Abstract
Nostoc (Anabaena) sp. PCC 7120 is a filamentous cyanobacterial species that fixes N2 to nitrogenous compounds using specialised heterocyst cells. Changes in the intracellular ratio of carbon to nitrogen (C/N balance) is known to trigger major transcriptional reprogramming of the cell, including initiating the differentiation of vegetative cells to heterocysts. Substantial transcriptional analysis has been performed on Nostoc sp. PCC 7120 during N stepdown (low to high C/N), but not during C stepdown (high to low C/N). In the current study, we shifted the metabolic balance of Nostoc sp. PCC 7120 cultures grown at 3% CO2 by introducing them to atmospheric conditions containing 0.04% CO2 for 1 h, after which the changes in gene expression were measured using RNAseq transcriptomics. This analysis revealed strong upregulation of carbon uptake, while nitrogen uptake and metabolism and early stages of heterocyst development were downregulated in response to the shift to low CO2. Furthermore, gene expression changes revealed a decrease in photosynthetic electron transport and increased photoprotection and reactive oxygen metabolism, as well a decrease in iron uptake and metabolism. Differential gene expression was largely attributed to change in the abundances of the metabolites 2-phosphoglycolate and 2-oxoglutarate, which signal a rapid shift from fluent photoassimilation to glycolytic metabolism of carbon after transition to low CO2. This work shows that the C/N balance in Nostoc sp. PCC 7120 rapidly adjusts the metabolic strategy through transcriptional reprogramming, enabling survival in the fluctuating environment.
1. Introduction
Cyanobacteria use light energy to fix inorganic carbon (Ci) and nitrogen (N), harvested from their aquatic environment, into the metabolic components required for growth and propagation. Environmental sources of Ci include dissolved CO2 and bicarbonate (HCO3−), while N can be supplied by nitrate (NO3−), nitrite (NO2−), ammonium (NH4+), urea or N2 (in diazotrophic cyanobacteria; reviewed in [1]). The tight coupling of the concentrations of Ci and N taken up from the environment prevents metabolic imbalance within the cell, which allows cyanobacteria to thrive amidst varying nutritional conditions. This is achieved in large part by transcriptional modifications that are triggered by fluctuations in the cellular homeostasis of organic carbon (C) and N, which are represented by changes in the relative abundances of key metabolite signals (reviewed in [2,3,4]). One such metabolite is 2-oxoglutarate (2OG), also known as α-ketoglutarate (αKG), which is a product of the tricarboxylic acid (TCA) cycle. The metabolite 2OG provides the inorganic carbohydrate skeleton for glutamate synthesis that occurs by the incorporation of NH4+ in the glutamine synthetase/glutamine-oxoglutarate aminotransferase (GS/GOGAT) cycle. Cellular 2OG levels therefore represent the abundances of both C and N (reviewed in [5,6]), making 2OG a central signalling metabolite that triggers transcriptional adjustments to restore C/N balance [7,8,9]. 2-phosphoglycolate (2PG) is another metabolite that controls transcriptional reprogramming in response to C/N balance [10], and 2PG is formed when Rubisco catalyses the oxygenation of RuBP (photorespiration), instead of the favoured carboxylation reaction between RuBP and CO2 (reviewed in [4,11]). An increase in 2PG concentration therefore represents CO2 deficiency, triggering transcriptional reprogramming designed to upregulate Ci import into the cell [12,13,14,15].
Nostoc (Anabaena) sp. PCC 7120 is a filamentous, diazotrophic cyanobacterium wherein C/N balance controls the formation of heterocyst cells specialised for fixing N2 into NH4+, while photosynthetic CO2 fixation is restricted to vegetative cells (reviewed in [16,17]). In Nostoc sp. PCC 7120 and other heterocystous cyanobacteria, differentiation in cell structure and function is triggered by changes in C/N homeostasis and enacted by massive transcriptional reprogramming [1]. Emphasis on the impact of N concentration on heterocyst differentiation has revealed the central roles of 2OG and several regulatory proteins in instigating transcriptional and physiological responses to N deficit [6]. However, the transcriptional response of heterocystous cyanobacteria to Ci availability has drawn only little attention [18], in sharp contrast to that in non-diazotrophic species [12,13,15,19,20,21,22]. In the current study of the global transcriptome of Nostoc sp. PCC 7120, we found that a shift from 3% CO2 to 0.04% CO2 for 1 h can be largely attributed to changes in the levels of the metabolites 2PG and 2OG, which has a strong effect on genes involved in the import and metabolism of Ci and N. This study also identified that genes encoding factors involved in photosynthetic electron transport, glycolysis and iron homeostasis are regulated by C/N homeostasis, which is suggested to trigger a transition from efficient photoautotrophic growth and energy storage to photoinhibition and glycolysis.
2. Materials and Methods
2.1. Growth and CO2 Stepdown
Nostoc sp. PCC 7120 cultures were grown in BG11 medium [23] buffered with 10 mM TES-KOH (pH 8.0) at 30 °C under constant illumination of 50 μmol photons m−2 s−1 with 120 rpm agitation, in air enriched with 3% (v/v) CO2. During the exponential growth phase (OD750 = 1.0), 2 mL samples were taken from three individual replicate cultures and frozen for RNA isolation. For CO2 stepdown, the cultures were pelleted and the pellets washed once with fresh BG11, before resuspension in fresh BG11 and growth in air containing 0.04% (v/v) CO2. After 1 h, 2 mL samples were collected from three replicates and frozen for RNA isolation.
2.2. RNA Isolation and Transcriptomics
Total RNA was isolated as described in [24]. Total RNA samples were submitted to the Beijing Genomics Institute (China) for library construction and RNA sequencing using Illumina HiSeq2500. RNA reads were aligned using Strand NGS 2.7 software (Avadis) using the Nostoc sp. PCC 7120 reference genome and annotations downloaded from Ensembl (EBI). Aligned reads were normalised and quantified using the DESeq package (R). Significantly differentially expressed genes were identified using a 2-way ANOVA. p-Values were adjusted for false discovery rate (FDR) using the Benjamini–Hochberg procedure.
3. Results
The transcriptome of Nostoc sp. PCC 7120 grown in BG11 under 3% CO2-enriched air was compared with that of the same strain shifted from 3% CO2 to 0.04% CO2 for 1 h, revealing 230 genes to be upregulated >2-fold and 211 genes to be downregulated >2-fold, in the low CO2 condition. The RNAseq data are available at the NCBI Sequence Read Archive (submission SUB8244772). As expected, given the short period under a new metabolic condition, no differences in the growth rates, lengths of filaments or frequencies of heterocysts (approximately 4% of all cells under 3% CO2) were observed between the cultures exposed to 0.04% CO2 conditions, compared to those grown at 3% CO2. Therefore, statistical tests of these parameters in the different cultures were not performed.
3.1. Uptake and Metabolism of Carbon and Nitrogen Are Inversely Responsive to Low CO2 Conditions
The operons encoding three plasma membrane-localised HCO3− uptake systems were among the most strongly upregulated entities in Nostoc sp. PCC 7120 following CO2 stepdown (Table 1). In cyanobacteria, the Cmp (BCT1) system is powered by ATP hydrolysis [25], while the SbtA and BicA systems depend on Na+ ions for HCO3− symport [21,26]. The upregulation of the operon encoding the Mrp Na+:H+ antiporter upon CO2 stepdown may also be linked to HCO3− uptake through the extrusion of Na+ to support SbtA and BicA activity [27,28]. HCO3− uptake in cyanobacteria forms a major part of the carbon concentration mechanism (CCM), which also involves the concentration of cellular CO2 into HCO3− by a customised NAD(P)H dehydrogenase (NDH-1) complex [29]. Genes ndhF3, ndhD3 and cupA, which encode subunits that specialise the NDH1–MS complex for inducible CO2 uptake, were upregulated after CO2 stepdown (Table 1), as were several other genes encoding the core NDH–1M complex (see below). Notably, a putative cupS orthologue (alr1320), which is encoded separately from the ndhF3/ndhD3/cupA cluster in the Nostoc sp. PCC 7120 genome [30], was not differentially expressed (DE) in the current work. Genes encoding Rubisco and some carboxysome subunits were mildly upregulated in low CO2 (1.3 to 1.7 fold change (FC); not shown), while other CCM components were not DE, suggesting that these components were already in sufficient abundance before CO2 stepdown, whereas Ci uptake from the environment, especially HCO3− uptake, was apparently a primary concern for survival after 1 h under low CO2.

Table 1.
Differentially Expressed (DE) Genes Involved in Carbon and Nitrogen Metabolism.
The decrease in CO2 was found to cause downregulation of the nir operon, which encodes subunits of an ATP-dependent nitrate (NO3−) transporter, as well as NO3− and nitrite (NO2−) reductases [31,32]. The nir operon is responsible for NO3− uptake and reduction to ammonia (NH3), and is broadly conserved across cyanobacteria (reviewed in [5]). Unlike the nir operon, the majority of nif genes that encode subunits for the assembly and function of nitrogenase, which reduces atmospheric N2 to NH3, were not DE in the current work. Exceptions were nifB and nifH2, which were downregulated (Table 1). A gene that encodes a protein similar to the C-terminus of Mo-like nitrogenase (alr1713) was strongly downregulated, along with its neighbour (asr1714); however, the function of the encoded proteins is not known. Since heterocyst development is upregulated by a high C/N ratio (reviewed in [33]), it was not surprising to see many genes involved in the structural development of heterocysts repressed by the shift to low CO2. In particular, the hpd, hgl and dev gene clusters that encode many components for the synthesis and export of glycolipids, which form the oxygen-impermeable heterocyst envelope (reviewed in [34,35]), and were downregulated in the current data. Notably, the hep genes encoding heterocyst outer layer polysaccharides were only moderately downregulated by the shift to low CO2 (average FC −1.5, data not shown).
