Comparative Proteomic Analysis of Walnut (Juglans regia L.) Pellicle Tissues Reveals the Regulation of Nut Quality Attributes
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Physiological Maturity Sampling
2.3. Walnut Kernel Color Evaluations
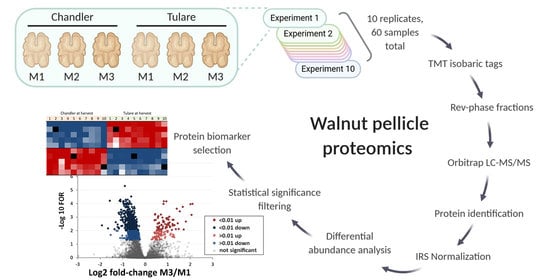
2.4. Walnut Pellicle Protein Extraction and Preparation of Samples for Mass Spectrometry
2.5. Proteome Data Preparation and Analysis
3. Results
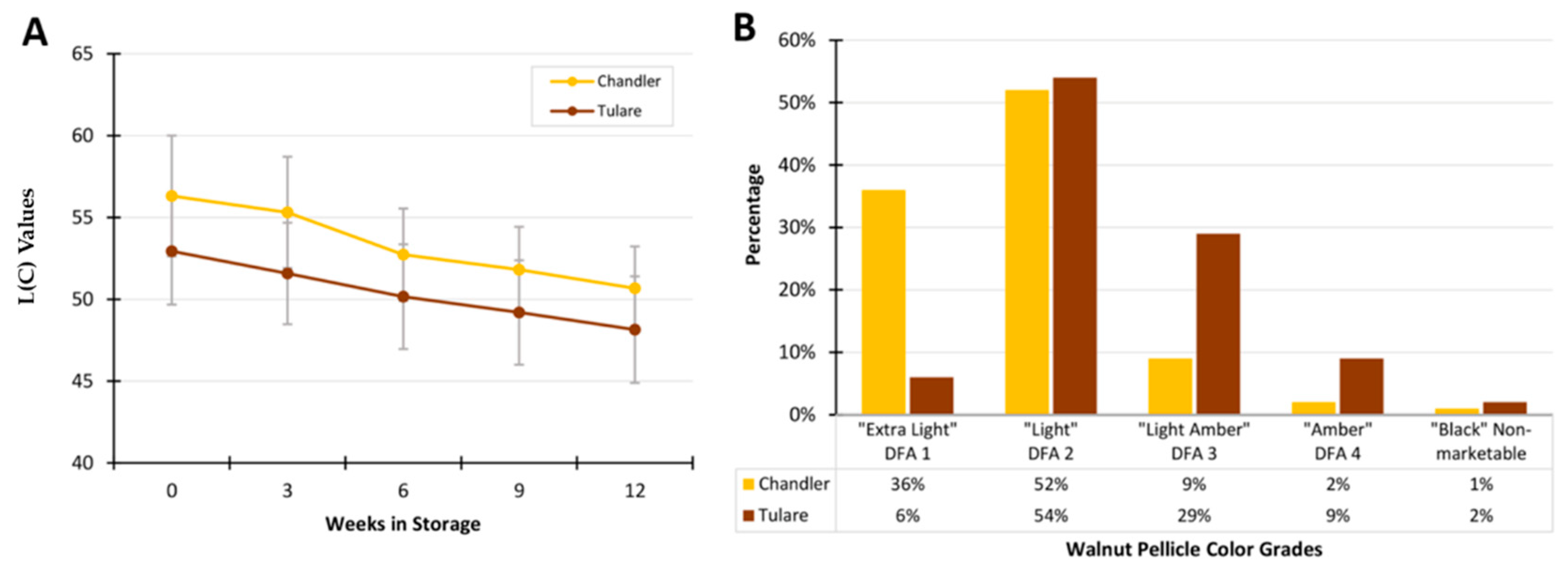
3.1. Analysis of Walnut Pellicle Coloration
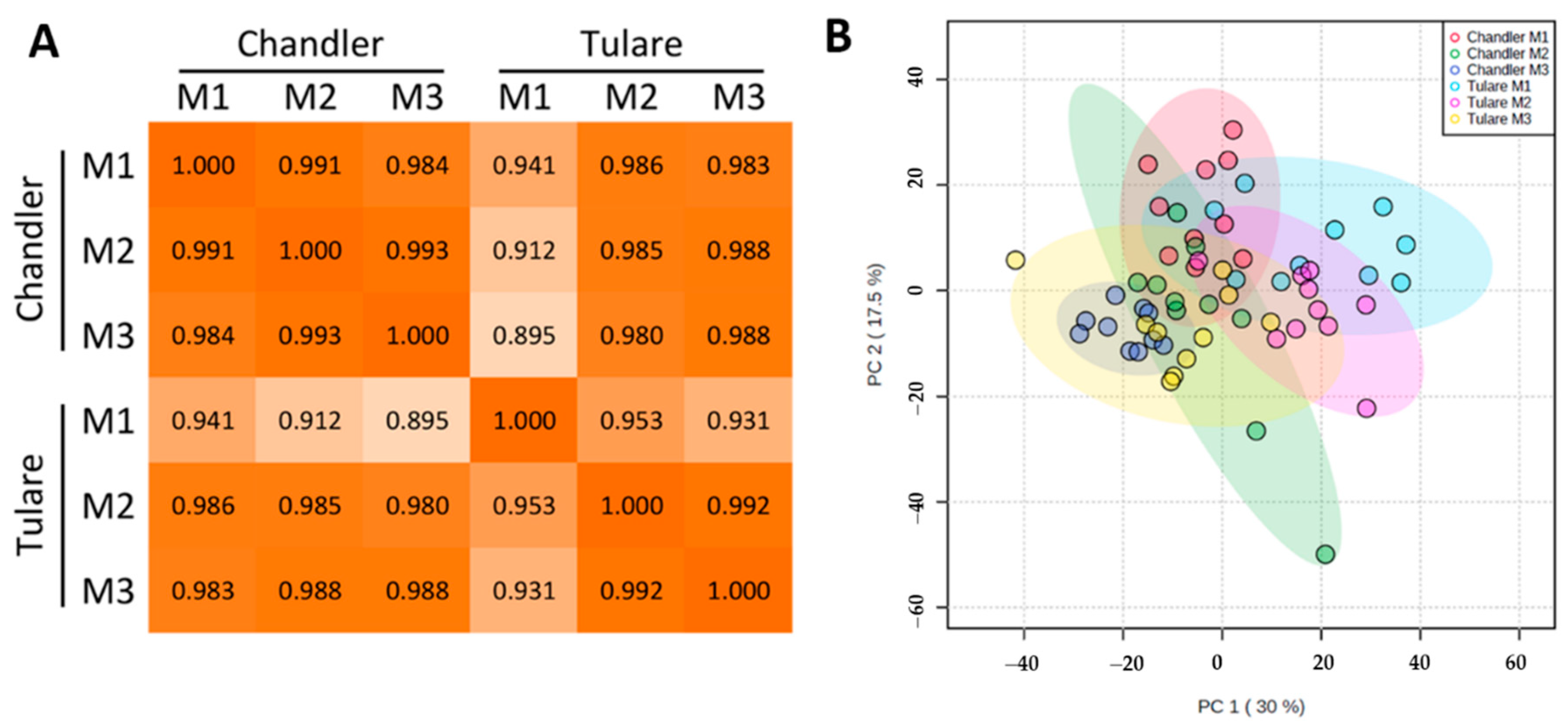
3.2. Proteomic Analysis of Walnut Pellicle Tissues
3.3. Effect of Walnut Fruit Maturity on Kernel Pellicle Proteome
3.4. Comparison of Cultivars ‘Chandler’ and ‘Tulare’ Pellicle Proteome at Nut Harvest Stage
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Abbey, M.; Noakes, M.; Belling, G.B.; Nestel, P.J. Partial replacement of saturated fatty acids with almonds or walnuts lowers total plasma cholesterol and low-density-lipoprotein cholesterol. Am. J. Clin. Nutr. 1994, 59, 995–999. [Google Scholar] [CrossRef]
- Chisholm, A.; Mann, J.; Skeaff, M.; Frampton, C.; Sutherland, W.; Duncan, A.; Tiszavari, S. A diet rich in walnuts favourably influences plasma fatty acid profile in moderately hyperlipidaemic subjects. Eur. J. Clin. Nutr. 1998, 52, 12–16. [Google Scholar] [CrossRef]
- Lavedrine, F.; Zmirou, D.; Ravel, A.; Balducci, F.; Alary, J. Blood Cholesterol and Walnut Consumption: A Cross-sectional Survey in France. Prev. Med. 1999, 28, 333–339. [Google Scholar] [CrossRef]
- Shahidi, F.; Ambigaipalan, P. Phenolics and polyphenolics in foods, beverages and spices: Antioxidant activity and health effects—A review. J. Funct. Food 2015, 18, 820–897. [Google Scholar] [CrossRef]
- Alasalvar, C.; Bolling, B.W. Review of nut phytochemicals, fat-soluble bioactives, antioxidant components and health effects. Br. J. Nutr. 2015, 113 (Suppl. 2), S68–S78. [Google Scholar] [CrossRef]
- Bamberger, C.; Rossmeier, A.; Lechner, K.; Wu, L.; Waldmann, E.; Stark, R.G.; Altenhofer, J.; Henze, K.; Parhofer, K.G. A Walnut-Enriched Diet Reduces Lipids in Healthy Caucasian Subjects, Independent of Recommended Macronutrient Replacement and Time Point of Consumption: A Prospective, Randomized, Controlled Trial. Nutrients 2017, 9, 1097. [Google Scholar] [CrossRef]
- Bitok, E.; Rajaram, S.; Jaceldo-Siegl, K.; Oda, K.; Sala-Vila, A.; Serra-Mir, M.; Ros, E.; Sabaté, J. Effects of Long-Term Walnut Supplementation on Body Weight in Free-Living Elderly: Results of a Randomized Controlled Trial. Nutrients 2018, 10, 1317. [Google Scholar] [CrossRef]
- Ros, E. Health Benefits of Nut Consumption. Nutrients 2010, 2, 652–682. [Google Scholar] [CrossRef]
