Novel Modular Rhodopsins from Green Algae Hold Great Potential for Cellular Optogenetic Modulation Across the Biological Model Systems
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification of Rhodopsin Domain, Homology and Structural Analysis
2.2. Evolutionary Analysis of Rhodopsin Domains of Modular Proteins
2.3. Protein-Protein Interaction Analysis of Novel Domains from Modular Algal Rhodopsins
3. Results and Discussion
3.1. Microbial Rhodopsins With Modular Domain Organization
3.2. Modular Channelrhodopsins and Their Optogenetic Potential
3.3. Modular Sensory-Type Rhodopsins and Their Optogenetic Potential
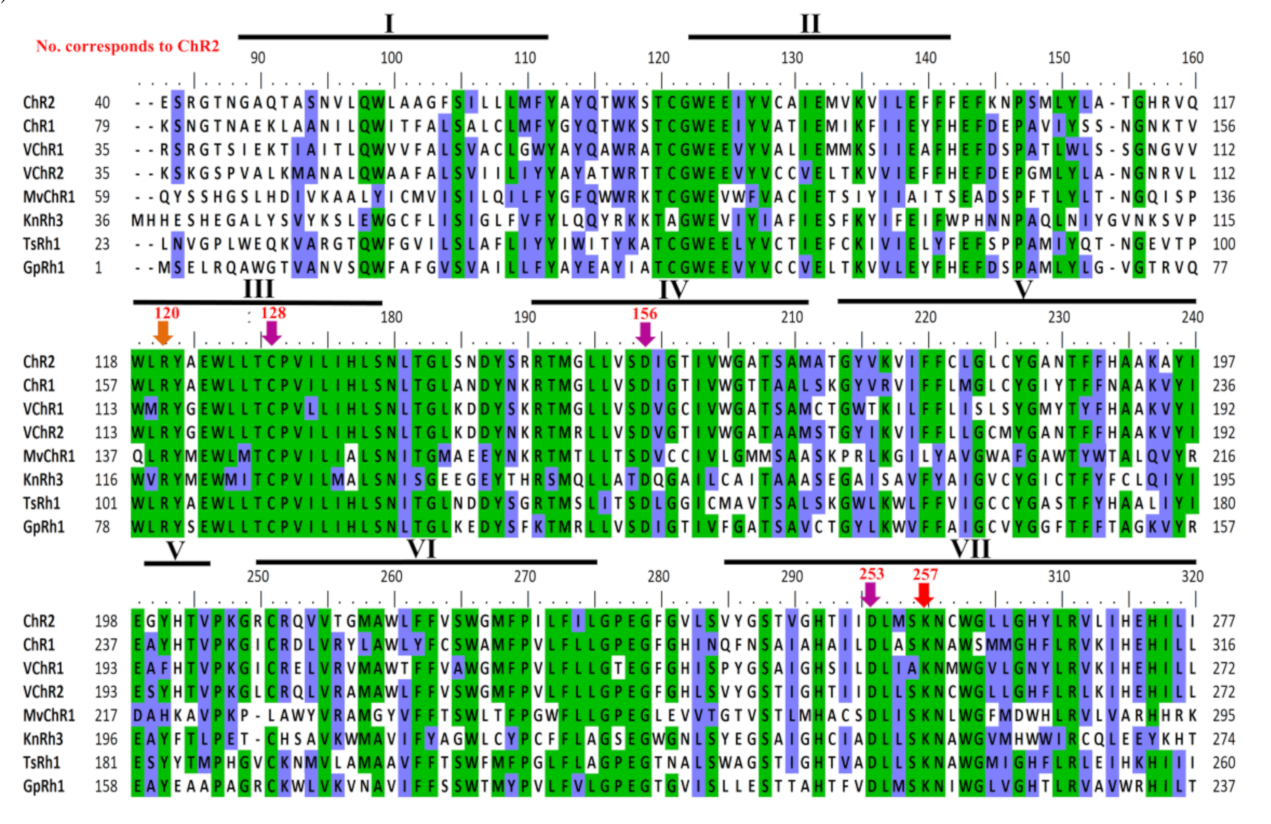
3.4. Light-Gated Ion Pump and Photo-Sensory Function Prediction Based on Conserved Residues of Rhodopsins
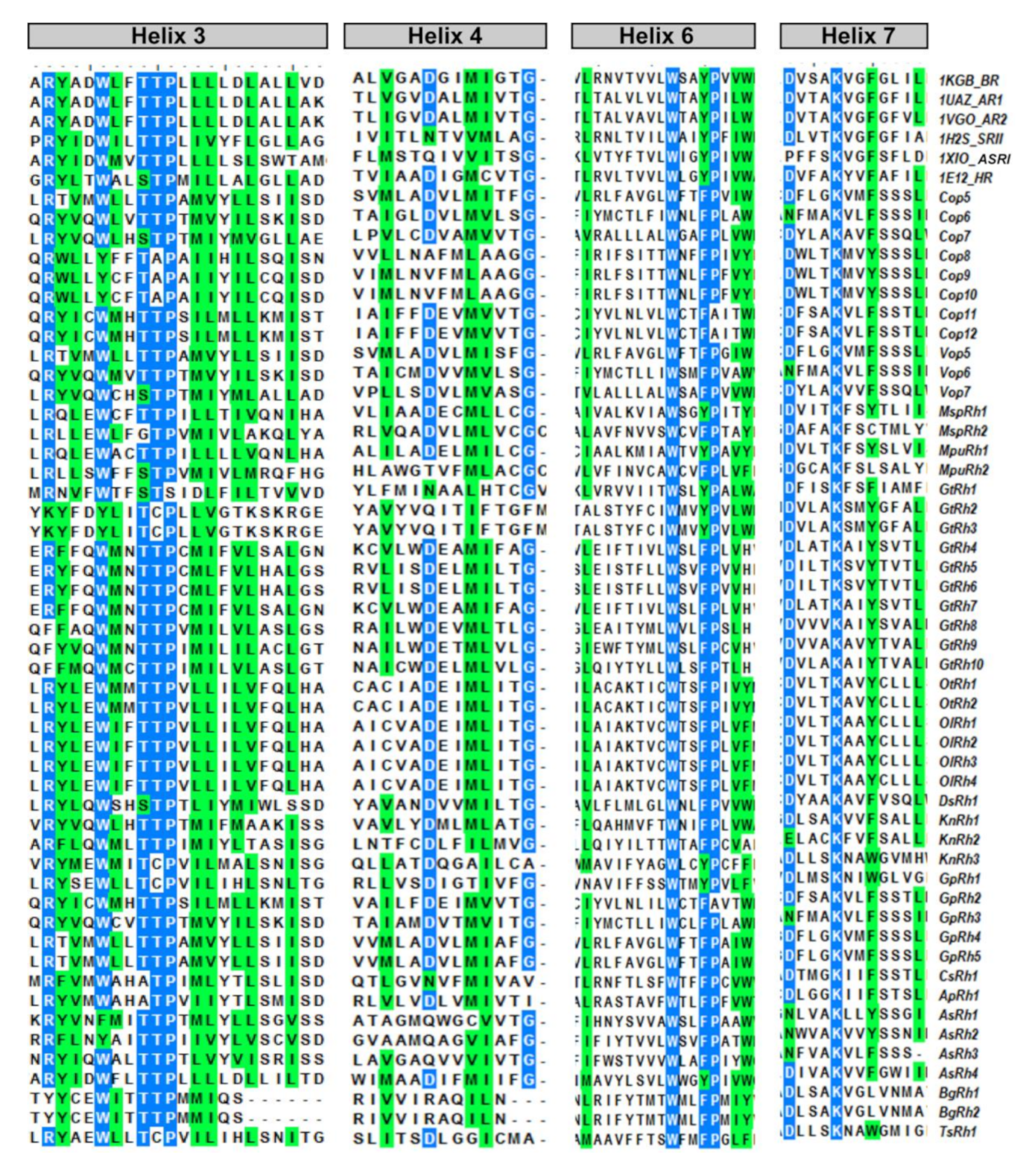
3.5. Spectral Tuning of the New Microbial Rhodopsins
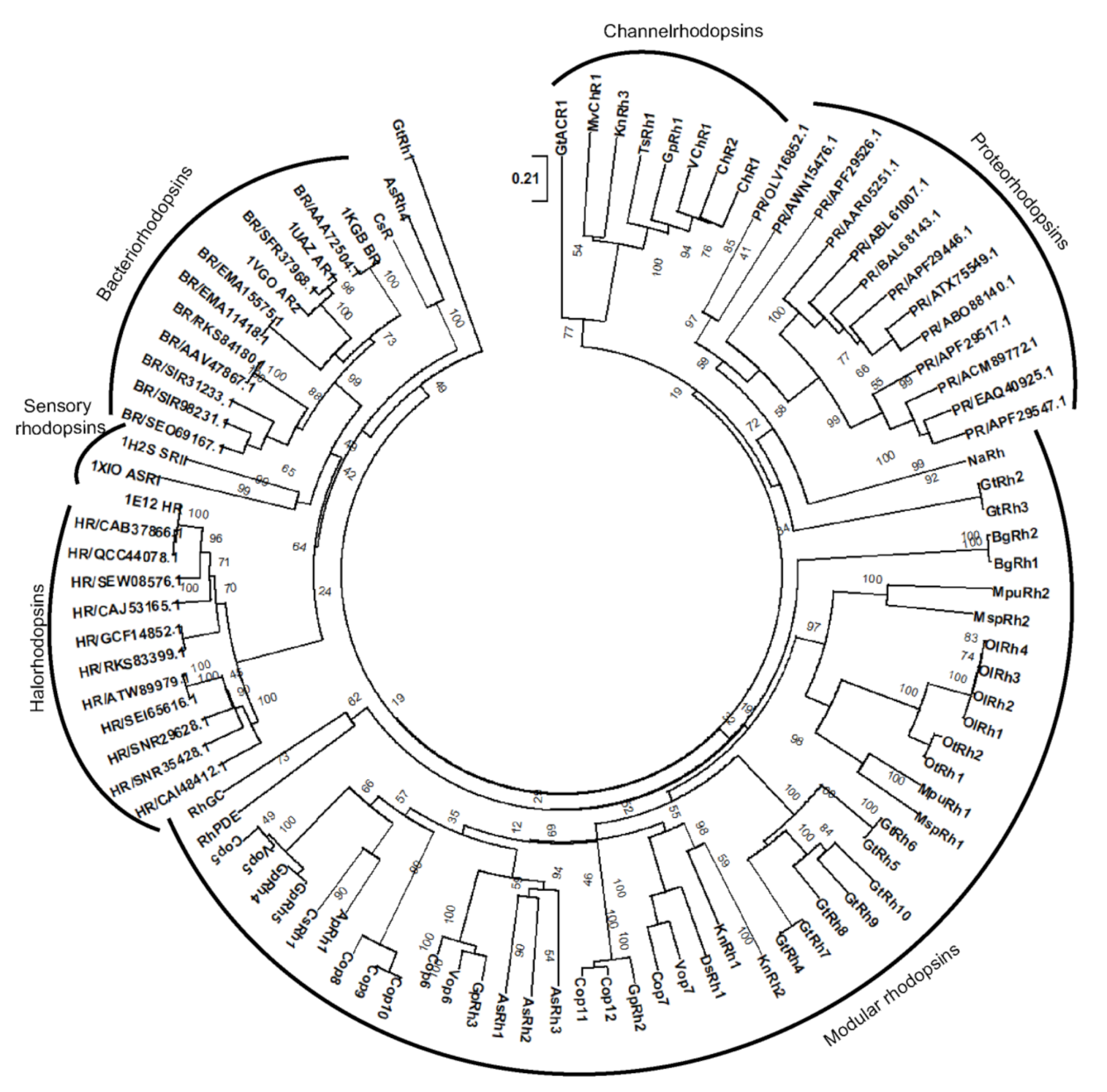
3.6. Evolutionary Pattern of the Modular Microbial Rhodopsins
3.7. Cyclase Domain is a Canonical Secondary Messenger of Modular Sensory Type Rhodopsin
3.8. Optogenetic Potential of the Novel Modular Rhodopsins
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| Cop-Chlamyopsin | rhodopsin from Chlamydomonas reinhardtii |
| Vop-Volvoxopsin | rhodopsin from Volvox carteri |
| GpRh 1−5 | rhodopsin from Gonium pectorale |
| AsRh1−4 | Asterochloris sp. |
| KnRh1−3 | Klebsormidium nitens |
| OtRh1−2 | Ostreococcus tauri |
| MpuRh1&2 | Micromonas pusilla |
| MspRh1&2 | Micromonas species |
| OlRh1−4 | Ostreococcus lucimarinus |
| CsRh1 | Chlorella sorokiniana |
| ApRh1 | Auxenochlorella protothecoides |
| BgRh1&2 | Bigelowiella natans |
| GtRh1−10 | Guillardia theta |
| DsRh1 | Dunaliella salina |
| TsRh1 | Tetraselmis subcordiformis |