3.2. Expression of Genes Encoding Photosynthetic and Respiratory Components Responds to Low CO2 Conditions
Expression of genes encoding photosystem II (PSII) core proteins D1 and D2 was upregulated after the shift to low CO2 (Table 2), indicating an increase in the damage and turnover of PSII reaction centres [36,37]. In keeping with this, the genes encoding the two FtsH proteases involved in the degradation and turnover of damaged D1 protein were also upregulated [38,39]. We also found strongly induced expression of the flv2–flv4 operon, as well as several genes encoding orange carotenoid proteins (OCPs) and early light-inducible proteins (ELIPs), all of which are associated with PSII photoprotection [40,41,42,43,44]. These expression data suggest that the shift to low CO2 led to the over-reduction, damage and repair of PSII. Given this evidence for PSII over-reduction, it was surprising to find the gene encoding the “plastid” terminal oxidase (PTOX) as one of the most strongly downregulated in the current study (Table 2), being highly expressed under 3% CO2 and strongly repressed in 0.04% CO2. PTOX is part of a water–water cycle that moves electrons from reduced plastoquinone (PQ) to O2, and is thought to be an electron valve for balancing the photosynthetic redox state [45], which would presumably be important under low CO2 (discussed below).

Table 2.
Differentially Expressed (DE) Genes Involved in Photosynthesis and Respiration.
Virtually all genes encoding subunits of photosystem I (PSI) were substantially downregulated in the current study, which is in contrast to their increased expression in Synechocystis sp. PCC 6803 and unchanged expression Synechococcus elongatus PCC 7942 in low CO2 [13,20]. PSI downregulation in Nostoc sp. PCC 7120 points towards a decrease in PSI electron transport that may be related to the diminution of the terminal electron acceptor CO2. These conditions would also be expected to upregulate O2 reduction and the subsequent formation of toxic superoxide radicals (O2•−); indeed, genes encoding a superoxide dismutase (SodB; alr2938) and peroxiredoxin (all2375), involved in reactive oxygen species (ROS) scavenging, were upregulated 3.2-fold and 2.3-fold, respectively (not shown). The expression of isiB (alr2405), which encodes a flavodoxin (Fld) that accepts electrons from PSI via a flavin mononucleotide cofactor [46], was also upregulated after low CO2 treatment (Table 2), suggesting a shortage of oxidised ferredoxin (Fd) acceptors [47]. The upregulated expression of genes encoding F0-F1 ATP synthase points to an increased demand for energy in low CO2, required for HCO3− import and CO2 hydration (reviewed in [2]). Notably, the gene alr1004 encoding an enzyme that converts glyoxylate to glycine for the detoxification of 2PG [18] was found to be downregulated after CO2 stepdown, while other enzymes in the glycolate metabolism pathway were not DE.
The downregulation of PSI abundance in response to low CO2 can partially clarify the apparent PSII over-reduction discussed above. Lower PSI levels may also be linked to a decrease in the number of heterocysts, which contain a higher PSI:PSII ratio than in vegetative cells [16]. Although such a decrease in heterocysts was not observed after 1 h in low CO2, suppressed heterocyst development was evident in the downregulation of hgl and dev clusters (Table 1), and this can also explain the suppression of genes encoding cytochrome c6, Flv1B and Flv3B (Table 2) that operate predominately [48] or exclusively [49] in heterocysts. The gene encoding the small subunit of the heterocyst-specific uptake hydrogenase (HupS) was also downregulated here, reflecting the downregulation of the heterocystous nitrogenase activity under low CO2.
We observed downregulation of the pec cluster that encodes the phycoerythrocyanin (PEC) parts of the light-harvesting phycobilisome (PBS) complex [50,51], while genes encoding the other components of the PBS were not DE, indicating that the light-harvesting cross-section of PBS in Nostoc sp. PCC 7120 is altered in response to low CO2. An operon encoding a subunit of the light-independent protochlorophyllide reductase (DPOR) was also downregulated after 1 h in low CO2, while the expression of chlG and hemH genes, involved in later stages of chlorophyll and haem synthesis, respectively, were upregulated (Table 2).
Most genes encoding subunits of the NDH-1 complex were upregulated under low CO2 (Table 2), probably to fulfil their role in CO2 uptake as part of the NDH–1MS complex described above. In contrast, ndbA encoding NDH-2 was downregulated in the current study, suggesting a decrease in NDH-2-mediated respiration after the shift to low CO2. In addition, the gene encoding phosphoenolpyruvate (PEP) synthase, which converts pyruvate to PEP that is consumed in the TCA cycle, was strongly downregulated (Table 2). Many genes encoding subunits of the bidirectional hydrogenase (Hox) were among the most strongly downregulated in response to low CO2 conditions (Table 2), being both highly expressed in 3% CO2 and strongly repressed in low CO2. Hox reversibly catalyses the reduction of H+ to form H2, powered by reduced Fd/Fld with electrons derived from either PSI or pyruvate, the latter route by way of pyruvate ferredoxin/flavodoxin oxidoreductase (PFOR), which converts pyruvate to acetyl-CoA (reviewed in [52]). The nifJ gene encoding PFOR followed a similar expression profile to Hox subunits, being another of the most strongly downregulated genes in the current study. The physiological role of the Hox enzyme is not known, but has been described as an electron valve that can maintain redox balance and store reducing power as H2 during excess photosynthesis or fermentation [52,53,54,55]. The expression profile of Hox and PFOR genes suggests that pyruvate-powered hydrogen production is active under 3% CO2 and inactivated by the shift to low CO2.
3.3. Expression of Transcription Regulators Responds to Changes in CO2 Conditions
The current study revealed substantial changes in the expression of several genes encoding transcription regulators, providing evidence of an ongoing cascade of transcriptional reprogramming after 1 h under low CO2 (Table 3). Upregulated expression of the LysR-type regulator (LTTR) cmpR corresponds to the strong upregulation of its target, the cmp cluster encoding the BCT1 HCO3− transporter (Table 1), as previously shown in Synechocystis sp. PCC 6803, Synechococcus sp. PCC 7942 [56] and Nostoc sp. PCC 7120 [57]. Two sigB-type group 2 sigma factors, which have roles in the transcriptional response to low CO2 and C/N balance [58,59,60], were upregulated after the shift to low CO2 (Table 3). SigB has also been implicated in response to environmental stress and resistance to photoinhibition in Synechocystis sp. PCC 6803 [61,62], which is in line with the upregulation in this study of groES and groEL genes (4.3 to 5.5 FC; not shown) and photoprotective factors such as OCPs and the flv2–flv4 operon (Table 2). Two homologous two-component response regulator clusters, which each comprise a histidine kinase and a DNA-binding regulator, were found to be upregulated by low CO2. Of the two, the chromosomal hik31 (C-hik31) operon was more highly upregulated, and has been found to be involved in the responses to oxygen concentration, light and metabolism [63,64]. A TetR-family transcription regulator with unknown function was also upregulated by low CO2 (Table 3).

Table 3.
Differentially Expressed (DE) Genes Encoding Transcription Regulators.
Another LTTR gene that was highly expressed under high CO2 and was strongly downregulated after low CO2 transition (Table 3) shared substantial sequence homology with the ndhR transcription repressor (also called ccmR) of Synechocystis sp. PCC 6803 [12,27]. In other cyanobacteria, NdhR represses the expression of CCM genes, including the sbt and bicA HCO3− importers, the mrp cluster and the ndhF3/ndhD3/cupA cluster [12,15,20,27,65]. The rapid downregulation of a putative ndhR orthologue in Nostoc sp. PCC 7120 may reveal the mechanism behind the strong upregulation of CCM genes after CO2 stepdown in the current study (Table 1). Downregulation of the transcription enhancer ntcB, which increases N metabolism through upregulation of the nir operon [66], also correlates with downregulation of other N-related genes in the current work (Table 1). Expression of ntcB is controlled by NtcA [1]. As ntcA was not DE in the current study, downregulation of ntcB and other NtcA regulons may be due to the low CO2-induced inactivation of NtcA (discussed below). Similarly, downregulated transcriptional regulator genes patB (also called cnfR), devH and nrrA can also be attributed to inhibited NtcA activity [67,68,69,70,71]. These genes are expressed in heterocysts, where PatB upregulates the expression of nifB [72], DevH regulates heterocyst glycolipids [48,68,73] and NrrA induces expression of both the heterocyst regulator hetR and fraF encoding a filament integrity protein [70,74]. Both nifB and the hgl cluster were downregulated in this study (Table 1), while hetR and fraF were not DE (not shown).