- Sze-Tao, K.W.C.; Sathe, S.K. Walnuts (Juglans regia L.): Proximate composition, protein solubility, protein amino acid composition and protein in vitro digestibility. J. Sci. Food Agric. 2000, 80, 1393–1401. [Google Scholar] [CrossRef]
- Kornsteiner, M.; Wagner, K.-H.; Elmadfa, I. Tocopherols and total phenolics in 10 different nut types. Food Chem. 2006, 98, 381–387. [Google Scholar] [CrossRef]
- Sabate, J.; Fraser, G.E.; Burke, K.; Knutsen, S.F.; Bennett, H.; Lindsted, K.D. Effects of Walnuts on Serum Lipid Levels and Blood Pressure in Normal Men. N. Engl. J. Med. 1993, 328, 603–607. [Google Scholar] [CrossRef] [PubMed]
- Global Walnut Consumption Has Softened Its Robust Growth. Available online: https://www.producereport.com/article/global-walnut-consumption-has-softened-its-robust-growth (accessed on 27 October 2020).
- USDA/NASS 2019 State Agriculture Overview for California. Available online: https://www.nass.usda.gov/Quick_Stats/Ag_Overview/stateOverview.php?state=CALIFORNIA (accessed on 27 October 2020).
- Ward, M.; Inouye, A.U.S. Tree Nuts to Lose Market Share in China Due to Additional Tariffs. Available online: https://apps.fas.usda.gov/newgainapi/api/report/downloadreportbyfilename?filename=Tree%20Nuts%20Annual_Beijing_China%20-%20Peoples%20Republic%20of_9-12-2019.pdf (accessed on 22 October 2020).
- Publication, Tree Nuts: World Markets and Trade, ID: tm70mv16z, USDA Economics, Statistics and Market Information System. Available online: https://usda.library.cornell.edu/concern/publications/tm70mv16z?locale=en (accessed on 27 October 2020).
- Ortiz, C.M.; Vicente, A.R.; Fields, R.P.; Grilo, F.; Labavitch, J.M.; Donis-Gonzalez, I.; Crisosto, C.H. Walnut (Juglans regia L.) kernel postharvest deterioration as affected by pellicle integrity, cultivar and oxygen concentration. Postharvest Biol. Technol. 2019, 156, 110948. [Google Scholar] [CrossRef]
- Harris, L.J.; Ferguson, L. 15—Improving the safety of almonds and pistachios. In Improving the Safety and Quality of Nuts; Harris, L.J., Ed.; Woodhead Publishing Series in Food Science, Technology and Nutrition; Woodhead Publishing: Southton, UK, 2013; pp. 350–378. ISBN 978-0-85709-266-3. [Google Scholar]
- Radchuk, V.; Borisjuk, L. Physical, metabolic and developmental functions of the seed coat. Front. Plant Sci. 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Pinney, K.; Polito, V.S. English walnut fruit growth and development. Sci. Hortic. 1983, 21, 19–28. [Google Scholar] [CrossRef]
- Perry, E. Harvesting and Storing Your Home Orchard’s Nut Crop; University of California Agriculture and Natural Resources Publication: Davis, CA, USA, 1998; p. 9. [Google Scholar] [CrossRef]
- United States Department of Agriculture United States Standards for Grades of Shelled Walnuts. 2018. Available online: https://www.ams.usda.gov/sites/default/files/media/WalnutColorChart.pdf (accessed on 22 October 2020).
