemistry 2015, 54, 7063–7066. [Google Scholar] [CrossRef] [PubMed]
- Horlbeck, M.A.; Witkowsky, L.B.; Guglielmi, B.; Replogle, J.M.; Gilbert, L.A.; Villalta, J.E.; Torigoe, S.E.; Tjian, R.; Weissman, J.S. Nucleosomes impede cas9 access to DNA in vivo and in vitro. Elife 2016, 5, e12677. [Google Scholar] [CrossRef]
- Isaac, R.S.; Jiang, F.; Doudna, J.A.; Lim, W.A.; Narlikar, G.J.; Almeida, R. Nucleosome breathing and remodeling constrain crispr-cas9 function. Elife 2016, 5, e13450. [Google Scholar] [CrossRef]
- Kallimasioti-Pazi, E.M.; Thelakkad Chathoth, K.; Taylor, G.C.; Meynert, A.; Ballinger, T.; Kelder, M.J.E.; Lalevee, S.; Sanli, I.; Feil, R.; Wood, A.J. Heterochromatin delays crispr-cas9 mutagenesis but does not influence the outcome of mutagenic DNA repair. Plos Biol. 2018, 16, e2005595. [Google Scholar] [CrossRef]
- DePolo, N.J.; Reed, J.D.; Sheridan, P.L.; Townsend, K.; Sauter, S.L.; Jolly, D.J.; Dubensky, T.W., Jr. Vsv-g pseudotyped lentiviral vector particles produced in human cells are inactivated by human serum. Mol. Ther. 2000, 2, 218–222. [Google Scholar] [CrossRef]