References
- Oesterhelt, D.; Stoeckenius, W. Functions of a new photoreceptor membrane. Proc. Natl. Acad. Sci. USA 1973, 70, 2853–2857. [Google Scholar] [CrossRef] [PubMed]
- Schobert, B.; Lanyi, J.K. Halorhodopsin is a light-driven chloride pump. J. Biol. Chem. 1982, 257, 10306–10313. [Google Scholar] [PubMed]
- Nagel, G.; Ollig, D.; Fuhrmann, M.; Kateriya, S.; Musti, A.M.; Bamberg, E.; Hegemann, P. Channelrhodopsin-1: A light-gated proton channel in green algae. Science 2002, 296, 2395–2398. [Google Scholar] [CrossRef] [PubMed]
- Nagel, G.; Szellas, T.; Huhn, W.; Kateriya, S.; Adeishvili, N.; Berthold, P.; Ollig, D.; Hegemann, P.; Bamberg, E. Channelrhodopsin-2: A directly light-gated cation-selective membrane channel. Proc. Natl. Acad. Sci. USA 2003, 100, 13940–13945. [Google Scholar] [CrossRef]
- Hoff, W.D.; Jung, K.-H.; Spudich, J.L. Molecular mechanism of photosignaling by archaeal sensory rhodopsins. Annu. Rev. Biophys. Biomol. Struct. 1997, 26, 223–258. [Google Scholar] [CrossRef]
- Nakagawa, M.; Iwasa, T.; Kikkawa, S.; Tsuda, M.; Ebrey, T.G. How vertebrate and invertebrate visual pigments differ in their mechanism of photoactivation. Proc. Natl. Acad. Sci. USA 1999, 96, 6189–6192. [Google Scholar] [CrossRef]
- Awasthi, M.; Ranjan, P.; Sharma, K.; Veetil, S.K.; Kateriya, S. The trafficking of bacterial type rhodopsins into the Chlamydomonas eyespot and flagella is IFT mediated. Sci. Rep. 2016, 6, 34646. [Google Scholar] [CrossRef]
- Zhang, F.; Vierock, J.; Yizhar, O.; Fenno, L.E.; Tsunoda, S.; Kianianmomeni, A.; Prigge, M.; Berndt, A.; Cushman, J.C.; Polle, J.; et al. The microbial opsin family of optogenetic tools. Cell 2011, 147, 1446–1457. [Google Scholar] [CrossRef]