- American Pomological Society—Journal Volume 74 Number 3 Late Maturity and Excess Irrigation Trigger Kernel Darkening in ‘Howard’ English Walnut (Juglans regia L.) at Harvest, But Not in ‘Chandler’. Available online: http://www.pubhort.org/aps/74/v74_n3_a2.htm (accessed on 27 October 2020).
- Wu, G.L.; Liu, Q.L.; Teixeira da Silva, J.A. Ultrastructure of pericarp and seed capsule cells in the developing walnut (Juglans regia L.) fruit. S. Afr. J. Bot. 2009, 75, 128–136. [Google Scholar] [CrossRef]
- Colarič, M.; Štampar, F.; Hudina, M.; Solar, A. Sensory evaluation of different walnut cultivars (Juglans regia L.). Acta Agric. Slov. 2006, 87, 403–411. [Google Scholar]
- Arcan, I.; Yemenicioğlu, A. Antioxidant activity and phenolic content of fresh and dry nuts with or without the seed coat. J. Food Compos. Anal. 2009, 22, 184–188. [Google Scholar] [CrossRef]
- Marrano, A.; Martínez-García, P.J.; Bianco, L.; Sideli, G.M.; Pierro, E.A.D.; Leslie, C.A.; Stevens, K.A.; Crepeau, M.W.; Troggio, M.; Langley, C.H.; et al. A new genomic tool for walnut (Juglans regia L.): Development and validation of the high-density AxiomTM J. regia 700K SNP genotyping array. Plant. Biotechnol. J. 2019, 17, 1027–1036. [Google Scholar] [CrossRef]
- Arab, M.M.; Marrano, A.; Abdollahi-Arpanahi, R.; Leslie, C.A.; Askari, H.; Neale, D.B.; Vahdati, K. Genome-wide patterns of population structure and association mapping of nut-related traits in Persian walnut populations from Iran using the Axiom J. regia 700K SNP array. Sci. Rep. 2019, 9, 6376. [Google Scholar] [CrossRef]
- Marrano, A.; Sideli, G.M.; Leslie, C.A.; Cheng, H.; Neale, D.B. Deciphering of the Genetic Control of Phenology, Yield, and Pellicle Color in Persian Walnut (Juglans regia L.). Front. Plant. Sci. 2019, 10. [Google Scholar] [CrossRef]
- Martínez-García, P.J.; Crepeau, M.W.; Puiu, D.; Gonzalez-Ibeas, D.; Whalen, J.; Stevens, K.A.; Paul, R.; Butterfield, T.S.; Britton, M.T.; Reagan, R.L.; et al. The walnut (Juglans regia) genome sequence reveals diversity in genes coding for the biosynthesis of non-structural polyphenols. Plant. J. 2016, 87, 507–532. [Google Scholar] [CrossRef]
- Marrano, A.; Britton, M.; Zaini, P.A.; Zimin, A.V.; Workman, R.E.; Puiu, D.; Bianco, L.; Pierro, E.A.D.; Allen, B.J.; Chakraborty, S.; et al. High-quality chromosome-scale assembly of the walnut (Juglans regia L.) reference genome. Gigascience 2020, 9. [Google Scholar] [CrossRef] [PubMed]
- Rao, G.; Sui, J.; Zhang, J. Metabolomics reveals significant variations in metabolites and correlations regarding the maturation of walnuts (Juglans regia L.). Biol. Open 2016, 5, 829–836. [Google Scholar] [CrossRef] [PubMed]
- Hasey, J. Managing Walnut Harvest for Quality. University of California Cooperative Extension. Available online: http://cebutte.ucanr.edu/files/43205.pdf (accessed on 20 October 2020).
- Butterfield, T.; Phu, M.; Chakraborty, S.; Walawage, S.; Reagan, R.; Leslie, A.; Jacobson, A.; Escobar, M.; Dandekar, A.M. Identifying the Genetic Determinant(S) of Pellicle Coloration in Walnut; University of California Agriculture and Natural Resources Publication: Davis, CA, USA, 2016; p. 17. [Google Scholar]
- Evans, K.J.; Lang, M.D. Advancing Hull Split to Maximize Yield and Quality of Walnuts; Horticultural Australia: North Sydney, Australia, 2010; ISBN 978-0-7341-2475-3. [Google Scholar]
- The Art of Ethephon—West Coast Nut. Available online: http://www.wcngg.com/2020/08/06/the-art-of-ethephon/ (accessed on 27 October 2020).
- McGuire, R.G. Reporting of Objective Color Measurements. HortScience 1992, 27, 1254–1255. [Google Scholar] [CrossRef]
- Donis-González, I.R.; Bergman, S.M.; Sideli, G.M.; Slaughter, D.C.; Crisosto, C.H. Color vision system to assess English walnut (Juglans regia) kernel pellicle color. Postharvest Biol. Technol. 2020, 167, 111199. [Google Scholar] [CrossRef]
- R: The R Project for Statistical Computing. Available online: https://www.r-project.org/ (accessed on 27 October 2020).