- Brown, B.D.; Sitia, G.; Annoni, A.; Hauben, E.; Sergi, L.S.; Zingale, A.; Roncarolo, M.G.; Guidotti, L.G.; Naldini, L. In vivo administration of lentiviral vectors triggers a type i interferon response that restricts hepatocyte gene transfer and promotes vector clearance. Blood 2007, 109, 2797–2805. [Google Scholar] [CrossRef]
- Schauber-Plewa, C.; Simmons, A.; Tuerk, M.J.; Pacheco, C.D.; Veres, G. Complement regulatory proteins are incorporated into lentiviral vectors and protect particles against complement inactivation. Gene Ther. 2005, 12, 238–245. [Google Scholar] [CrossRef] [PubMed]
- Milani, M.; Annoni, A.; Moalli, F.; Liu, T.; Cesana, D.; Calabria, A.; Bartolaccini, S.; Biffi, M.; Russo, F.; Visigalli, I.; et al. Phagocytosis-shielded lentiviral vectors improve liver gene therapy in nonhuman primates. Sci. Transl. Med. 2019, 11, eaav7325. [Google Scholar] [CrossRef] [PubMed]
- Milani, M.; Annoni, A.; Bartolaccini, S.; Biffi, M.; Russo, F.; Di Tomaso, T.; Raimondi, A.; Lengler, J.; Holmes, M.C.; Scheiflinger, F.; et al. Genome editing for scalable production of alloantigen-free lentiviral vectors for in vivo gene therapy. Embo Mol. Med. 2017, 9, 1558–1573. [Google Scholar] [CrossRef] [PubMed]
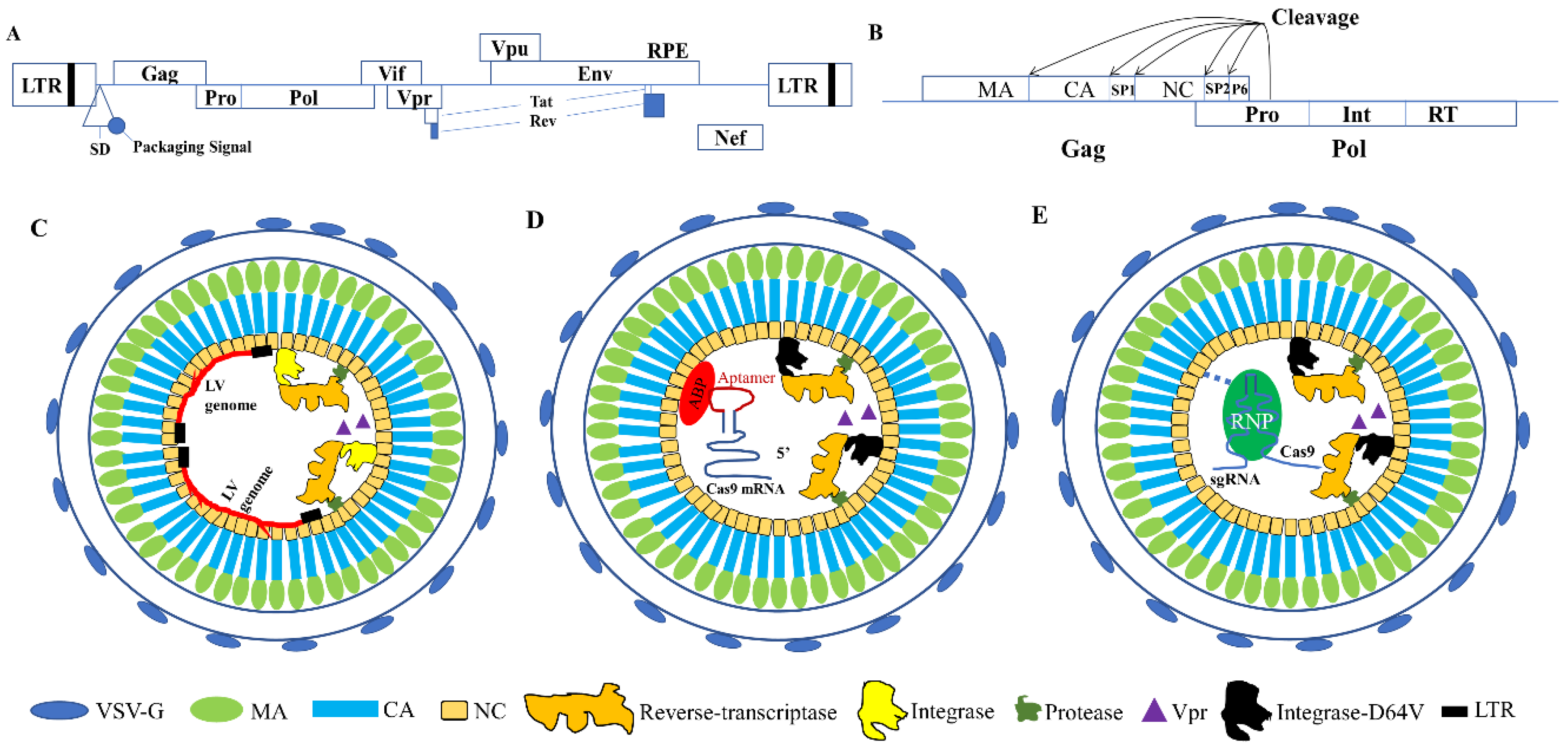
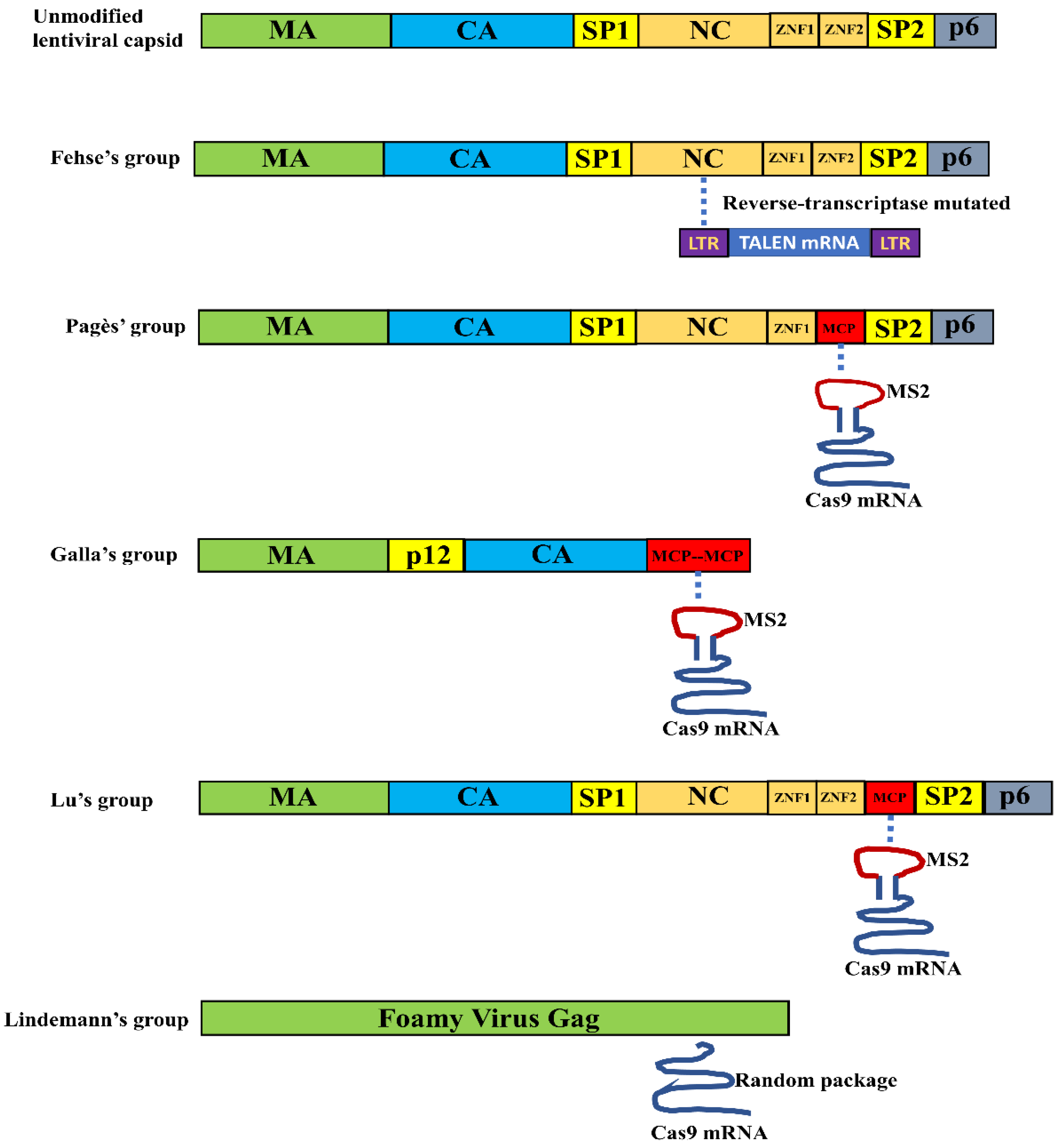
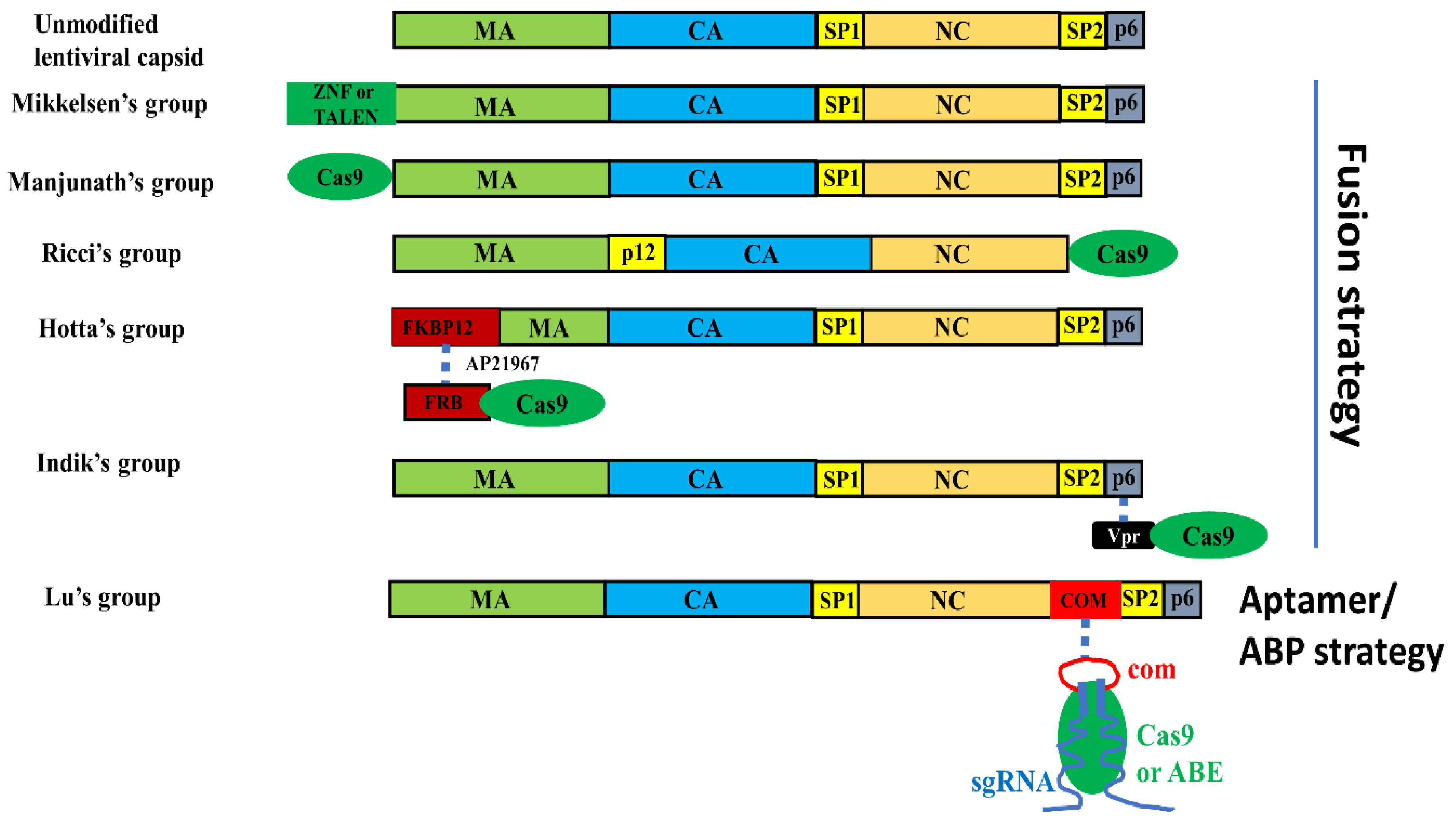
| Virus Type | Capsid Modification | RNA Package | Copy Number | Addgene Plasmids | Reference |