3.4. Low CO2 Conditions Influence the Expression of Metal Homeostasis Genes
A number of genes and gene clusters related to cellular homeostasis of iron (Fe) and other metals were found to be DE after CO2 stepdown (Table 4). A gene cluster encoding subunits of a periplasmic ferrous Fe (Fe(II)) transporter [75,76] was strongly downregulated in the current study (Table 4), indicating a lower uptake of Fe from the environment under low CO2. The suf cluster, encoding proteins involved in Fe mobilization and Fe–S cluster assembly, was also downregulated. The expression of both the Fe(II) transporter and the suf cluster is upregulated by Fe deprivation [77,78,79], suggesting a surplus of cellular Fe after low CO2 treatment. Several neighbouring clusters of genes encoding subunits of metal cation efflux systems such as copper, nickel, zinc, cadmium and cobalt, were upregulated after low CO2 treatment (Table 4). These genes have been implicated in heavy metal resistance [80,81], although the link to the current conditions is not clear. Taken together, low CO2 appears to induce an active decrease in cellular metal content, which may be a strategy to avoid oxidative stress during the reducing conditions induced by an insufficient availability of photosynthetic electron acceptors.

Table 4.
Differentially Expressed (DE) Genes Involved in the Transport and Metabolism of Metals.
4. Discussion
4.1. Transcriptional Regulation in Response to CO2 Stepdown is Triggered by Metabolites
Induction of the most strongly upregulated genes, the HCO3− transporters (Table 1), after CO2 stepdown, suggests a rapid transcriptional response that is highly sensitive to cellular Ci levels. In some unicellular cyanobacteria, repression of the CCM genes by NdhR (also called CcmR) can be modulated by both 2OG and 2PG [12,15,65,82]. Increased cellular 2PG concentration caused by increased photorespiration in low CO2 leads to increased abundance of the NdhR–2PG complex that is unable to bind DNA to repress expression [4]. Additionally, declining 2OG levels due to lower CO2 fixation and potentially lower TCA cycle activity decrease the abundance of the NdhR-2OG repressor complex, although it is unclear whether 2OG levels would actually decrease after only 1 h in low CO2 due to the mobilisation of stored glycogen into the TCA cycle [2,15,82,83]. NADP+, another co-repressor of NdhR [82], is also theoretically far less abundant after CO2 stepdown, due to decreased CO2 fixation and lower NADPH consumption despite constant light conditions. Although a putative NdhR in Nostoc sp. PCC 7120 has not been studied, it appears that the LTTR encoded by all4986 represents such an orthologue, and that the strong downregulation of all4986 after CO2 stepdown led to de-repression of the NdhR regulon, which includes the ndhR gene itself [84]. Previous transcriptomics studies have shown ndhR expression to be upregulated in Synechocystis sp. PCC 6803 after >3 h in low Ci conditions [12,13], which is in contrast to the strong downregulation of all4986 seen here after 1 h (Table 3). This suggests that NdhR de-repression in response to CO2 stepdown may be transient, and/or that expression of the NdhR regulon is also controlled by other transcription factors [18]. In the current study, de-repression by the putative NdhR is proposed to have caused a rapid and strong increase in HCO3− and CO2 uptake under C limitation, with the upregulated cmp operon (Table 1) inducing further increase in HCO3− uptake. The cmp inducer CmpR is activated by 2PG or RuBP [82,85], both of which are in higher concentrations after CO2 stepdown due to decreased CO2 fixation. Furthermore, cmpR expression is also auto-upregulated (Table 3) [57]. In Synechococcus sp. PCC 7942 CmpR additionally upregulates the expression of PSII core subunits [86,87], found upregulated also in the current study alongside factors for PSII photoprotection and turnover, and downregulation of most PSI genes (Table 2). The overlap between cellular responses to either low CO2 or high light stress is well documented [12,20,86,88], highlighting insufficient electron sinks similarly created by both high light and low CO2, and resulting in the over-reduction of photosynthetic electron carriers [2]. Notably, several PSII photoprotection factors encoded by genes that were DE in the current study, including psbAIII, flv4 and sodB, were shown to be regulated together with Rubisco and other CCM genes by another LTTR in Nostoc sp. PCC 7120 called PacR [18], suggesting the likely involvement of PacR in the transcriptional reprogramming seen here.
In addition to the LTTR transcription factors, the transcriptional response to low CO2 is also regulated by LexA and the cyAbrB paralogues [89,90,91,92]. The vast change in expression of the hox operon after CO2 stepdown (Table 2) may be related to the activity of LexA [93] and/or cyAbrB [90,94,95,96]. In Synechocystis sp. PCC 6803, cyAbrB2 controls the expression of many CCM components that were likewise found to be upregulated in this study (Table 1) [90], while the cyAbrB2 orthologue in Nostoc sp. PCC 7120 regulates the expression of FeSOD [96], also upregulated here. Nostoc sp. PCC 7120 cyAbrB1 has been recently implicated in transcriptional regulation of heterocyst differentiation [97], demonstrating a role close to the interface of Ci and N availability that suggests cyAbrB1/2 were likely active in the transcription regulation observed in the current study.
Given the transcriptional activation of NtcA, the master regulator of N metabolism, by high levels of 2OG upon a shift to low N (high C/N ratio) [7,98,99], it is widely assumed that a decrease in CO2 leads to a decline in NtcA activity by decreasing the abundance of the 2OG–NtcA–PipX activator complex [100]. In the current study, lower NtcA activity was indeed evident in the downregulation of nir genes encoding NO3− uptake and metabolism (Table 1), as well as downregulation of the nir co-activator ntcB (Table 3; reviewed in [101]). This transcriptional regulation would effectively bring N metabolism into alignment with decreased C metabolism after CO2 stepdown, despite the presence of N sources in the BG11 media. Overall, nearly 50% of genes downregulated in the current study have NtcA-binding promoters [71], although many NtcA-regulated genes, such as those involved in the uptake of NH3 and urea, regulation of GS-GOGAT enzymes, as well as NtcA itself [71,102,103,104], were not DE after 1 h in low CO2. The current work may therefore include only the early NtcA regulon. The NtcA-activated differentiation of vegetative cells to heterocysts occurs over approximately 24 h, via a cascade of transcriptional regulation that includes early upregulation of the co-activator nrrA [33]. The rapid downregulation in the current work of some heterocyst regulators, including nrrA, may block the commencement of heterocyst differentiation in response to relative N excess over C after CO2 stepdown, and may signal an eventual decrease in the small number of heterocysts that are known to occur under high C/N [105]. Downregulation of NtcA activity in the current study appears to indicate a decline in 2OG levels after only 1 h in low CO2, although an increased concentration of NH3 derived from 2PG metabolism can also explain suppression the NtcA regulon under low CO2 (low C/N ratio) [83]. N excess under low CO2 is also evident in the −2.2 FC downregulated expression of cyanophycinase chb2 (all0571; not shown), which is regulated by NrrA, suggesting a decrease in the mobilisation of stored N under low CO2 (reviewed in [106]). It can also be argued that an increase in the ADP/ATP ratio under CO2 stepdown, caused by decreased photosynthetic electron transport and rapid changes in metabolism, increases the abundance of both the ADP–PII–PipX complex and the inactive form of NtcA [6].
The strong downregulation of operons involved in ferrous Fe import and Fe–S cluster assembly in the current work (Table 4) suggests a connection between cellular Fe homeostasis and C/N balance in Nostoc sp. PCC 7120, which has been explored [107]. The current results indicate a CO2 stepdown-induced cellular excess of Fe and other transition metals, which may be due to downregulation of Fe-rich PSI complexes (Table 2) [108] and/or an excess of reductant caused by insufficient electron sinks. Both conditions pose the danger of ROS formation, evidenced by upregulated expression of SOD, peroxiredoxin and protein chaperones.
4.2. Altered C/N Balance Modulates the Energetic Strategy of Nostoc sp. PCC 7120
The current study shows that a stepdown from 3% CO2 in enriched air to 0.04% CO2 (atmospheric) for just 1 h led to substantial reprogramming of global gene expression in Nostoc sp. PCC 7120 cultures. As discussed above, most of the transcriptional changes observed here can be directly attributed to metabolite signalling instigated by the alteration of the cellular C/N balance (Figure 1), initiated by the decrease in CO2 concentration. Furthermore, these results highlight rapid transcriptional reprogramming of photosynthesis and energy metabolism in Nostoc sp. PCC 7120 in response to CO2 levels (Figure 2). High CO2 in light promotes a high rate of photosynthesis and the storage of photosynthate in the form of glycogen [13,109,110,111]. Strong expression of PEP synthase and PFOR under these conditions indicate glycolysis/gluconeogenesis through the metabolism of pyruvate, which is consumed in the TCA cycle to facilitate respiratory electron transport and to provide 2OG for amino acid synthesis (reviewed in [112,113]). Therefore, growth under 3% CO2 somewhat resembles photomixotrophy, with photosynthetic and glycolytic metabolisms occurring concomitantly, even though glucose was not externally provided to cultures. Under these conditions, the cells experience a high C/N, which is evident in the relatively high expression of N metabolic genes, reflecting a cellular excess of 2OG [7,98,99]. In high CO2, strong expression of Hox, which is important under mixotrophy and N deprivation [55], and PTOX, may provide a system to maintain redox poise [45], and in the case of Hox, also store surplus energy as H2 [53]. The transfer of Synechocystis sp. PCC 6803 and Synechococcus elongatus PCC 7942 cultures from high to low CO2 showed that the toxic effects of 2PG transiently block Calvin–Benson–Bassham (CBB) activity [15,20,83] and this is also evident here in the upregulated expression of PSII repair, photoprotection and ROS scavenging enzymes in Nostoc sp. PCC 7120, which indicate over-reduction of the photosynthetic electron transport chain. Interestingly, detoxification of 2PG appeared to be downregulated through repression of alr1004 during CO2 stepdown (Table 2), perhaps highlighting the importance of the metabolite for signalling during the early stages of Ci deprivation. We also observed downregulation of the terminal proteins in the phycobilisomes, suggesting modified harvesting of light energy to alleviate excitation pressure on the photosynthetic system. Under these conditions, inhibition of photoassimilation is compensated by glycolysis of stored glucose and CBB intermediates, providing an important supply of substrates for anaplerosis of the CBB and TCA cycles during acclimation to the transition [83,114,115]. In the current work, enhanced glycolytic activity is indicated by the upregulation of NDH-1, suggesting an increased reliance on respiratory electron transport for ATP generation, while strong downregulation of PEP synthase and PFOR after CO2 stepdown may prevent diversion of pyruvate away from the TCA cycle. Notably, PEP abundance increased substantially in Synechocystis sp. PCC 6803 and Synechococcus elongatus PCC 7942 after a shift from high to low CO2 [20,83], while genes encoding both pyruvate kinase and PFOR were highly expressed in Synechococcus elongatus PCC 7942 after long-term acclimation to low CO2, but not directly after the transition [20]. These findings support the results of this study and suggest that the metabolism of PEP and pyruvate are tightly regulated after the transition to low CO2. This may be linked to the role of PEP and pyruvate as substrates to anaplerotic carbon fixation to produce TCA cycle intermediates oxaloacetate and malate (reviewed in [116]). During the adjustment to a low C/N ratio, TCA cycle activity generates 2OG for amino acid synthesis, utilising excess NH4+ generated through 2PG detoxification [11,83].