- Faurobert, M.; Pelpoir, E.; Chaïb, J. Phenol Extraction of Proteins for Proteomic Studies of Recalcitrant Plant Tissues. In Plant Proteomics: Methods and Protocols; Thiellement, H., Zivy, M., Damerval, C., Méchin, V., Eds.; Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 2007; pp. 9–14. ISBN 978-1-59745-227-4. [Google Scholar]
- HD Sagawa, C.; de AB Assis, R.; Zaini, P.A.; Wilmarth, P.A.; Phinney, B.S.; Moreira, L.M.; Dandekar, A.M. Proteome Analysis of Walnut Bacterial Blight Disease. Int. J. Mol. Sci. 2020, 21, 7453. [Google Scholar] [CrossRef]
- Chambers, M.C.; Maclean, B.; Burke, R.; Amodei, D.; Ruderman, D.L.; Neumann, S.; Gatto, L.; Fischer, B.; Pratt, B.; Egertson, J.; et al. A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol. 2012, 30, 918–920. [Google Scholar] [CrossRef]
- Wilmarth, P.A.; Riviere, M.A.; David, L.L. Techniques for accurate protein identification in shotgun proteomic studies of human, mouse, bovine, and chicken lenses. J. Ocul. Biol. Dis. Inform. 2009, 2, 223–234. [Google Scholar] [CrossRef]
- McDonald, W.H.; Tabb, D.L.; Sadygov, R.G.; MacCoss, M.J.; Venable, J.; Graumann, J.; Johnson, J.R.; Cociorva, D.; Yates, J.R. MS1, MS2, and SQT-three unified, compact, and easily parsed file formats for the storage of shotgun proteomic spectra and identifications. Rapid Commun. Mass Spectrom. 2004, 18, 2162–2168. [Google Scholar] [CrossRef]
- Eng, J.K.; Jahan, T.A.; Hoopmann, M.R. Comet: An open-source MS/MS sequence database search tool. Proteomics 2013, 13, 22–24. [Google Scholar] [CrossRef]
- Elias, J.E.; Gygi, S.P. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 2007, 4, 207–214. [Google Scholar] [CrossRef] [PubMed]
- Plubell, D.L.; Wilmarth, P.A.; Zhao, Y.; Fenton, A.M.; Minnier, J.; Reddy, A.P.; Klimek, J.; Yang, X.; David, L.L.; Pamir, N. Extended Multiplexing of Tandem Mass Tags (TMT) Labeling Reveals Age and High Fat Diet Specific Proteome Changes in Mouse Epididymal Adipose Tissue. Mol. Cell Proteom. 2017, 16, 873–890. [Google Scholar] [CrossRef] [PubMed]
- MassIVE, Mass Spectrometry Interactive Virtual Environment. Available online: https://massive.ucsd.edu/ProteoSAFe/static/massive.jsp (accessed on 15 November 2020).
- Deutsch, E.W.; Csordas, A.; Sun, Z.; Jarnuczak, A.; Perez-Riverol, Y.; Ternent, T.; Campbell, D.S.; Bernal-Llinares, M.; Okuda, S.; Kawano, S.; et al. The ProteomeXchange consortium in 2017: Supporting the cultural change in proteomics public data deposition. Nucleic Acids Res. 2017, 45, D1100–D1106. [Google Scholar] [CrossRef] [PubMed]
- Mi, H.; Muruganujan, A.; Casagrande, J.T.; Thomas, P.D. Large-scale gene function analysis with the PANTHER classification system. Nat. Protoc. 2013, 8, 1551–1566. [Google Scholar] [CrossRef] [PubMed]
