Origin of Life: The Point of No Return
Abstract
“All my life I have wondered about the possibility of life elsewhere. What would it be like? Of what would it be made? …Is extraterrestrial life, if it exists, based on the same organic molecules as life on Earth? Do the beings of other worlds look much like life on Earth? … What else is possible? The nature of life on Earth and the search for life elsewhere are two sides of the same question - the search for who we are.” Carl Sagan’s Cosmos
1. Introduction
2. The Mechanism of Darwinian Evolution
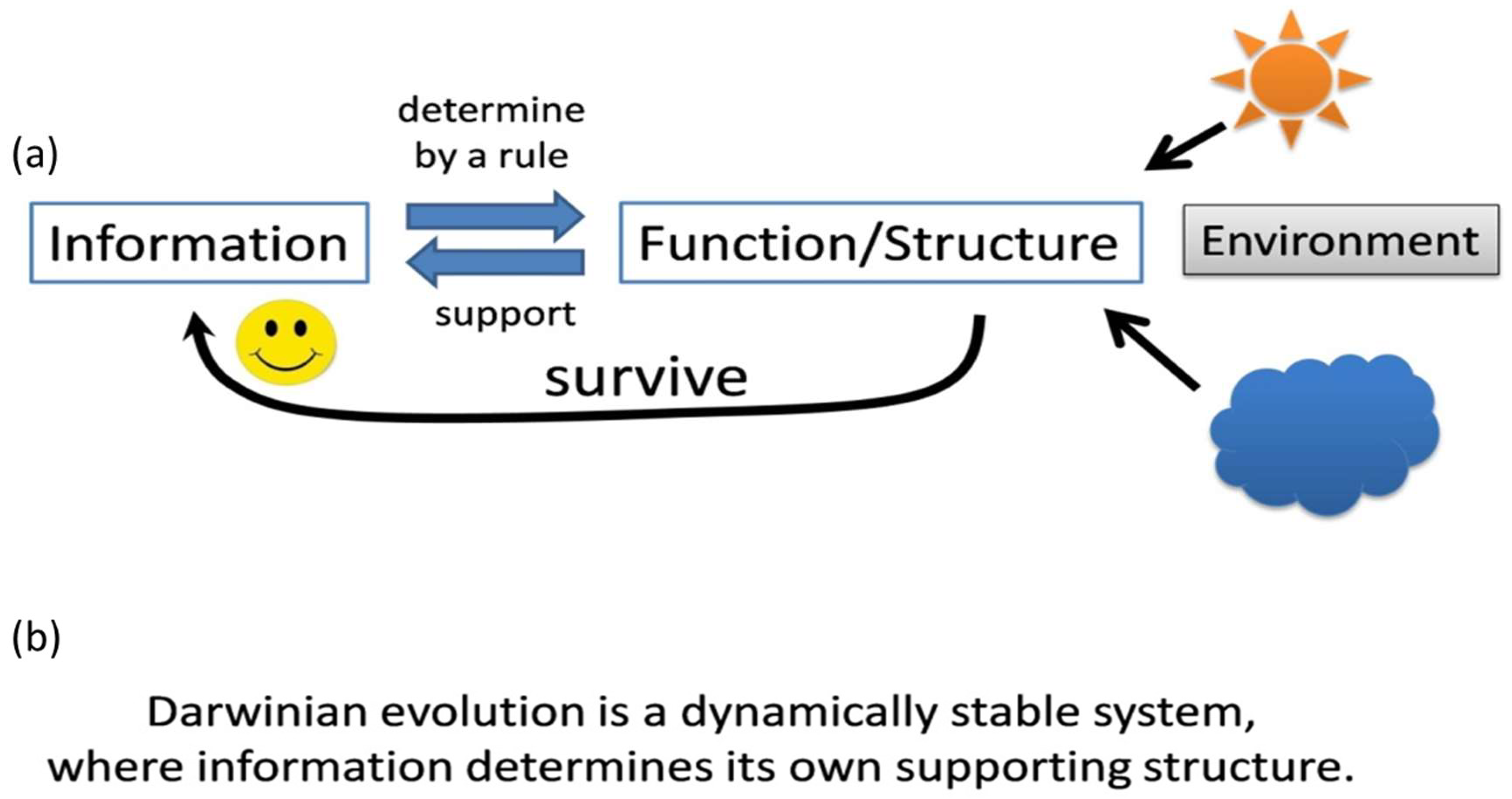
“Darwinian Evolution is a Dynamically Stable System, Where Information Determines Its Own Supporting Structure.”