- Yoshida, K.; Tsunoda, S.; Brown, L.S.; Kandori, H. A unique choanoflagellate enzyme rhodopsin exhibits light-dependent cyclic nucleotide phosphodiesterase activity. J. Biol. Chem. 2017, 292, 7531–7541. [Google Scholar] [CrossRef]
- Tian, Y.; Gao, S.; Yang, S.; Nagel, G. A novel rhodopsin phosphodiesterase from Salpingoeca rosetta shows light-enhanced substrate affinity. Biochem. J. 2018, 475, 1121–1128. [Google Scholar] [CrossRef]
- Lamarche, L.B.; Kumar, R.P.; Trieu, M.M.; Devine, E.L.; Cohen-Abeles, L.E.; Theobald, D.L.; Oprian, D.D. Purification and characterization of RhoPDE, a retinylidene/phosphodiesterase fusion protein and potential optogenetic tool from the choanoflagellate salpingoeca rosetta. Biochemistry 2017, 56, 5812–5822. [Google Scholar] [CrossRef] [PubMed]
- Scheib, U.; Stehfest, K.; Gee, C.E.; Körschen, H.G.; Fudim, R.; Oertner, T.G.; Hegemann, P. The rhodopsin–guanylyl cyclase of the aquatic fungus Blastocladiella emersonii enables fast optical control of cGMP signaling. Optogenetics 2015, 8, rs8. [Google Scholar]
- Scheib, U.; Broser, M.; Constantin, O.M.; Yang, S.; Gao, S.; Mukherjee, S.; Stehfest, K.; Nagel, G.; Gee, C.E.; Ehegemann, P. Rhodopsin-cyclases for photocontrol of cGMP/cAMP and 2.3 Å structure of the adenylyl cyclase domain. Nat. Commun. 2018, 9, 2046. [Google Scholar] [CrossRef] [PubMed]
- Butryn, A.; Raza, H.; Rada, H.; Moraes, I.; Owens, R.J.; Orville, A.M. Molecular basis for GTP recognition by light-activated guanylate cyclase Rh GC. FEBS J. 2019. [Google Scholar] [CrossRef] [PubMed]
- Trieu, M.M.; Devine, E.L.; Lamarche, L.B.; Ammerman, A.E.; Greco, J.A.; Birge, R.R.; Theobald, D.L.; Oprian, D.D. Expression, purification, and spectral tuning of RhoGC, a retinylidene/guanylyl cyclase fusion protein and optogenetics tool from the aquatic fungus Blastocladiella emersonii. J. Biol. Chem. 2017, 292, 10379–10389. [Google Scholar] [CrossRef]