- Chong, J.; Soufan, O.; Li, C.; Caraus, I.; Li, S.; Bourque, G.; Wishart, D.S.; Xia, J. MetaboAnalyst 4.0: Towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018, 46, W486–W494. [Google Scholar] [CrossRef] [PubMed]
- McGranahan, G.; Leslie, C. ‘Robert Livermore’, a Persian Walnut Cultivar with a Red Seedcoat. HortScience 2004, 39, 1772. [Google Scholar] [CrossRef]
- Miernyk, J.A.; Johnston, M.L. Proteomic analysis of the testa from developing soybean seeds. J. Proteom. 2013, 89, 265–272. [Google Scholar] [CrossRef]
- Robotham, J.M.; Xia, L.; Willison, L.N.; Teuber, S.S.; Sathe, S.K.; Roux, K.H. Characterization of a cashew allergen, 11S globulin (Ana o 2), conformational epitope. Mol. Immunol. 2010, 47, 1830–1838. [Google Scholar] [CrossRef]
- Kshirsagar, H.H.; Fajer, P.; Sharma, G.M.; Roux, K.H.; Sathe, S.K. Biochemical and spectroscopic characterization of almond and cashew nut seed 11S legumins, amandin and anacardein. J. Agric. Food Chem. 2011, 59, 386–393. [Google Scholar] [CrossRef]
- Marcus, J.P.; Green, J.L.; Goulter, K.C.; Manners, J.M. A family of antimicrobial peptides is produced by processing of a 7S globulin protein in Macadamia integrifolia kernels. Plant. J. 1999, 19, 699–710. [Google Scholar] [CrossRef]
- Chardin, H.; Mayer, C.; Sénéchal, H.; Wal, J.M.; Poncet, P.; Desvaux, F.X.; Peltre, G. Lipid transfer protein 1 is a possible allergen in Arabidopsis thaliana. Int. Arch. Allergy Immunol. 2003, 131, 85–90. [Google Scholar] [CrossRef] [PubMed]
- Segura, A.; Moreno, M.; García-Olmedo, F. Purification and antipathogenic activity of lipid transfer proteins (LTPs) from the leaves of Arabidopsis and spinach. FEBS Lett. 1993, 332, 243–246. [Google Scholar] [CrossRef]
- Simon-Nobbe, B.; Denk, U.; Schneider, P.B.; Radauer, C.; Teige, M.; Crameri, R.; Hawranek, T.; Lang, R.; Richter, K.; Schmid-Grendelmeier, P.; et al. NADP-dependent mannitol dehydrogenase, a major allergen of Cladosporium herbarum. J. Biol. Chem. 2006, 281, 16354–16360. [Google Scholar] [CrossRef] [PubMed]
- Sels, J.; Mathys, J.; De Coninck, B.M.A.; Cammue, B.P.A.; De Bolle, M.F.C. Plant pathogenesis-related (PR) proteins: A focus on PR peptides. Plant. Physiol. Biochem. 2008, 46, 941–950. [Google Scholar] [CrossRef] [PubMed]
- Elmer, A.; Chao, W.; Grimes, H. Protein sorting and expression of a unique soybean cotyledon protein, GmSBP, destined for the protein storage vacuole. Plant. Mol. Biol. 2003, 52, 1089–1106. [Google Scholar] [CrossRef]
- Eastmond, P.J.; Graham, I.A. Re-examining the role of the glyoxylate cycle in oilseeds. Trends Plant. Sci. 2001, 6, 72–78. [Google Scholar] [CrossRef]
- Graham, I.A. Seed Storage Oil Mobilization. Annu. Rev. Plant. Biol. 2008, 59, 115–142. [Google Scholar] [CrossRef]