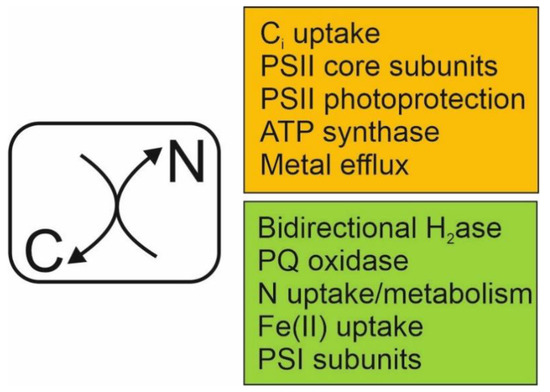
Figure 1.
The transcriptional response of Nostoc sp. PCC 7120 cells to a change in the cellular concentrations of carbon and nitrogen (C/N balance).Decrease in the external CO2 concentration of cultures causes a decline in the cellular C/N balance signalled by an increased production of 2PG and decreased 2OG levels relative to N. This metabolic change leads to upregulation of genes encoding processes depicted in orange, and downregulation of genes encoding processes depicted in green.
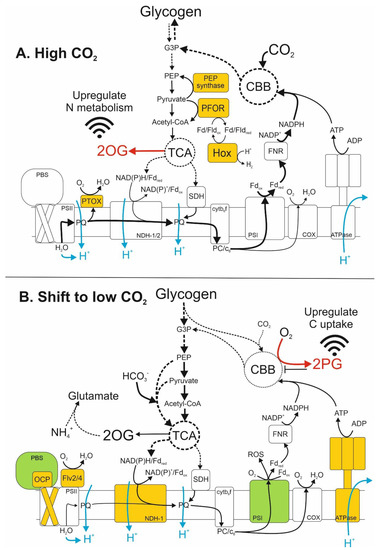
Figure 2.
Schematic representation of the interactions between photosynthetic/respiratory electron transport and accumulation of 2OG and 2PG for signalling the C/N balance in Nostoc sp. PCC 7120 cells. Scheme is based on global transcriptomic profiling of Nostoc sp. PCC 7120 cultures under high CO2 conditions and after CO2 stepdown. (A) Under 3% CO2, efficient photosynthetic electron transport and Calvin–Benson–Bassham (CBB) cycle activity enables gluconeogenesis of glyceraldehyde-3-phosphate (G3P), leading to accumulation of carbohydrate storage in the form of glycogen. Glycolysis of glycogen stores and/or photosynthate supplies pyruvate to the incomplete tricarboxylic acid (TCA) cycle, which produces reductant, ATP and succinate to drive respiratory electron transport through NAD(P)H-dehydrogenase (NDH) and succinate dehydrogenase (SDH), as well as other cellular processes. The TCA cycle also generates 2-oxoglutarate (2OG), which accumulates under high CO2 due to a relative shortage of NH4+ and glutamine. 2OG operates as a signal for upregulating genes involved in N uptake and metabolism. Strong expression of phosphoenolpyruvate (PEP) synthase converts pyruvate to PEP. Pyruvate:ferredoxin/flavodoxin oxidoreductase (PFOR) converts pyruvate to acetyl-CoA, reducing oxidised ferredoxin/flavodoxin (Fd/Flvox) that is consumed by bidirectional hydrogenase (Hox) for storage of excess energy as H2. Strong expression of plastoquinone terminal oxidase (PTOX) maintains redox homeostasis of the plastoquinone (PQ) pool during high photosynthetic electron transport, while cytochrome c6 oxidase (COX) maintains the redox poise of lumenal electron carriers cytochrome c6 (c6) and plastocyanin (PC). Genes encoding factors coloured orange are highly expressed under 3% CO2 and strongly downregulated by the shift to 0.04% CO2. (B) After 1 h in 0.04% CO2, a deficiency of CO2 electron acceptors leads to oxygenation of Rubisco, causing photorespiration that produces 2-phosphoglycolate (2PG). The CBB cycle and other metabolic pathways are inhibited by 2PG, which also signals upregulation of the transcriptomic response to low CO2. Low CBB activity causes over-reduction of the photosynthetic electron transport chain, leading to the production of reactive oxygen species (ROS) at photosystem I (PSI) and downregulation of PSI subunits. Increased reducing pressure on photosystem II (PSII) also causes upregulation of the PSII repair cycle and upregulation of PSII photoprotection by flavoproteins (Flv2/4) and orange carotenoid proteins (OCPs), as well as downregulation of phycoerythrocyanin in the phycobilisome (PBS). Glycolysis triggered by decreased CO2 assimilation provides substrates for anaplerotic supplementation of the CBB and TCA cycles, including the production of oxaloacetate from PEP and bicarbonate (HCO3−). TCA cycle activity also produces 2OG for glutamate production from excess ammonium (NH4+) resulting from the shift from 3% to 0.04% CO2. Genes encoding factors coloured orange or green are upregulated or downregulated, respectively, 1 h after the shift from 3% to 0.04% CO2. Black arrows indicate the movement of electrons or ATP, dashed arrows summarise multiple enzymatic reactions in carbohydrate metabolism, blue arrows indicate the movement of protons. The red arrow in (A) shows the production of 2OG by TCA cycle activity, the red arrow in (B) shows the production of 2PG by Rubisco oxygenation in the CBB cycle and the black bar in (B) indicates CBB inhibition by 2PG.
This work has revealed rapid transcriptional reprogramming in Nostoc sp. PCC 7120 in response to a decrease in Ci availability, namely strong upregulation of CCM components and photoprotection, and downregulation of N uptake and early stages of heterocyst differentiation. Despite the vast increase in HCO3− uptake, glycolysis of stored C apparently plays an important role in energy metabolism at low CO2, likely due to 2PG-induced inhibition of the CBB cycle. A majority of the transcriptional effects induced by low CO2 in Nostoc sp. PCC 7120 can be attributed to 2PG modulation of CmpR and a putative NdhR homologue; however, the effects of changing abundance of 2OG and NtcA activity after 1 h in 0.04% CO2, as well as the roles of other transcriptional regulators cannot be discounted. Finally, this work highlights the sensitivity of Nostoc sp. PCC 7120 to factors that influence the cellular C/N balance and demonstrates the speed at which genetic and metabolic reprogramming can take place, allowing rapid acclimation for surviving and thriving in the fluctuating environment.
Author Contributions
Conceptualization, P.J.G., E.-M.A.; investigation and analysis, P.J.G.; original draft preparation, P.J.G.; writing—review and editing, P.J.G., D.M.-P., E.-M.A.; funding acquisition, P.J.G., E.-M.A. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Academy of Finland, projects 26080341 (P.J.G.) and 307335 (E.-M.A.), and the Jane and Aatos Erkko Foundation (E.-M.A.).