3. The Information, the Structure, and the Corresponding Rule Are Locked in Its Origin
“During Origin of Life, the Information, the Structure, and the Corresponding Rule Are Locked in Their Origin”.
3.1. The Beginning of the Biological Information
3.2. The Beginning of the Biological Structure
3.3. The Beginning of the Genetic Code
4. Too Many “Evolutions”: We Need Just One
5. The Existing Hypotheses for the Origin of Life in Respect to Darwinian Evolution
5.1. Systems Before Genetic Information
5.2. RNA World Scenario(s)
5.3. RNA-Peptide World Scenario
“Darwinian evolution is a dynamically stable system where the sequence of one hetero-polymer (e.g., RNA carrier of information) determines the sequence of another type of hetero-polymer (e.g., protein or peptide carrier of structure/function).”
Funding
Acknowledgments
Conflicts of Interest
References
- Oro, J.; Lazcano, A. A minimal living system and the origin of a protocell. Adv. Space Res. 1984, 4, 167–176. [Google Scholar] [CrossRef]
- Longo, A.; Damer, B. Factoring Origin of Life Hypotheses into the Search for Life in the Solar System and Beyond. Life 2020, 10, 52. [Google Scholar] [CrossRef] [PubMed]
- Weiss, M.C.; Preiner, M.; Xavier, J.C.; Zimorski, V.; Martin, W.F. The last universal common ancestor between ancient Earth chemistry and the onset of genetics. PLoS Genet. 2018, 14, e1007518. [Google Scholar] [CrossRef] [PubMed]
- Szostak, J.W. Attempts to define life do not help to understand the origin of life. J. Biomol. Struct. Dyn. 2012, 29, 599–600. [Google Scholar] [CrossRef]
- Trifonov, E.N. Vocabulary of definitions of life suggests a definition. J. Biomol. Struct. Dyn. 2011, 29, 259–266. [Google Scholar] [CrossRef]
- Eigen, M. Selforganization of matter and the evolution of biological macromolecules. Naturwissenschaften 1971, 58, 465–523. [Google Scholar] [CrossRef]
- Wills, P.R.; Carter, C.W., Jr. Insuperable problems of the genetic code initially emerging in an RNA world. Biosystems 2018, 164, 155–166. [Google Scholar] [CrossRef] [PubMed]
- de Duve, C. The onset of selection. Nature 2005, 433, 581–582. [Google Scholar] [CrossRef]
- Higgs, P.G. Chemical Evolution and the Evolutionary Definition of Life. J. Mol. Evol. 2017, 84, 225–235. [Google Scholar] [CrossRef]
- Tessera, M. Is pre-Darwinian evolution plausible? Biol. Direct 2018, 13, 18. [Google Scholar] [CrossRef]
- Auboeuf, D. Physicochemical Foundations of Life that Direct Evolution: Chance and Natural Selection are not Evolutionary Driving Forces. Life 2020, 10, 7. [Google Scholar] [CrossRef] [PubMed]
- Basener, W.F.; Sanford, J.C. The fundamental theorem of natural selection with mutations. J. Math. Biol. 2018, 76, 1589–1622. [Google Scholar] [CrossRef] [PubMed]
- Caetano-Anolles, D.; Caetano-Anolles, G. Piecemeal Buildup of the Genetic Code, Ribosomes, and Genomes from Primordial tRNA Building Blocks. Life 2016, 6, 43. [Google Scholar] [CrossRef]
- Pascal, R.; Pross, A.; Sutherland, J.D. Towards an evolutionary theory of the origin of life based on kinetics and thermodynamics. Open Biol. 2013, 3, 130156. [Google Scholar] [CrossRef]