- Iglesias-Fernández, R.; Matilla, A. After-ripening alters the gene expression pattern of oxidases involved in the ethylene and gibberellin pathways during early imbibition of Sisymbrium officinale L. seeds. J. Exp. Bot. 2009, 60, 1645–1661. [Google Scholar] [CrossRef]
- Qin, Y.-M.; Hu, C.-Y.; Pang, Y.; Kastaniotis, A.J.; Hiltunen, J.K.; Zhu, Y.-X. Saturated Very-Long-Chain Fatty Acids Promote Cotton Fiber and Arabidopsis Cell Elongation by Activating Ethylene Biosynthesis. Plant. Cell 2007, 19, 3692–3704. [Google Scholar] [CrossRef]
- Sauter, M.; Moffatt, B.; Saechao, M.C.; Hell, R.; Wirtz, M. Methionine salvage and S-adenosylmethionine: Essential links between sulfur, ethylene and polyamine biosynthesis. Biochem. J. 2013, 451, 145–154. [Google Scholar] [CrossRef]
- Hornung, E.; Pernstich, C.; Feussner, I. Formation of conjugated Delta11Delta13-double bonds by Delta12-linoleic acid (1,4)-acyl-lipid-desaturase in pomegranate seeds. Eur. J. Biochem. 2002, 269, 4852–4859. [Google Scholar] [CrossRef] [PubMed]
- Murata, M. Browning and pigmentation in food through the Maillard reaction. Glycoconj. J. 2020. [Google Scholar] [CrossRef] [PubMed]



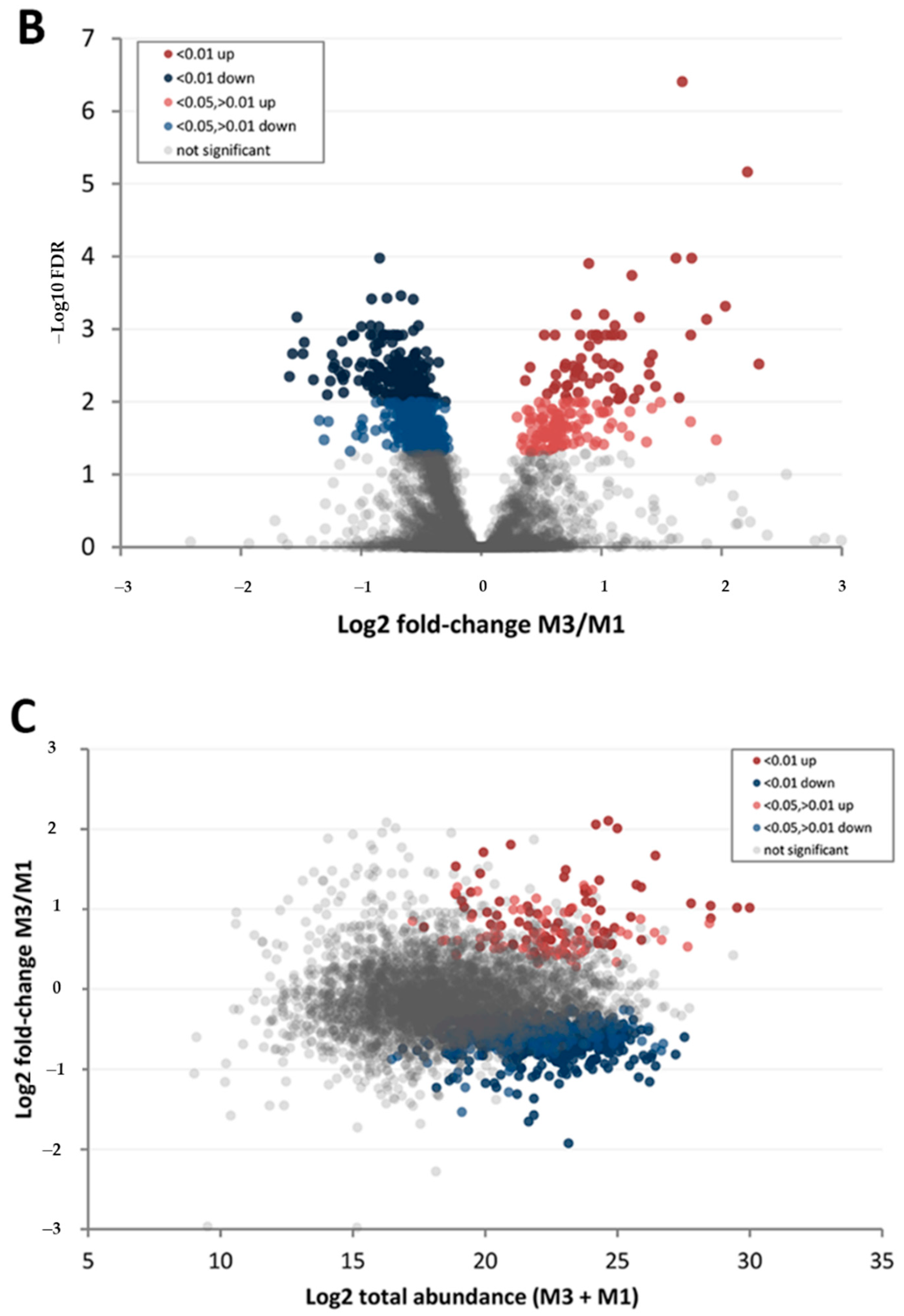
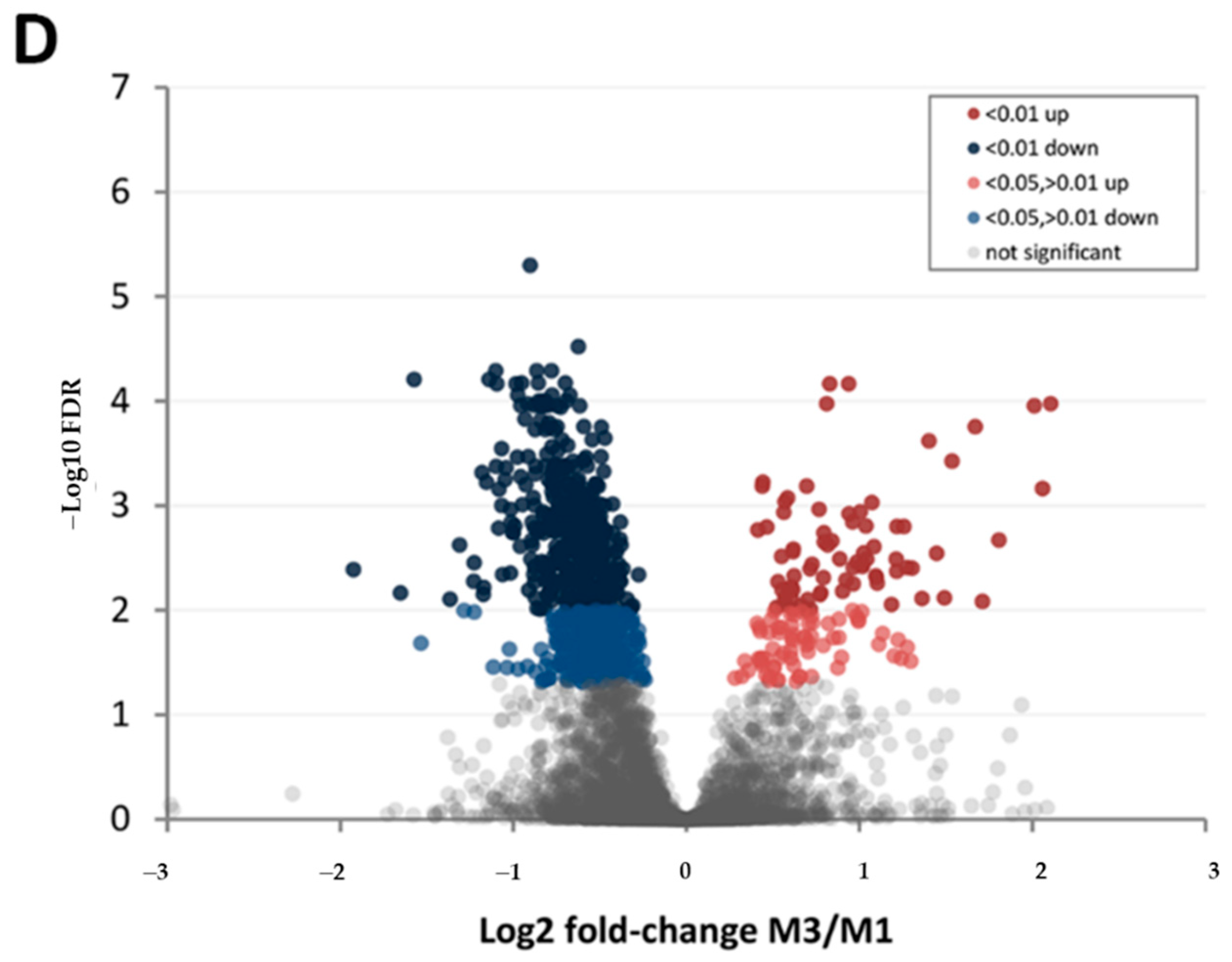

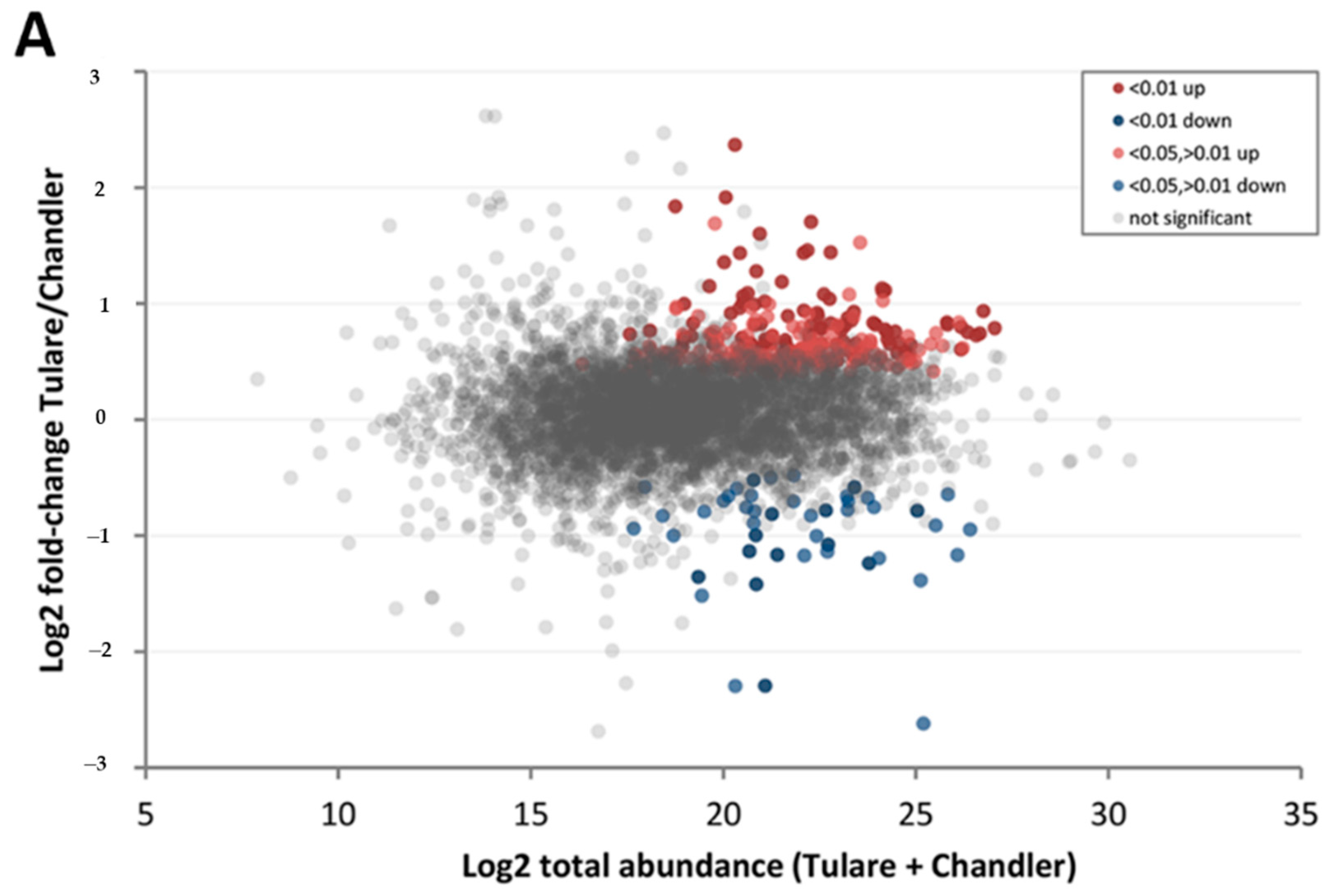
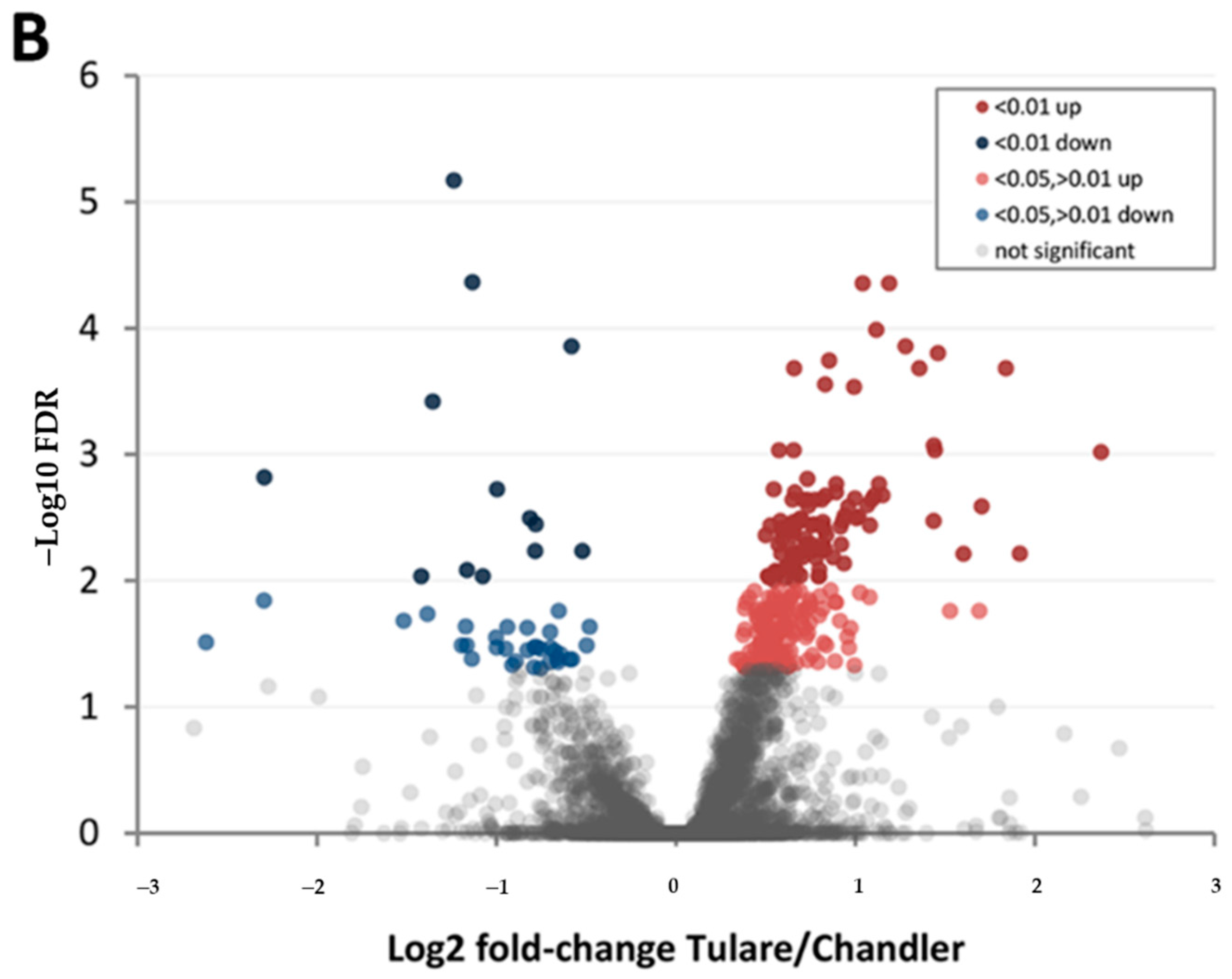

| Protein Category | ‘Chandler’ M3/M1 1 | ‘Tulare’ M3/M1 | ‘Tulare’/’Chandler’ M2 |
|---|---|---|---|
| FDR < 0.01 up | 66 | 76 | 103 |
| FDR < 0.05, >0.01 up | 114 | 71 | 172 |
| FDR < 0.01 down | 241 | 388 | 13 |
| FDR < 0.05, >0.01 down | 374 | 263 | 33 |
| Difference not significant | 4142 | 4139 | 4616 |
| PANTHER GO-Slim Biological Process | Observed | Expected | Fold Change | p-Value | FDR |
|---|---|---|---|---|---|
| secondary metabolic process (GO:0019748) | 5 | 0.08 | 66.18 | 2.39 × 10−8 | 3.57 × 10−5 |
| proteolysis (GO:0006508) | 7 | 1.17 | 5.96 | 1.78 × 10−4 | 3.33 × 10−2 |
| cellular catabolic process (GO:0044248) | 8 | 1.59 | 5.04 | 1.86 × 10−4 | 3.09 × 10−2 |
| PANTHER GO-Slim Molecular Function | |||||
| carboxypeptidase activity (GO:0004180) | 6 | 0.12 | 50.66 | 3.80 × 10−9 | 1.69 × 10−6 |
| hydrolase activity (GO:0016825) | 6 | 0.21 | 28.25 | 1.01 × 10−7 | 1.50 × 10−5 |
| acyl transferase activity (GO:0016746) | 6 | 0.45 | 13.42 | 6.64 × 10−6 | 4.92 × 10−4 |
| PANTHER GO-Slim Cellular Component | |||||
| anchored on plasma membrane (GO:0046658) | 4 | 0.22 | 17.97 | 8.51 × 10−5 | 3.49 × 10−2 |
| PANTHER Protein Class | |||||
| serine protease (PC00203) | 6 | 0.54 | 11.13 | 1.87 × 10−5 | 3.19 × 10−3 |
| PANTHER GO-Slim Biological Process | Observed | Expected | Fold Change | p-Value | FDR |