Acknowledgments
The authors acknowledge Julia Walter for the growth of cultures and the isolation of RNA used in this study.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Herrero, A.; Flores, E. Genetic responses to carbon and nitrogen availability in Anabaena. Environ. Microbiol. 2019, 21, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Burnap, R.L.; Hagemann, M.; Kaplan, A. Regulation of CO2 Concentrating Mechanism in Cyanobacteria. Life 2015, 5, 348–371. [Google Scholar] [CrossRef] [PubMed]
- Huergo, L.F.; Dixon, R. The Emergence of 2-Oxoglutarate as a Master Regulator Metabolite. Microbiol. Mol. Biol. Rev. 2015, 79, 419–435. [Google Scholar] [CrossRef]
- Zhang, C.-C.; Zhou, C.-Z.; Burnap, R.L.; Peng, L. Carbon/Nitrogen Metabolic Balance: Lessons from Cyanobacteria. Trends Plant Sci. 2018, 23, 1116–1130. [Google Scholar] [CrossRef]
- Flores, E.; Frías, J.E.; Rubio, L.M.; Herrero, A. Photosynthetic nitrate assimilation in cyanobacteria. Photosynth. Res. 2005, 83, 117–133. [Google Scholar] [CrossRef]
- Forchhammer, K.; Selim, K.A. Carbon/nitrogen homeostasis control in cyanobacteria. FEMS Microbiol. Rev. 2019, 44, 33–53. [Google Scholar] [CrossRef]
- Muro-Pastor, M.I.; Reyes, J.C.; Florencio, F.J. Cyanobacteria perceive nitrogen status by sensing intracellular 2-oxoglutarate levels. J. Biol. Chem. 2001, 276, 38320–38328. [Google Scholar] [PubMed]
- Vázquez-Bermúdez, M.F.; Herrero, A.; Flores, E. 2-Oxoglutarate increases the binding affinity of the NtcA (nitrogen control) transcription factor for the Synechococcus glnA promoter. FEBS Lett. 2002, 512, 71–74. [Google Scholar] [CrossRef]
- Li, J.-H.; Laurent, S.; Konde, V.; Bédu, S.; Zhang, C.-C. An increase in the level of 2-oxoglutarate promotes heterocyst development in the cyanobacterium Anabaena sp. strain PCC 7120. Microbiology 2003, 149, 3257–3263. [Google Scholar] [CrossRef]
- Haimovich-Dayan, M.; Lieman-Hurwitz, J.; Orf, I.; Hagemann, M.; Kaplan, A. Does 2-phosphoglycolate serve as an internal signal molecule of inorganic carbon deprivation in the cyanobacterium Synechocystis sp. PCC 6803? Environ. Microbiol. 2015, 17, 1794–1804. [Google Scholar] [CrossRef]
- Hagemann, M.; Bauwe, H. Photorespiration and the potential to improve photosynthesis. Curr. Opin. Chem. Biol. 2016, 35, 109–116. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.-L.; Postier, B.L.; Burnap, R.L. Alterations in Global Patterns of Gene Expression in Synechocystis sp. PCC 6803 in Response to Inorganic Carbon Limitation and the Inactivation of ndhR, a LysR Family Regulator. J. Biol. Chem. 2004, 279, 5739–5751. [Google Scholar] [CrossRef] [PubMed]
- Eisenhut, M.; Von Wobeser, E.A.; Jonas, L.; Schubert, H.; Ibelings, B.W.; Bauwe, H.; Matthijs, H.C.; Hagemann, M. Long-Term Response toward Inorganic Carbon Limitation in Wild Type and Glycolate Turnover Mutants of the Cyanobacterium Synechocystis sp. Strain PCC 6803. Plant Physiol. 2007, 144, 1946–1959. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, A.; Hagemann, M.; Bauwe, H.; Kahlon, S.; Ogawa, T. Carbon acquisition by cyanobacteria: Mechanisms, comparative genomics and evolution. In The Cyanobacteria: Molecular Biology, Genomics and Evolution; Herrero, A., Flores, E., Eds.; Caister Academic Press: Norwich, UK, 2008; pp. 305–323. [Google Scholar]
- Klähn, S.; Orf, I.; Schwarz, D.; Matthiessen, J.K.; Kopka, J.; Hess, W.R.; Hagemann, M. Integrated Transcriptomic and Metabolomic Characterization of the Low-Carbon Response Using an ndhR Mutant of Synechocystis sp. PCC 68031. Plant Physiol. 2015, 169, 1540–1556. [Google Scholar] [CrossRef]
- Wolk, C.P.; Ernst, A.; Elhai, J. Heterocyst Metabolism and Development. In The Molecular Biology of Cyanobacteri; Springer: Dordrecht, The Netherlands, 1994; pp. 769–823. [Google Scholar] [CrossRef]
- Herrero, A.; Muro-Pastor, A.M.; Valladares, A.; Flores, E. Cellular differentiation and the NtcA transcription factor in filamentous cyanobacteria. FEMS Microbiol. Rev. 2004, 28, 469–487. [Google Scholar] [CrossRef]
- Picossi, S.; Flores, E.; Herrero, A. The LysR-type transcription factor PacR is a global regulator of photosynthetic carbon assimilation inAnabaena. Environ. Microbiol. 2015, 17, 3341–3351. [Google Scholar] [CrossRef]
- Allahverdiyeva, Y.; Vainonen, J.P.; Vorontsova, N.; Keränen, M.; Carmel, D.; Aro, E.-M. Dynamic Changes in the Proteome of Synechocystis 6803 in Response to CO2 Limitation Revealed by Quantitative Proteomics. J. Proteome Res. 2010, 9, 5896–5912. [Google Scholar] [CrossRef]
- Schwarz, D.; Nodop, A.; Hüge, J.; Purfürst, S.; Forchhammer, K.; Michel, K.-P.; Bauwe, H.; Kopka, J.; Hagemann, M. Metabolic and Transcriptomic Phenotyping of Inorganic Carbon Acclimation in the Cyanobacterium Synechococcus elongatus PCC 7942. Plant Physiol. 2011, 155, 1640–1655. [Google Scholar] [CrossRef]
- Price, G.D.; Woodger, F.J.; Badger, M.R.; Howitt, S.M.; Tucker, L. Identification of a SulP-type bicarbonate transporter in marine cyanobacteria. Proc. Natl. Acad. Sci. USA 2004, 101, 18228–18233. [Google Scholar] [CrossRef]
- McGinn, P.J.; Price, G.D.; Maleszka, R.; Badger, M.R. Inorganic Carbon Limitation and Light Control the Expression of Transcripts Related to the CO2-Concentrating Mechanism in the Cyanobacterium Synechocystis sp. Strain PCC6803. Plant Physiol. 2003, 132, 218–229. [Google Scholar] [CrossRef]
- Rippka, R.; Stanier, R.Y.; Deruelles, J.; Herdman, M.; Waterbury, J.B. Generic Assignments, Strain Histories and Properties of Pure Cultures of Cyanobacteria. Microbiology 1979, 111, 1–61. [Google Scholar] [CrossRef]
- Walter, J.; Lynch, F.; Battchikova, N.; Aro, E.-M.; Gollan, P.J. Calcium impacts carbon and nitrogen balance in the filamentous cyanobacterium Anabaena sp. PCC 7120. J. Exp. Bot. 2016, 67, 3997–4008. [Google Scholar] [CrossRef] [PubMed]
- Omata, T.; Price, G.D.; Badger, M.R.; Okamura, M.; Gohta, S.; Ogawa, T. Identification of an ATP-binding cassette transporter involved in bicarbonate uptake in the cyanobacterium Synechococcus sp. strain PCC 7942. Proc. Natl. Acad. Sci. USA 1999, 96, 13571–13576. [Google Scholar] [CrossRef] [PubMed]
- Shibata, M.; Katoh, H.; Sonoda, M.; Ohkawa, H.; Shimoyama, M.; Fukuzawa, H.; Kaplan, A.; Ogawa, T. Genes Essential to Sodium-dependent Bicarbonate Transport in Cyanobacteria: Function and phylogenetic analysis. J. Biol. Chem. 2002, 277, 18658–18664. [Google Scholar] [CrossRef] [PubMed]
- Blanco-Rivero, A.; Leganés, F.; Fernández-Valiente, E.; Calle, P.; Fernández-Piñas, F. mrpA, a gene with roles in resistance to Na+ and adaptation to alkaline pH in the cyanobacterium Anabaena sp. PCC7120. Microbiology 2005, 151, 1671–1682. [Google Scholar] [CrossRef] [PubMed]
- Fukaya, F.; Promden, W.; Hibino, T.; Tanaka, Y.; Nakamura, T.; Takabe, T. An Mrp-like cluster in the halotolerant cyanobacterium Aphanothece halophytica functions as a Na+/H+ antiporter. Appl. Environ. Microbiol. 2009, 75, 6626–6629. [Google Scholar] [CrossRef]
- Allahverdiyeva, Y.; Aro, E.-M.; Nixon, P.J. Structure and Physiological Function of NDH-1 Complexes in Cyanobacteria. In Bioenergetic Processes of Cyanobacteria; Springer: Dordrecht, The Netherlands, 2011; pp. 445–467. [Google Scholar] [CrossRef]
- Kaneko, T.; Nakamura, Y.; Wolk, C.P.; Kuritz, T.; Sasamoto, S.; Watanabe, A.; Iriguchi, M.; Ishikawa, A.; Kawashima, K.; Kimura, T.; et al. Complete Genomic Sequence of the Filamentous Nitrogen-fixing Cyanobacterium Anabaena sp. Strain PCC 7120. DNA Res. 2001, 8, 205–213. [Google Scholar] [CrossRef]