- Cruz-Rosas, H.I.; Riquelme, F.; Ramírez-Padrón, A.; Buhse, T.; Cocho, G.; Miramontes, P. Molecular shape as a key source of prebiotic information. J. Theor. Biol. 2020, 499, 110316. [Google Scholar] [CrossRef]
- Gross, R.; Fouxon, I.; Lancet, D.; Markovitch, O. Quasispecies in population of compositional assemblies. BMC Evol. Biol. 2014, 14, 265. [Google Scholar] [CrossRef]
- Walker, S.I.; Davies, P.C. The algorithmic origins of life. J. R. Soc. Interface 2013, 10, 20120869. [Google Scholar] [CrossRef]
- Lancet, D.; Zidovetzki, R.; Markovitch, O. Systems protobiology: Origin of life in lipid catalytic networks. J. R. Soc. Interface 2018, 15. [Google Scholar] [CrossRef]
- Markovitch, O.; Lancet, D. Multispecies population dynamics of prebiotic compositional assemblies. J. Theor. Biol. 2014, 357, 26–34. [Google Scholar] [CrossRef]
- Pulselli, R.M.; Simoncini, E.; Tiezzi, E. Self-organization in dissipative structures: A thermodynamic theory for the emergence of prebiotic cells and their epigenetic evolution. Biosystems 2009, 96, 237–241. [Google Scholar] [CrossRef]
- Harrison, S.A.; Lane, N. Life as a guide to prebiotic nucleotide synthesis. Nat. Commun. 2018, 9, 5176. [Google Scholar] [CrossRef]
- West, T.; Sojo, V.; Pomiankowski, A.; Lane, N. The origin of heredity in protocells. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2017, 372. [Google Scholar] [CrossRef] [PubMed]
- Damer, B. A Field Trip to the Archaean in Search of Darwin’s Warm Little Pond. Life 2016, 6, 21. [Google Scholar] [CrossRef]
- Goto, Y.; Suga, H. Translation initiation with initiator tRNA charged with exotic peptides. J. Am. Chem. Soc. 2009, 131, 5040–5041. [Google Scholar] [CrossRef]
- Lee, N.; Bessho, Y.; Wei, K.; Szostak, J.W.; Suga, H. Ribozyme-catalyzed tRNA aminoacylation. Nat. Struct. Biol. 2000, 7, 28–33. [Google Scholar] [CrossRef]
- Muller, S. Engineering of ribozymes with useful activities in the ancient RNA world. Ann. N. Y. Acad. Sci. 2015, 1341, 54–60. [Google Scholar] [CrossRef]
- Suga, H.; Hayashi, G.; Terasaka, N. The RNA origin of transfer RNA aminoacylation and beyond. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2011, 366, 2959–2964. [Google Scholar] [CrossRef] [PubMed]
- Ikehara, K. Evolutionary Steps in the Emergence of Life Deduced from the Bottom-Up Approach and GADV Hypothesis (Top-Down Approach). Life 2016, 6, 6. [Google Scholar] [CrossRef]
- Milner-White, E.J. Protein three-dimensional structures at the origin of life. Interface Focus 2019, 9, 20190057. [Google Scholar] [CrossRef]
- van der Gulik, P.; Massar, S.; Gilis, D.; Buhrman, H.; Rooman, M. The first peptides: The evolutionary transition between prebiotic amino acids and early proteins. J. Theor. Biol. 2009, 261, 531–539. [Google Scholar] [CrossRef] [PubMed]
- van der Gulik, P.T.; Speijer, D. How amino acids and peptides shaped the RNA world. Life 2015, 5, 230–246. [Google Scholar] [CrossRef]
- Kim, Y.; Opron, K.; Burton, Z.F. A tRNA- and Anticodon-Centric View of the Evolution of Aminoacyl-tRNA Synthetases, tRNAomes, and the Genetic Code. Life 2019, 9, 37. [Google Scholar] [CrossRef]