|---|---|---|---|---|---|
| branched-chain amino acid biosynthesis (GO:0009082) | 5 | 0.13 | 39.51 | 6.69 × 10−7 | 2.33 × 10−5 |
| glycolytic process (GO:0006096) | 8 | 0.41 | 19.45 | 2.84 × 10−8 | 1.09 × 10−6 |
| aromatic amino acid family biosynthesis (GO:0009073) | 4 | 0.26 | 15.17 | 2.25 × 10−4 | 4.26 × 10−3 |
| transmembrane import into organelle (GO:0044743) | 5 | 0.35 | 14.37 | 4.54 × 10−5 | 1.01 × 10−3 |
| tricarboxylic acid cycle (GO:0006099) | 3 | 0.23 | 12.93 | 2.18 × 10−3 | 3.14 × 10−2 |
| chaperone-mediated protein folding (GO:0061077) | 6 | 0.52 | 11.61 | 2.32 × 10−5 | 5.88 × 10−4 |
| purine ribonucleotide biosynthetic process (GO:0009152) | 5 | 0.71 | 7.08 | 9.47 × 10−4 | 1.56 × 10−2 |
| cofactor biosynthetic process (GO:0051188) | 5 | 0.93 | 5.39 | 2.95 × 10−3 | 3.83 × 10−2 |
| translational elongation (GO:0006414) | 10 | 3.35 | 2.98 | 2.51 × 10−3 | 3.44 × 10−2 |
| PANTHER GO-Slim Molecular Function | |||||
| mRNA 3′-UTR binding (GO:0003730) | 2 | 0.08 | 23.71 | 4.62 × 10−3 | 4.03 × 10−2 |
| heat shock protein binding (GO:0031072) | 5 | 0.27 | 18.24 | 1.63 × 10−5 | 2.79 × 10−4 |
| ATP binding (GO:0005524) | 5 | 0.41 | 12.16 | 9.32 × 10−5 | 1.38 × 10−3 |
| endopeptidase activity (GO:0004175) | 11 | 1.96 | 5.61 | 7.86 × 10−6 | 1.52 × 10−4 |
| ligase activity (GO:0016874) | 6 | 1.22 | 4.91 | 1.81 × 10−3 | 1.87 × 10−2 |
| PANTHER GO-Slim Cellular Component | |||||
| proteasome core complex (GO:0019773) | 5 | 0.08 | 59.27 | 1.44 × 10−7 | 3.69 × 10−6 |
| respiratory chain complex I (GO:0045271) | 3 | 0.32 | 9.48 | 4.87 × 10−3 | 4.34 × 10−2 |
| mitochondrial matrix (GO:0005759) | 4 | 0.5 | 8.07 | 1.98 × 10−3 | 2.19 × 10−2 |
| plastid part (GO:0044435) | 7 | 1.78 | 3.93 | 2.59 × 10−3 | 2.59 × 10−2 |
| cytosolic ribosome (GO:0022626) | 9 | 2.43 | 3.71 | 9.74 × 10−4 | 1.29 × 10−2 |
| PANTHER Protein Class | |||||
| chaperonin (PC00073) | 8 | 0.2 | 39.93 | 2.40 × 10−10 | 8.22 × 10−9 |
| acetyltransferase (PC00038) | 4 | 0.3 | 13.55 | 3.32 × 10−4 | 4.06 × 10−3 |
| epimerase/racemase (PC00096) | 3 | 0.26 | 11.38 | 3.04 × 10−3 | 3.25 × 10−2 |
| isomerase (PC00135) | 7 | 0.62 | 11.25 | 5.79 × 10−6 | 9.91 × 10−5 |
| chaperone (PC00072) | 14 | 1.31 | 10.71 | 2.25 × 10−10 | 9.60 × 10−9 |
| dehydrogenase (PC00092) | 17 | 2.35 | 7.23 | 7.56 × 10−10 | 2.15 × 10−8 |
| transaminase (PC00216) | 4 | 0.56 | 7.16 | 2.97 × 10−3 | 3.39 × 10−2 |
| translation factor (PC00223) | 8 | 1.21 | 6.6 | 4.69 × 10−5 | 6.68 × 10−4 |
| lyase (PC00144) | 12 | 2.34 | 5.13 | 7.27 × 10−6 | 1.13 × 10−4 |
| PANTHER GO-Slim Biological Process | Observed | Expected | Fold Change | p-Value | FDR |
|---|---|---|---|---|---|
| tricarboxylic acid cycle (GO:0006099) | 4 | 0.18 | 21.73 | 6.00 × 10−5 | 1.72 × 10−3 |