- Frías, J.E.; Flores, E.; Herrero, A. Nitrate assimilation gene cluster from the heterocyst-forming cyanobacterium Anabaena sp. strain PCC 7120. J. Bacteriol. 1997, 179, 477–486. [Google Scholar] [CrossRef]
- Frías, J.E.; Flores, E. Negative Regulation of Expression of the Nitrate Assimilation nirA Operon in the Heterocyst-Forming Cyanobacterium Anabaena sp. Strain PCC 7120. J. Bacteriol. 2010, 192, 2769–2778. [Google Scholar] [CrossRef]
- Flores, E.; Picossi, S.; Valladares, A.; Herrero, A. Transcriptional regulation of development in heterocyst-forming cyanobacteria. Biochim. Biophys. Acta Bioenergy 2019, 1862, 673–684. [Google Scholar] [CrossRef]
- Kumar, K.; Mella-Herrera, R.A.; Golden, J.W. Cyanobacterial Heterocysts. Cold Spring Harb. Perspect. Biol. 2010, 2, a000315. [Google Scholar] [CrossRef] [PubMed]
- Maldener, I.; Muro-Pastor, A.M. Cyanobacterial Heterocysts. In Encyclopedia of Life Sciences; John Wiley & Sons, Ltd.: Chichester, UK, 2010. [Google Scholar] [CrossRef]
- Aro, E.-M.; Virgin, I.; Andersson, B. Photoinhibition of Photosystem II. Inactivation, protein damage and turnover. Biochim. Biophys. Acta Bioenergy 1993, 1143, 113–134. [Google Scholar] [CrossRef]
- Komenda, J.; Hassan, H.A.; Diner, B.A.; Debus, R.J.; Barber, J.; Nixon, P.J. Degradation of the Photosystem II D1 and D2 proteins in different strains of the cyanobacterium Synechocystis PCC 6803 varying with respect to the type and level of psbA transcript. Plant Mol. Biol. 2000, 42, 635–645. [Google Scholar] [CrossRef]
- Silva, P.; Thompson, E.P.; Bailey, S.; Kruse, O.; Mullineaux, C.W.; Robinson, C.; Mann, N.H.; Nixon, P.J. FtsH Is Involved in the Early Stages of Repair of Photosystem II in Synechocystis sp PCC 6803. Plant Cell 2003, 15, 2152–2164. [Google Scholar] [CrossRef] [PubMed]
- Komenda, J.; Barker, M.; Kuviková, S.; De Vries, R.; Mullineaux, C.W.; Tichý, M.; Nixon, P.J. The FtsH Protease slr0228 Is Important for Quality Control of Photosystem II in the Thylakoid Membrane of Synechocystis sp. PCC 6803. J. Biol. Chem. 2006, 281, 1145–1151. [Google Scholar] [CrossRef] [PubMed]
- Eisenhut, M.; Georg, J.; Klähn, S.; Sakurai, I.; Mustila, H.; Zhang, P.; Hess, W.R.; Aro, E.-M. The Antisense RNA As1_flv4 in the Cyanobacterium Synechocystis sp. PCC 6803 Prevents Premature Expression of the flv4-2 Operon upon Shift in Inorganic Carbon Supply. J. Biol. Chem. 2012, 287, 33153–33162. [Google Scholar] [CrossRef]
- Bersanini, L.; Battchikova, N.; Jokel, M.; Rehman, A.; Vass, I.; Allahverdiyeva, Y.; Aro, E.M. Flavodiiron protein Flv2/Flv4-related photoprotective mechanism dissipates excitation pressure of PSII in cooperation with phycobilisomes in cyanobacteria. Plant Physiol. 2014, 164, 805–818. [Google Scholar] [CrossRef]
- Gwizdala, M.; Wilson, A.; Kirilovsky, D. In Vitro Reconstitution of the Cyanobacterial Photoprotective Mechanism Mediated by the Orange Carotenoid Protein in Synechocystis PCC 6803. Plant Cell 2011, 23, 2631–2643. [Google Scholar] [CrossRef]
- López-Igual, R.; Wilson, A.; Leverenz, R.L.; Melnicki, M.R.; De Carbon, C.B.; Sutter, M.; Turmo, A.; Perreau, F.; Kerfeld, C.A.; Kirilovsky, D. Different Functions of the Paralogs to the N-Terminal Domain of the Orange Carotenoid Protein in the Cyanobacterium Anabaena sp. PCC 7120. Plant Physiol. 2016, 171, 1852–1866. [Google Scholar] [CrossRef]
- Kufryk, G.; Hernandez-Prieto, M.A.; Kieselbach, T.; Miranda, H.; Vermaas, W.; Funk, C. Association of small CAB-like proteins (SCPs) of Synechocystis sp. PCC 6803 with Photosystem II. Photosynth. Res. 2008, 95, 135–145. [Google Scholar] [CrossRef]
- Lea-smith, D.J.; Bombelli, P.; Vasudevan, R.; Howe, C.J. Photosynthetic, Respiratory and extracellular electron transport pathways in cyanobacteria. Biochim. Biophys. Acta Bioenergy 2016, 1857, 247–255. [Google Scholar] [CrossRef] [PubMed]
- González, A.; Fillat, M.F. Overexpression, immunodetection, and site-directed mutagenesis of Anabaena sp. PCC 7120 flavodoxin: A comprehensive laboratory practice on molecular biology. Biochem. Mol. Biol. Educ. 2018, 46, 493–501. [Google Scholar] [CrossRef]
- Laudenbach, D.E.; Straus, N.A. Characterization of a cyanobacterial iron stress-induced gene similar to psbC. J. Bacteriol. 1988, 170, 5018–5026. [Google Scholar] [CrossRef] [PubMed]
- Torrado, A.; Ramírez-Moncayo, C.; Navarro, J.A.; Mariscal, V.; Molina-Heredia, F.P. Cytochrome c6 is the main respiratory and photosynthetic soluble electron donor in heterocysts of the cyanobacterium Anabaena sp. PCC 7120. Biochim. Biophys. Acta Bioenergy 2019, 1860, 60–68. [Google Scholar] [CrossRef] [PubMed]
- Ermakova, M.; Allahverdiyeva, Y.; Allahverdiyeva, Y.; Aro, E.-M.; Allahverdiyeva, Y. Novel heterocyst-specific flavodiiron proteins in Anabaena sp. PCC 7120. FEBS Lett. 2013, 587, 82–87. [Google Scholar] [CrossRef]
- Swanson, R.V.; De Lorimier, R.; Glazer, A.N. Genes encoding the phycobilisome rod substructure are clustered on the Anabaena chromosome: Characterization of the phycoerythrocyanin operon. J. Bacteriol. 1992, 174, 2640–2647. [Google Scholar] [CrossRef]
- Ducret, A.; Sidler, W.; Wehrli, E.; Frank, G.; Zuber, H. Isolation, Characterization and Electron Microscopy Analysis of A Hemidiscoidal Phycobilisome Type from the Cyanobacterium Anabaena sp. PCC 7120. JBIC J. Biol. Inorg. Chem. 1996, 236, 1010–1024. [Google Scholar] [CrossRef]
- Khanna, N.; Lindblad, P. Cyanobacterial Hydrogenases and Hydrogen Metabolism Revisited: Recent Progress and Future Prospects. Int. J. Mol. Sci. 2015, 16, 10537–10561. [Google Scholar] [CrossRef]
- Appel, J.; Phunpruch, S.; Steinmüller, K.; Schulz, R. The bidirectional hydrogenase of Synechocystis sp. PCC 6803 works as an electron valve during photosynthesis. Arch. Microbiol. 2000, 173, 333–338. [Google Scholar] [CrossRef]
- Carrieri, D.; Wawrousek, K.; Eckert, C.; Yu, J.; Maness, P. The role of the bidirectional hydrogenase in cyanobacteria. Bioresour. Technol. 2011, 102, 8368–8377. [Google Scholar] [CrossRef]
- Gutekunst, K.; Chen, X.; Schreiber, K.; Kaspar, U.; Makam, S.; Appel, J. The Bidirectional NiFe-hydrogenase in Synechocystis sp. PCC 6803 Is Reduced by Flavodoxin and Ferredoxin and Is Essential under Mixotrophic, Nitrate-limiting Conditions. J. Biol. Chem. 2014, 289, 1930–1937. [Google Scholar] [CrossRef] [PubMed]
- Omata, T.; Gohta, S.; Takahashi, Y.; Harano, Y.; Maeda, S.-I. Involvement of a CbbR Homolog in Low CO2-Induced Activation of the Bicarbonate Transporter Operon in Cyanobacteria. J. Bacteriol. 2001, 183, 1891–1898. [Google Scholar] [CrossRef] [PubMed]
- López-Igual, R.; Picossi, S.; López-Garrido, J.; Flores, E.; Herrero, A. N and C control of ABC-type bicarbonate transporter Cmp and its LysR-type transcriptional regulator CmpR in a heterocyst-forming cyanobacterium, Anabaena sp. Environ. Microbiol. 2012, 14, 1035–1048. [Google Scholar] [CrossRef] [PubMed]
- Brahamsha, B.; Haselkorn, R. Identification of multiple RNA polymerase sigma factor homologs in the cyanobacterium Anabaena sp. strain PCC 7120: Cloning, expression, and inactivation of the sigB and sigC genes. J. Bacteriol. 1992, 174, 7273–7282. [Google Scholar] [CrossRef]
- Caslake, L.F.; Gruber, T.M.; Bryant, N.A. Expression of two alternative sigma factors of Synechococcus sp. strain PCC 7002 is modulated by carbon and nitrogen stress. Microbiology 1997, 143, 3807–3818. [Google Scholar] [CrossRef]