- Bergs, A.; Schultheis, C.; Fischer, E.; Tsunoda, S.P.; Erbguth, K.; Husson, S.J.; Govorunova, E.G.; Spudich, J.L.; Nagel, G.; Gottschalk, A.; et al. Rhodopsin optogenetic toolbox v2.0 for light-sensitive excitation and inhibition in Caenorhabditis elegans. PLoS ONE 2018, 13, e0191802. [Google Scholar] [CrossRef]
- Mukherjee, S.; Hegemann, P.; Broser, M. Enzymerhodopsins: Novel photoregulated catalysts for optogenetics. Curr. Opin. Struct. Biol. 2019, 57, 118–126. [Google Scholar] [CrossRef]
- Greiner, A.; Kelterborn, S.; Evers, H.; Kreimer, G.; Sizova, I.; Hegemann, P. Targeting of photoreceptor genes in chlamydomonas reinhardtii via zinc-finger nucleases and CRISPR/Cas9. Plant Cell 2017, 29, 2498–2518. [Google Scholar] [CrossRef]
- Thompson, J.D. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997, 25, 4876–4882. [Google Scholar] [CrossRef]
- Geer, L.Y.; Domrachev, M.; Lipman, D.J.; Bryant, S.H. CDART: Protein homology by domain architecture. Genome Res. 2002, 12, 1619–1623. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Bo, Y.; Han, L.; He, J.; Lanczycki, C.J.; Lu, S.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; et al. CDD/SPARCLE: Functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2016, 45, D200–D203. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef]
- Perrière, G.; Gouy, M. WWW-query: An on-line retrieval system for biological sequence banks. Biochimie 1996, 78, 364–369. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Morris, J.H.; Cook, H.; Kuhn, M.; Wyder, S.; Simonovic, M.; Santos, A.; Doncheva, N.T.; Roth, A.; Bork, P.; et al. The STRING database in 2017: Quality-controlled protein–protein association networks, made broadly accessible. Nucleic Acids Res. 2016, 45, D362–D368. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Salomé, P.A.; Merchant, S.S. A series of fortunate events: Introducing chlamydomonas as a reference organism. Plant Cell 2019, 31, 1682–1707. [Google Scholar] [CrossRef]
- Ranjan, P.; Awasthi, M.; Snell, W.J. Transient internalization and microtubule-dependent trafficking of a ciliary signaling receptor from the plasma membrane to the cilium. Curr. Biol. 2019, 29, 2942–2947. [Google Scholar] [CrossRef]
- Govorunova, E.G.; Sineshchekov, O.A.; Li, H.; Janz, R.; Spudich, J.L. Characterization of a highly efficient blue-shifted channelrhodopsin from the marine alga platymonas subcordiformis. J. Biol. Chem. 2013, 288, 29911–29922. [Google Scholar] [CrossRef]
- Volkov, O.; Kovalev, K.; Polovinkin, V.; Borshchevskiy, V.; Bamann, C.; Astashkin, R.; Marin, E.; Popov, A.; Balandin, T.; Willbold, D.; et al. Structural insights into ion conduction by channelrhodopsin 2. Science 2017, 358, eaan8862. [Google Scholar] [CrossRef]
- Nack, M.; Radu, I.; Gossing, M.; Bamann, C.; Bamberg, E.; Von Mollard, G.F.; Heberle, J. The DC gate in Channelrhodopsin-2: Crucial hydrogen bonding interaction between C128 and D156. Photochem. Photobiol. Sci. 2010, 9, 194–198. [Google Scholar] [CrossRef]
- Stehfest, K.; Ritter, E.; Berndt, A.; Bartl, F.J.; Hegemann, P. The branched photocycle of the slow-cycling channelrhodopsin-2 Mutant C128T. J. Mol. Biol. 2010, 398, 690–702. [Google Scholar] [CrossRef]