| reactive oxygen species metabolic process (GO:0072593) | 5 | 0.25 | 19.91 | 1.00 × 10−5 | 4.28 × 10−4 |
| glycolytic process (GO:0006096) | 5 | 0.33 | 15.32 | 3.15 × 10−5 | 1.07 × 10−3 |
| protein targeting to mitochondrion (GO:0006626) | 5 | 0.37 | 13.58 | 5.36 × 10−5 | 1.60 × 10−3 |
| PANTHER GO-Slim Molecular Function | |||||
| ATP binding (GO:0005524) | 4 | 0.33 | 12.26 | 4.43 × 10−4 | 7.59 × 10−3 |
| unfolded protein binding (GO:0051082) | 10 | 0.98 | 10.21 | 1.16 × 10−7 | 1.03 × 10−5 |
| endopeptidase activity (GO:0004175) | 11 | 1.56 | 7.07 | 8.70 × 10−7 | 4.30 × 10−5 |
| small molecule binding (GO:0036094) | 13 | 1.92 | 6.75 | 1.43 × 10−7 | 1.06 × 10−5 |
| oxidoreductase activity (GO:0016491) | 19 | 7.64 | 2.49 | 3.17 × 10−4 | 5.89 × 10−3 |
| PANTHER GO-Slim Cellular Component | |||||
| proteasome core complex (GO:0019773) | 6 | 0.07 | 89.62 | 8.74 × 10−10 | 7.17 × 10−8 |
| PANTHER Protein Class | |||||
| peroxidase (PC00180) | 3 | 0.11 | 27.57 | 2.92 × 10−4 | 6.24 × 10−3 |
| isomerase (PC00135) | 5 | 0.49 | 10.13 | 1.94 × 10−4 | 4.73 × 10−3 |
| lyase (PC00144) | 13 | 1.86 | 7 | 9.71 × 10−8 | 3.32 × 10−6 |
| dehydrogenase (PC00092) | 11 | 1.87 | 5.89 | 4.62 × 10−6 | 1.32 × 10−4 |
| oxidoreductase (PC00176) | 29 | 8.85 | 3.28 | 3.57 × 10−8 | 1.52 × 10−6 |
| PANTHER Pathways | |||||
| Glycolysis (P00024) | 4 | 0.19 | 20.78 | 7.00 × 10−5 | 3.75 × 10−3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zaini, P.A.; Feinberg, N.G.; Grilo, F.S.; Saxe, H.J.; Salemi, M.R.; Phinney, B.S.; Crisosto, C.H.; Dandekar, A.M. Comparative Proteomic Analysis of Walnut (Juglans regia L.) Pellicle Tissues Reveals the Regulation of Nut Quality Attributes. Life 2020, 10, 314. https://doi.org/10.3390/life10120314
Zaini PA, Feinberg NG, Grilo FS, Saxe HJ, Salemi MR, Phinney BS, Crisosto CH, Dandekar AM. Comparative Proteomic Analysis of Walnut (Juglans regia L.) Pellicle Tissues Reveals the Regulation of Nut Quality Attributes. Life. 2020; 10(12):314. https://doi.org/10.3390/life10120314
Chicago/Turabian StyleZaini, Paulo A., Noah G. Feinberg, Filipa S. Grilo, Houston J. Saxe, Michelle R. Salemi, Brett S. Phinney, Carlos H. Crisosto, and Abhaya M. Dandekar. 2020. "Comparative Proteomic Analysis of Walnut (Juglans regia L.) Pellicle Tissues Reveals the Regulation of Nut Quality Attributes" Life 10, no. 12: 314. https://doi.org/10.3390/life10120314
APA StyleZaini, P. A., Feinberg, N. G., Grilo, F. S., Saxe, H. J., Salemi, M. R., Phinney, B. S., Crisosto, C. H., & Dandekar, A. M. (2020). Comparative Proteomic Analysis of Walnut (Juglans regia L.) Pellicle Tissues Reveals the Regulation of Nut Quality Attributes. Life, 10(12), 314. https://doi.org/10.3390/life10120314