|---|---|---|---|---|---|
| LV | Not modified | TALEN mRNA | 2 copies | LeGO-iG2-wPRE-pA (60489) | [45] |
| LV | MCP replaced the second zinc finger domain of NC | SpCas9 mRNA | ~6 copies | Not available | [46] |
| Murine Leukemia Virus | Two copies of MCP replaced NC | SpCas9 mRNA and sgRNA | Not available | Not available | [47] |
| LV | MCP inserted after the second zinc finger domain of NC | SaCas9 mRNA | 50~100 copies | pSaCas9-1xms2-2x3′UTR (122946) | [48] |
| pSaCas9-1xPP7-2x3′UTR(122947) | |||||
| psPAX2-D64V-NC-PP7(122945) | |||||
| psPAX2-D64V-NC-MS2(122944) | |||||
| Foamy Viruses | Not modified | SpCas9 mRNA | 60 copies | Not available | [49] |
| Capsid Type | Mechanism of Nuclease Recruitment | Editing Effectors Delivered | Addgene Plasmids | Reference |
|---|---|---|---|---|
| LV | Fusing editing effector to the N-terminus of Gag | ZNF and TALEN | Not available | [50] |
| LV | Fusing Cas9 protein to the N-terminus of Gag | SpCas9 | Not available | [51] |
| MLV | Fusing Cas9 to the C-terminus of MLV Gag | SpCas9 | BIC-Gag-CAS9(119942) | [52] |
| LV | Fusing FKBP12 to Gag, fusing FRB to SpCas9. FKBP12/AP21967/FRB interaction brings SpCas9 to Gag | SpCas9 | pHLS-EF1a-FRB-SpCas9-A(138477) | [53] |