- Scheffers, D.J.; Pinho, M.G. Bacterial cell wall synthesis: New insights from localization studies. Microbiol. Mol. Biol. Rev. 2005, 69, 585–607. [Google Scholar] [CrossRef]
- Gulik, P.T. On the Origin of Sequence. Life 2015, 5, 1629–1637. [Google Scholar] [CrossRef]
- Li, F.; Fitz, D.; Fraser, D.G.; Rode, B.M. Catalytic effects of histidine enantiomers and glycine on the formation of dileucine and dimethionine in the salt-induced peptide formation reaction. Amino Acids 2010, 38, 287–294. [Google Scholar] [CrossRef]
- Plankensteiner, K.; Reiner, H.; Rode, B.M. Catalytic effects of glycine on prebiotic divaline and diproline formation. Peptides 2005, 26, 1109–1112. [Google Scholar] [CrossRef]
- Plankensteiner, K.; Righi, A.; Rode, B.M. Glycine and diglycine as possible catalytic factors in the prebiotic evolution of peptides. Orig. Life Evol. Biosph. 2002, 32, 225–236. [Google Scholar] [CrossRef] [PubMed]
- Thomas, B.R. The origin of the genetic code. Biochem. Biophys. Res. Commun. 1970, 40, 1289–1296. [Google Scholar] [CrossRef]
- Di Giulio, M. Reflections on the origin of the genetic code: A hypothesis. J. Theor. Biol. 1998, 191, 191–196. [Google Scholar] [CrossRef]
- Koonin, E.V.; Novozhilov, A.S. Origin and evolution of the genetic code: The universal enigma. IUBMB Life 2009, 61, 99–111. [Google Scholar] [CrossRef]
- Guimarães, R.C.; Moreira, C.H.; de Farias, S.T. A self-referential model for the formation of the genetic code. Theory BioSci. 2008, 127, 249–270. [Google Scholar] [CrossRef] [PubMed]
- Rodin, A.S.; Szathmáry, E.; Rodin, S.N. On origin of genetic code and tRNA before translation. Biol. Direct 2011, 6, 14. [Google Scholar] [CrossRef]
- Koonin, E.V. Frozen Accident Pushing 50: Stereochemistry, Expansion, and Chance in the Evolution of the Genetic Code. Life 2017, 7, 22. [Google Scholar] [CrossRef]
- Koonin, E.V.; Novozhilov, A.S. Origin and Evolution of the Universal Genetic Code. Annu. Rev. Genet. 2017, 51, 45–62. [Google Scholar] [CrossRef]
- Lei, L.; Burton, Z.F. Evolution of Life on Earth: tRNA, Aminoacyl-tRNA Synthetases and the Genetic Code. Life 2020, 10, 21. [Google Scholar] [CrossRef]
- Gospodinov, A.; Kunnev, D. Universal Codons with Enrichment from GC to AU Nucleotide Composition Reveal a Chronological Assignment from Early to Late Along with LUCA Formation. Life 2020, 10, 81. [Google Scholar] [CrossRef]
- Wolf, Y.I.; Koonin, E.V. On the origin of the translation system and the genetic code in the RNA world by means of natural selection, exaptation, and subfunctionalization. Biol. Direct 2007, 2, 14. [Google Scholar] [CrossRef]
- Higgs, P.G.; Pudritz, R.E. A thermodynamic basis for prebiotic amino acid synthesis and the nature of the first genetic code. Astrobiology 2009, 9, 483–490. [Google Scholar] [CrossRef]
- Higgs, P.G. A four-column theory for the origin of the genetic code: Tracing the evolutionary pathways that gave rise to an optimized code. Biol. Direct 2009, 4, 16. [Google Scholar] [CrossRef]
- Pak, D.; Kim, Y.; Burton, Z.F. Aminoacyl-tRNA synthetase evolution and sectoring of the genetic code. Transcription 2018, 9, 205–224. [Google Scholar] [CrossRef]