- Muro-Pastor, A.M.; Herrero, A.; Flores, E. Nitrogen-Regulated Group 2 Sigma Factor from Synechocystis sp. Strain PCC 6803 Involved in Survival under Nitrogen Stress. J. Bacteriol. 2001, 183, 1090–1095. [Google Scholar] [CrossRef]
- Tuominen, I.; Pollari, M.; Tyystjärvi, E.; Tyystjärvi, T. The SigB σ factor mediates high-temperature responses in the cyanobacterium Synechocystis sp. PCC6803. FEBS Lett. 2006, 580, 319–323. [Google Scholar] [CrossRef]
- Hakkila, K.; Antal, T.; Gunnelius, L.; Kurkela, J.; Matthijs, H.C.; Tyystjärvi, E.; Tyystjärvi, T. Group 2 Sigma Factor Mutant sigCDE of the Cyanobacterium Synechocystis sp. PCC 6803 Reveals Functionality of Both Carotenoids and Flavodiiron Proteins in Photoprotection of Photosystem II. Plant Cell Physiol. 2013, 54, 1780–1790. [Google Scholar] [CrossRef]
- Summerfield, T.C.; Nagarajan, S.; Sherman, L.A. Gene expression under low-oxygen conditions in the cyanobacterium Synechocystis sp. PCC 6803 demonstrates Hik31-dependent and -independent responses. Microbiology 2011, 157, 301–312. [Google Scholar] [CrossRef]
- Nagarajan, S.; Sherman, D.M.; Shaw, I.; Sherman, L.A. Functions of the Duplicated hik31 Operons in Central Metabolism and Responses to Light, Dark, and Carbon Sources in Synechocystis sp. Strain PCC 6803. J. Bacteriol. 2011, 194, 448–459. [Google Scholar] [CrossRef]
- Jiang, Y.-L.; Wang, X.-P.; Sun, H.; Han, S.-J.; Li, W.-F.; Cui, N.; Lin, G.-M.; Zhang, J.-Y.; Cheng, W.; Cao, D.-D.; et al. Coordinating carbon and nitrogen metabolic signaling through the cyanobacterial global repressor NdhR. Proc. Natl. Acad. Sci. USA 2017, 115, 403–408. [Google Scholar] [CrossRef]
- Frías, J.E.; Flores, E.; Herrero, A. Activation of the Anabaena nir operon promoter requires both NtcA (CAP family) and NtcB (LysR family) transcription factors. Mol. Microbiol. 2000, 38, 613–625. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Scappino, L.; Haselkorn, R. The patB gene product, required for growth of the cyanobacterium Anabaena sp. strain PCC 7120 under nitrogen-limiting conditions, contains ferredoxin and helix-turn-helix domains. J. Bacteriol. 1993, 175, 1697–1704. [Google Scholar] [CrossRef] [PubMed]
- Hebbar, P.B.; Curtis, S.E. Characterization of devH, a Gene Encoding a Putative DNA Binding Protein Required for Heterocyst Function in Anabaena sp. Strain PCC 7120. J. Bacteriol. 2000, 182, 3572–3581. [Google Scholar] [CrossRef] [PubMed]
- Jones, K.M.; Buikema, W.J.; Haselkorn, R. Heterocyst-Specific Expression of patB, a Gene Required for Nitrogen Fixation in Anabaena sp. Strain PCC 7120. J. Bacteriol. 2003, 185, 2306–2314. [Google Scholar] [CrossRef]
- Ehira, S.; Ohmori, M. NrrA, a nitrogen-responsive response regulator facilitates heterocyst development in the cyanobacterium Anabaena sp. strain PCC 7120. Mol. Microbiol. 2006, 59, 1692–1703. [Google Scholar] [CrossRef]
- Picossi, S.; Flores, E.; Herrero, A. ChIP analysis unravels an exceptionally wide distribution of DNA binding sites for the NtcA transcription factor in a heterocyst-forming cyanobacterium. BMC Genom. 2014, 15, 22. [Google Scholar] [CrossRef]
- Tsujimoto, R.; Kamiya, N.; Fujita, Y. Identification of a cis-acting element in nitrogen fixation genes recognized by CnfR in the nonheterocystous nitrogen-fixing cyanobacterium Leptolyngbya boryana. Mol. Microbiol. 2016, 101, 411–424. [Google Scholar] [CrossRef]
- Ramírez, M.E.; Hebbar, P.B.; Zhou, R.; Wolk, C.P.; Curtis, S.E. Anabaena sp. Strain PCC 7120 Gene devH Is Required for Synthesis of the Heterocyst Glycolipid Layer. J. Bacteriol. 2005, 187, 2326–2331. [Google Scholar] [CrossRef]
- Ehira, S.; Ohmori, M. NrrA directly regulates expression of the fraF gene and antisense RNAs for fraE in the heterocyst-forming cyanobacterium Anabaena sp. strain PCC 7120. Microbiology 2014, 160, 844–850. [Google Scholar] [CrossRef]
- Katoh, H.; Hagino, N.; Grossman, A.R.; Ogawa, T. Genes Essential to Iron Transport in the Cyanobacterium Synechocystis sp. Strain PCC 6803. J. Bacteriol. 2001, 183, 2779–2784. [Google Scholar] [CrossRef] [PubMed]
- Fresenborg, L.S.; Graf, J.; Schätzle, H.; Schleiff, E. Iron homeostasis of cyanobacteria: Advancements in siderophores and metal transporters. In Advances in Cyanobacterial Biology; Academic Press: Cambridge, MA, USA, 2020. [Google Scholar] [CrossRef]
- Shen, G.; Balasubramanian, R.; Wang, T.; Wu, Y.; Hoffart, L.M.; Krebs, C.; Bryant, N.A.; Golbeck, J.H. SufR Coordinates Two [4Fe-4S]2+, 1+ Clusters and Functions as a Transcriptional Repressor of the sufBCDS Operon and an Autoregulator of sufR in Cyanobacteria. J. Biol. Chem. 2007, 282, 31909–31919. [Google Scholar] [CrossRef] [PubMed]
- Shcolnick, S.; Summerfield, T.C.; Reytman, L.; Sherman, L.A.; Keren, N. The Mechanism of Iron Homeostasis in the Unicellular Cyanobacterium Synechocystis sp. PCC 6803 and Its Relationship to Oxidative Stress. Plant Physiol. 2009, 150, 2045–2056. [Google Scholar] [CrossRef]
- Vuorijoki, L.; Tiwari, A.; Kallio, P.T.; Aro, E.-M. Inactivation of iron-sulfur cluster biogenesis regulator SufR in Synechocystis sp. PCC 6803 induces unique iron-dependent protein-level responses. Biochim. Biophys. Acta Gen. Subj. 2017, 1861, 1085–1098. [Google Scholar] [CrossRef]
- Garcia-Dominguez, M.; López-Maury, L.; Florencio, F.J.; Reyes, J.C.; Braunstein, M.; Brown, A.M.; Kurtz, S.; Jacobs, W.R. A Gene Cluster Involved in Metal Homeostasis in the Cyanobacterium Synechocystis sp. Strain PCC 6803. J. Bacteriol. 2000, 182, 1507–1514. [Google Scholar] [CrossRef]
- Huertas, M.; López-Maury, L.; Giner-Lamia, J.; Riego, A.M.S.; Florencio, F.J. Metals in Cyanobacteria: Analysis of the Copper, Nickel, Cobalt and Arsenic Homeostasis Mechanisms. Life 2014, 4, 865–886. [Google Scholar] [CrossRef]
- Daley, S.M.E.; Kappell, A.D.; Carrick, M.J.; Burnap, R.L. Regulation of the Cyanobacterial CO2-Concentrating Mechanism Involves Internal Sensing of NADP+ and α-Ketogutarate Levels by Transcription Factor CcmR. PLoS ONE 2012, 7, e41286. [Google Scholar] [CrossRef]
- Eisenhut, M.; Huege, J.; Schwarz, D.; Bauwe, H.; Kopka, J.; Hagemann, M. Metabolome Phenotyping of Inorganic Carbon Limitation in Cells of the Wild Type and Photorespiratory Mutants of the Cyanobacterium Synechocystis sp. Strain PCC 6803. Plant Physiol. 2008, 148, 2109–2120. [Google Scholar] [CrossRef]
- Figge, R.M.; Cassier-Chauvat, C.; Chauvat, F.; Cerff, R. Characterization and analysis of an NAD(P)H dehydrogenase transcriptional regulator critical for the survival of cyanobacteria facing inorganic carbon starvation and osmotic stress. Mol. Microbiol. 2001, 39, 455–469. [Google Scholar] [CrossRef]
- Nishimura, T.; Takahashi, Y.; Yamaguchi, O.; Suzuki, H.; Maeda, S.-I.; Omata, T. Mechanism of low CO2-induced activation of the cmp bicarbonate transporter operon by a LysR family protein in the cyanobacterium Synechococcus elongatus strain PCC 7942. Mol. Microbiol. 2008, 68, 98–109. [Google Scholar] [CrossRef]
- Takahashi, Y.; Yamaguchi, O.; Omata, T. Roles of CmpR, a LysR family transcriptional regulator, in acclimation of the cyanobacterium Synechococcus sp. strain PCC 7942 to low-CO2 and high-light conditions. Mol. Microbiol. 2004, 52, 837–845. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, H.; Kitamura, M.; Nakano, Y.; Katayama, M.; Takahashi, Y.; Kondo, T.; Manabe, K.; Omata, T.; Kutsuna, S. CmpR is Important for Circadian Phasing and Cell Growth. Plant Cell Physiol. 2012, 53, 1561–1569. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Sicora, C.I.; Vorontsova, N.; Allahverdiyeva, Y.; Allahverdiyeva, Y.; Nixon, P.J.; Aro, E.-M. FtsH protease is required for induction of inorganic carbon acquisition complexes in Synechocystis sp. PCC 6803. Mol. Microbiol. 2007, 65, 728–740. [Google Scholar] [CrossRef] [PubMed]