| pHLS-EF1a-FKBP12-Gag(HIV)(138476) | ||||
| LV | Fusing Cas9 to the C-terminus of Vpr | SpCas9 | Not available | [54] |
| LV | Forming a three-component complex: Com-NC/aptamer-sgRNA/Cas9 protein. | SaCas9 | pSaCas9-sgRNA-Tetra-com- vector(131227) | [55] |
| psPAX2-D64V-NC-COM(131226) | ||||
| SpCas9 | pSpCas9-3′UTR-ST2-com-vector(136269) | [56] | ||
| ABE | pSpCas9-ABE-3′UTR-sgRNA-ST2-com- vector(136270) | [57] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lyu, P.; Wang, L.; Lu, B. Virus-Like Particle Mediated CRISPR/Cas9 Delivery for Efficient and Safe Genome Editing. Life 2020, 10, 366. https://doi.org/10.3390/life10120366
Lyu P, Wang L, Lu B. Virus-Like Particle Mediated CRISPR/Cas9 Delivery for Efficient and Safe Genome Editing. Life. 2020; 10(12):366. https://doi.org/10.3390/life10120366
Chicago/Turabian StyleLyu, Pin, Luxi Wang, and Baisong Lu. 2020. "Virus-Like Particle Mediated CRISPR/Cas9 Delivery for Efficient and Safe Genome Editing" Life 10, no. 12: 366. https://doi.org/10.3390/life10120366
APA StyleLyu, P., Wang, L., & Lu, B. (2020). Virus-Like Particle Mediated CRISPR/Cas9 Delivery for Efficient and Safe Genome Editing. Life, 10(12), 366. https://doi.org/10.3390/life10120366