- Pak, D.; Root-Bernstein, R.; Burton, Z.F. tRNA structure and evolution and standardization to the three nucleotide genetic code. Transcription 2017, 8, 205–219. [Google Scholar] [CrossRef]
- Wong, J.T. Coevolution theory of the genetic code at age thirty. Bioessays 2005, 27, 416–425. [Google Scholar] [CrossRef]
- Opron, K.; Burton, Z.F. Ribosome Structure, Function, and Early Evolution. Int. J. Mol. Sci. 2018, 20, 40. [Google Scholar] [CrossRef]
- Harish, A.; Caetano-Anollés, G. Ribosomal history reveals origins of modern protein synthesis. PLoS ONE 2012, 7, e32776. [Google Scholar] [CrossRef]
- Pearce, B.K.D.; Tupper, A.S.; Pudritz, R.E.; Higgs, P.G. Constraining the Time Interval for the Origin of Life on Earth. Astrobiology 2018, 18, 343–364. [Google Scholar] [CrossRef]
- Tashiro, T.; Ishida, A.; Hori, M.; Igisu, M.; Koike, M.; Mejean, P.; Takahata, N.; Sano, Y.; Komiya, T. Early trace of life from 3.95 Ga sedimentary rocks in Labrador, Canada. Nature 2017, 549, 516–518. [Google Scholar] [CrossRef]
- Ruiz-Mirazo, K.; Briones, C.; de la Escosura, A. Chemical roots of biological evolution: The origins of life as a process of development of autonomous functional systems. Open Biol. 2017, 7. [Google Scholar] [CrossRef]
- Vasas, V.; Fernando, C.; Santos, M.; Kauffman, S.; Szathmary, E. Evolution before genes. Biol. Direct 2012, 7, 1. [Google Scholar] [CrossRef]
- Hunding, A.; Kepes, F.; Lancet, D.; Minsky, A.; Norris, V.; Raine, D.; Sriram, K.; Root-Bernstein, R. Compositional complementarity and prebiotic ecology in the origin of life. Bioessays 2006, 28, 399–412. [Google Scholar] [CrossRef]
- Root-Bernstein, R.S.; Dillon, P.F. Molecular complementarity I: The complementarity theory of the origin and evolution of life. J. Theor. Biol. 1997, 188, 447–479. [Google Scholar] [CrossRef]
- Root-Bernstein, R. A modular hierarchy-based theory of the chemical origins of life based on molecular complementarity. Acc. Chem. Res. 2012, 45, 2169–2177. [Google Scholar] [CrossRef]
- Root-Bernstein, R.S. Peptide self-aggregation and peptide complementarity as bases for the evolution of peptide receptors: A review. J. Mol. Recognit. 2005, 18, 40–49. [Google Scholar] [CrossRef]
- de la Escosura, A. The Informational Substrate of Chemical Evolution: Implications for Abiogenesis. Life 2019, 9, 66. [Google Scholar] [CrossRef] [PubMed]
- Vincent, L.; Berg, M.; Krismer, M.; Saghafi, S.S.; Cosby, J.; Sankari, T.; Vetsigian, K.; Ii, H.J.C.; Baum, D.A. Chemical Ecosystem Selection on Mineral Surfaces Reveals Long-Term Dynamics Consistent with the Spontaneous Emergence of Mutual Catalysis. Life 2019, 9, 80. [Google Scholar] [CrossRef]
- Gilbert, W. Origin of life: The RNA world. Nature 1986, 319, 618. [Google Scholar] [CrossRef]
- Bernhardt, H.S.; Tate, W.P. Primordial soup or vinaigrette: Did the RNA world evolve at acidic pH? Biol. Direct 2012, 7, 4. [Google Scholar] [CrossRef]