- Domain, F.; Houot, L.; Chauvat, F.; Cassier-Chauvat, C. Function and regulation of the cyanobacterial genes lexA, recA and ruvB: LexA is critical to the survival of cells facing inorganic carbon starvation. Mol. Microbiol. 2004, 53, 65–80. [Google Scholar] [CrossRef] [PubMed]
- Lieman-Hurwitz, J.; Haimovich, M.; Shalev-Malul, G.; Ishii, A.; Hihara, Y.; Gaathon, A.; Lebendiker, M.; Kaplan, A. A cyanobacterial AbrB-like protein affects the apparent photosynthetic affinity for CO2 by modulating low-CO2-induced gene expression. Environ. Microbiol. 2009, 11, 927–936. [Google Scholar] [CrossRef]
- Kaniya, Y.; Kizawa, A.; Miyagi, A.; Kawai-Yamada, M.; Uchimiya, H.; Kaneko, Y.; Nishiyama, Y.; Hihara, Y. Deletion of the Transcriptional Regulator cyAbrB2 Deregulates Primary Carbon Metabolism in Synechocystis sp. PCC 68031[W]. Plant Physiol. 2013, 162, 1153–1163. [Google Scholar] [CrossRef]
- Orf, I.; Schwarz, D.; Kaplan, A.; Kopka, J.; Hess, W.R.; Hagemann, M.; Klähn, S. CyAbrB2 Contributes to the Transcriptional Regulation of Low CO2 Acclimation in Synechocystis sp. PCC 6803. Plant Cell Physiol. 2016, 57, 2232–2243. [Google Scholar] [CrossRef]
- Sjöholm, J.; Oliveira, P.; Lindblad, P. Transcription and Regulation of the Bidirectional Hydrogenase in the Cyanobacterium Nostoc sp. Strain PCC 7120. Appl. Environ. Microbiol. 2007, 73, 5435–5446. [Google Scholar] [CrossRef]
- Oliveira, P.; Lindblad, P. Transcriptional regulation of the cyanobacterial bidirectional Hox-hydrogenase. Dalton Trans. 2009, 9990–9996. [Google Scholar] [CrossRef]
- Ishii, A.; Hihara, Y. An AbrB-Like Transcriptional Regulator, Sll0822, Is Essential for the Activation of Nitrogen-Regulated Genes in Synechocystis sp. PCC 6803. Plant Physiol. 2008, 148, 660–670. [Google Scholar] [CrossRef]
- Agervald, Å.; Baebprasert, W.; Zhang, X.; Incharoensakdi, A.; Lindblad, P.; Stensjö, K. The CyAbrB transcription factor CalA regulates the iron superoxide dismutase in Nostoc sp. strain PCC 7120. Environ. Microbiol. 2010, 12, 2826–2837. [Google Scholar] [CrossRef] [PubMed]
- Higo, A.; Nishiyama, E.; Nakamura, K.; Hihara, Y.; Ehira, S. cyAbrB Transcriptional Regulators as Safety Devices To Inhibit Heterocyst Differentiation in Anabaena sp. Strain PCC 7120. J. Bacteriol. 2019, 201. [Google Scholar] [CrossRef] [PubMed]
- Tanigawa, R.; Shirokane, M.; Maeda, S.-I.; Omata, T.; Tanaka, K.; Takahashi, H. Transcriptional activation of NtcA-dependent promoters of Synechococcus sp. PCC 7942 by 2-oxoglutarate in vitro. Proc. Natl. Acad. Sci. USA 2002, 99, 4251–4255. [Google Scholar] [CrossRef]
- Zhao, M.-X.; Jiang, Y.-L.; He, Y.-X.; Chen, Y.-F.; Teng, Y.-B.; Chen, Y.; Zhang, C.-C.; Zhou, C.-Z. Structural basis for the allosteric control of the global transcription factor NtcA by the nitrogen starvation signal 2-oxoglutarate. Proc. Natl. Acad. Sci. USA 2010, 107, 12487–12492. [Google Scholar] [CrossRef]
- Espinosa, J.; Forchhammer, K.; Burillo, S.; Contreras, A. Interaction network in cyanobacterial nitrogen regulation: PipX, a protein that interacts in a 2-oxoglutarate dependent manner with PII and NtcA. Mol. Microbiol. 2006, 61, 457–469. [Google Scholar] [CrossRef] [PubMed]
- Ohashi, Y.; Shi, W.; Takatani, N.; Aichi, M.; Maeda, S.-I.; Watanabe, S.; Yoshikawa, H.; Omata, T. Regulation of nitrate assimilation in cyanobacteria. J. Exp. Bot. 2011, 62, 1411–1424. [Google Scholar] [CrossRef]
- Su, Z.; Olman, V.; Mao, F.; Xu, Y. Comparative genomics analysis of NtcA regulons in cyanobacteria: Regulation of nitrogen assimilation and its coupling to photosynthesis. Nucleic Acids Res. 2005, 33, 5156–5171. [Google Scholar] [CrossRef]
- Mitschke, J.; Vioque, A.; Haas, F.; Hess, W.R.; Muro-Pastor, A.M. Dynamics of transcriptional start site selection during nitrogen stress-induced cell differentiation in Anabaena sp. PCC7120. Proc. Natl. Acad. Sci. USA 2011, 108, 20130–20135. [Google Scholar] [CrossRef]
- Galmozzi, C.V.; Saelices, L.; Florencio, F.J.; Muro-Pastor, M.I. Posttranscriptional Regulation of Glutamine Synthetase in the Filamentous Cyanobacterium Anabaena sp. PCC 7120: Differential Expression between Vegetative Cells and Heterocysts. J. Bacteriol. 2010, 192, 4701–4711. [Google Scholar] [CrossRef]
- Kang, R.-J.; Shi, D.-J.; Cong, W.; Cai, Z.-L.; Ouyang, F. Regulation of CO2 on heterocyst differentiation and nitrate uptake in the cyanobacterium Anabaena sp. PCC 7120. J. Appl. Microbiol. 2005, 98, 693–698. [Google Scholar] [CrossRef]
- Flores, E.; Arévalo, S.; Burnat, M. Cyanophycin and arginine metabolism in cyanobacteria. Algal Res. 2019, 42, 101577. [Google Scholar] [CrossRef]
- Lopez-Gomollon, S.; Hernández, J.A.; Pellicer, S.; Angarica, V.E.; Peleato, M.L.; Fillat, M.F. Cross-talk Between Iron and Nitrogen Regulatory Networks in Anabaena (Nostoc) sp. PCC 7120: Identification of Overlapping Genes in FurA and NtcA Regulons. J. Mol. Biol. 2007, 374, 267–281. [Google Scholar] [CrossRef] [PubMed]
- Raven, J.A.; Evans, M.C.W.; Korb, R.E. The role of trace metals in photosynthetic electron transport in O2-evolving organisms. Photosynth. Res. 1999, 60, 111–150. [Google Scholar] [CrossRef]
- Gupta, J.K.; Rai, P.; Jain, K.K.; Srivastava, S. Overexpression of bicarbonate transporters in the marine cyanobacterium Synechococcus sp. PCC 7002 increases growth rate and glycogen accumulation. Biotechnol. Biofuels 2020, 13, 17. [Google Scholar] [CrossRef] [PubMed]
- Cano, M.; Holland, S.C.; Artier, J.; Burnap, R.L.; Ghirardi, M.; Morgan, J.A.; Yu, J. Glycogen Synthesis and Metabolite Overflow Contribute to Energy Balancing in Cyanobacteria. Cell Rep. 2018, 23, 667–672. [Google Scholar] [CrossRef]
- Schwarz, D.; Orf, I.; Kopka, J.; Hagemann, M. Effects of Inorganic Carbon Limitation on the Metabolome of the Synechocystis sp. PCC 6803 Mutant Defective in glnB Encoding the Central Regulator PII of Cyanobacterial C/N Acclimation. Metabolites 2014, 4, 232–247. [Google Scholar] [CrossRef]
- Vermaas, W.F. Photosynthesis and Respiration in Cyanobacteria. In Encyclopedia of Life Sciences; John Wiley & Sons, Ltd.: Chichester, UK, 2001. [Google Scholar] [CrossRef]
- Welkie, D.G.; Rubin, B.E.; Diamond, S.; Hood, R.D.; Savage, D.F.; Golden, S.S. A Hard Day’s Night: Cyanobacteria in Diel Cycles. Trends Microbiol. 2019, 27, 231–242. [Google Scholar] [CrossRef]
- Makowka, A.; Nichelmann, L.; Schulze, D.; Spengler, K.; Wittmann, C.; Forchhammer, K.; Gutekunst, K. Glycolytic Shunts Replenish the Calvin–Benson–Bassham Cycle as Anaplerotic Reactions in Cyanobacteria. Mol. Plant 2020, 13, 471–482. [Google Scholar] [CrossRef]
- Huege, J.; Goetze, J.; Schwarz, D.; Bauwe, H.; Hagemann, M.; Kopka, J. Modulation of the Major Paths of Carbon in Photorespiratory Mutants of Synechocystis. PLoS ONE 2011, 6, e16278. [Google Scholar] [CrossRef]
- Tang, J.K.-H.; Tang, Y.J.; Blankenship, R.E. Carbon Metabolic Pathways in Phototrophic Bacteria and Their Broader Evolutionary Implications. Front. Microbiol. 2011, 2, 165. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).