- Cech, T.R. Ribozymes, the first 20 years. Biochem. Soc. Trans. 2002, 30, 1162–1166. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.K.; Unrau, P.J. Closing the circle: Replicating RNA with RNA. Cold Spring Harb. Perspect. Biol. 2010, 2, a002204. [Google Scholar] [CrossRef]
- Jeffares, D.C.; Poole, A.M.; Penny, D. Relics from the RNA world. J. Mol. Evol. 1998, 46, 18–36. [Google Scholar] [CrossRef]
- Joyce, G.F.; Szostak, J.W. Protocells and RNA Self-Replication. Cold Spring Harb. Perspect. Biol. 2018, 10. [Google Scholar] [CrossRef] [PubMed]
- Inoue, T.; Orgel, L.E. Oligomerization of (guanosine 5’-phosphor)-2-methylimidazolide on poly(C). An RNA polymerase model. J. Mol. Biol. 1982, 162, 201–217. [Google Scholar] [CrossRef]
- Deck, C.; Jauker, M.; Richert, C. Efficient enzyme-free copying of all four nucleobases templated by immobilized RNA. Nat. Chem. 2011, 3, 603–608. [Google Scholar] [CrossRef] [PubMed]
- Prywes, N.; Blain, J.C.; Del Frate, F.; Szostak, J.W. Nonenzymatic copying of RNA templates containing all four letters is catalyzed by activated oligonucleotides. eLife 2016, 5. [Google Scholar] [CrossRef] [PubMed]
- Walton, T.; Zhang, W.; Li, L.; Tam, C.P.; Szostak, J.W. The Mechanism of Nonenzymatic Template Copying with Imidazole-Activated Nucleotides. Angew. Chem. Int. Ed. Engl. 2019, 58, 10812–10819. [Google Scholar] [CrossRef]
- Wu, T.; Orgel, L.E. Nonenzymatic template-directed synthesis on hairpin oligonucleotides. 3. Incorporation of adenosine and uridine residues. J. Am. Chem. Soc. 1992, 114, 7963–7969. [Google Scholar] [CrossRef]
- Zhang, W.; Pal, A.; Ricardo, A.; Szostak, J.W. Template-Directed Nonenzymatic Primer Extension Using 2-Methylimidazole-Activated Morpholino Derivatives of Guanosine and Cytidine. J. Am. Chem. Soc. 2019, 141, 12159–12166. [Google Scholar] [CrossRef]
- Higgs, P.G.; Lehman, N. The RNA World: Molecular cooperation at the origins of life. Nat. Rev. Genet. 2015, 16, 7–17. [Google Scholar] [CrossRef]
- Demongeot, J.; Norris, V. Emergence of a “Cyclosome” in a Primitive Network Capable of Building “Infinite” Proteins. Life 2019, 9, 51. [Google Scholar] [CrossRef]
- Shah, V.; de Bouter, J.; Pauli, Q.; Tupper, A.S.; Higgs, P.G. Survival of RNA Replicators is much Easier in Protocells than in Surface-Based, Spatial Systems. Life 2019, 9, 65. [Google Scholar] [CrossRef] [PubMed]
- Yarus, M. Getting past the RNA world: The initial Darwinian ancestor. Cold Spring Harb. Perspect. Biol. 2011, 3. [Google Scholar] [CrossRef]
- Bowman, J.C.; Hud, N.V.; Williams, L.D. The ribosome challenge to the RNA world. J. Mol. Evol. 2015, 80, 143–161. [Google Scholar] [CrossRef]
- Carter, C.W. What RNA World? Why a Peptide/RNA Partnership Merits Renewed Experimental Attention. Life 2015, 5, 294–320. [Google Scholar] [CrossRef]
- Carter, C.W., Jr. An Alternative to the RNA World. Nat. Hist 2016, 125, 28–33. [Google Scholar]
- Kunnev, D.; Gospodinov, A. Possible Emergence of Sequence Specific RNA Aminoacylation via Peptide Intermediary to Initiate Darwinian Evolution and Code Through Origin of Life. Life 2018, 8, 44. [Google Scholar] [CrossRef]
- Alva, V.; Lupas, A.N. From ancestral peptides to designed proteins. Curr. Opin. Struct. Biol. 2018, 48, 103–109. [Google Scholar] [CrossRef]
- Carter, C.W., Jr.; Wills, P.R. Interdependence, Reflexivity, Fidelity, Impedance Matching, and the Evolution of Genetic Coding. Mol. Biol. Evol. 2018, 35, 269–286. [Google Scholar] [CrossRef]
- Lanier, K.A.; Petrov, A.S.; Williams, L.D. The Central Symbiosis of Molecular Biology: Molecules in Mutualism. J. Mol. Evol. 2017, 85, 8–13. [Google Scholar] [CrossRef]
- Chatterjee, S.; Yadav, S. The Origin of Prebiotic Information System in the Peptide/RNA World: A Simulation Model of the Evolution of Translation and the Genetic Code. Life 2019, 9, 25. [Google Scholar] [CrossRef]
- Dila, G.; Ripp, R.; Mayer, C.; Poch, O.; Michel, C.J.; Thompson, J.D. Circular code motifs in the ribosome: A missing link in the evolution of translation? RNA 2019, 25, 1714–1730. [Google Scholar] [CrossRef]
- Caetano-Anollés, D.; Caetano-Anollés, K.; Caetano-Anollés, G. Evolution of macromolecular structure: A ‘double tale’ of biological accretion and diversification. Sci. Prog. 2018, 101, 360–383. [Google Scholar] [CrossRef]
- Cornell, C.E.; Black, R.A.; Xue, M.; Litz, H.E.; Ramsay, A.; Gordon, M.; Mileant, A.; Cohen, Z.R.; Williams, J.A.; Lee, K.K.; et al. Prebiotic amino acids bind to and stabilize prebiotic fatty acid membranes. Proc. Natl. Acad. Sci. USA 2019, 116, 17239–17244. [Google Scholar] [CrossRef]
- Gordon, K.H. Were RNA replication and translation directly coupled in the RNA (+protein?) world? J. Theor. Biol. 1995, 173, 179–193. [Google Scholar] [CrossRef]
- Shimizu, M. Specific aminoacylation of C4N hairpin RNAs with the cognate aminoacyl-adenylates in the presence of a dipeptide: Origin of the genetic code. J. Biochem. 1995, 117, 23–26. [Google Scholar] [CrossRef]
- Shimizu, M. Oligomer biology and polymer biology: Primitive RNA-protein interactions. Nucleic Acids Symp. Ser. 1995, 33, 179–181. [Google Scholar]
- Frenkel-Pinter, M.; Haynes, J.W.; Mohyeldin, A.M.; Martin, C.; Sargon, A.B.; Petrov, A.S.; Krishnamurthy, R.; Hud, N.V.; Williams, L.D.; Leman, L.J. Mutually stabilizing interactions between proto-peptides and RNA. Nat. Commun. 2020, 11, 3137. [Google Scholar] [CrossRef]
- Illangasekare, M.; Yarus, M. A tiny RNA that catalyzes both aminoacyl-RNA and peptidyl-RNA synthesis. Rna 1999, 5, 1482–1489. [Google Scholar] [CrossRef]
- Root-Bernstein, M.; Root-Bernstein, R. The ribosome as a missing link in the evolution of life. J. Theor. Biol. 2015, 367, 130–158. [Google Scholar] [CrossRef]
- Baross, J.A.; Martin, W.F. The Ribofilm as a Concept for Life’s Origins. Cell 2015, 162, 13–15. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kunnev, D. Origin of Life: The Point of No Return. Life 2020, 10, 269. https://doi.org/10.3390/life10110269
Kunnev D. Origin of Life: The Point of No Return. Life. 2020; 10(11):269. https://doi.org/10.3390/life10110269
Chicago/Turabian StyleKunnev, Dimiter. 2020. "Origin of Life: The Point of No Return" Life 10, no. 11: 269. https://doi.org/10.3390/life10110269
APA StyleKunnev, D. (2020). Origin of Life: The Point of No Return. Life, 10(11), 269. https://doi.org/10.3390/life10110269