- Luck, M.; Mathes, T.; Bruun, S.; Fudim, R.; Hagedorn, R.; Nguyen, T.M.T.; Kateriya, S.; Kennis, J.T.M.; Hildebrandt, P.; Ehegemann, P. A photochromic histidine kinase rhodopsin (HKR1) that is bimodally switched by ultraviolet and blue light. J. Biol. Chem. 2012, 287, 40083–40090. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Gao, S.; Von Der Heyde, E.L.; Hallmann, A.; Nagel, G. Two-component cyclase opsins of green algae are ATP-dependent and light-inhibited guanylyl cyclases. BMC Biol. 2018, 16, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Luck, M.; Escobar, F.V.; Glass, K.; Sabotke, M.-I.; Hagedorn, R.; Corellou, F.; Siebert, F.; Hildebrandt, P.; Ehegemann, P. Photoreactions of the histidine kinase rhodopsin Ot-HKR from the marine picoalga ostreococcus tauri. Biochemie 2019, 58, 1878–1891. [Google Scholar] [CrossRef] [PubMed]
- Man, D.; Wang, W.; Sabehi, G.; Aravind, L.; Post, A.F.; Massana, R.; Spudich, E.N.; Spudich, J.L.; Béjà, O. Diversification and spectral tuning in marine proteorhodopsins. EMBO J. 2003, 22, 1725–1731. [Google Scholar] [CrossRef] [PubMed]
- Shimono, K.; Iwamoto, M.; Sumi, M.; Kamo, N. Effects of three characteristic amino acid residues of pharaonis phoborhodopsin on the absorption maximum. Photochem. Photobiol. 2000, 72, 141–145. [Google Scholar] [PubMed]
- Shimono, K.; Ikeura, Y.; Sudo, Y.; Iwamoto, M.; Kamo, N. Environment around the chromophore in pharaonis phoborhodopsin: Mutation analysis of the retinal binding site. Biochim. Biophys. Acta BBA Biomembr. 2001, 1515, 92–100. [Google Scholar] [CrossRef]
- Luck, M.; Bruun, S.; Keidel, A.; Ehegemann, P.; Hildebrandt, P. Photochemical chromophore isomerization in histidine kinase rhodopsin HKR1. FEBS Lett. 2015, 589, 1067–1071. [Google Scholar] [CrossRef]
- Luck, M.; Ehegemann, P. The two parallel photocycles of the Chlamydomonas sensory photoreceptor histidine kinase rhodopsin 1. J. Plant Physiol. 2017, 217, 77–84. [Google Scholar] [CrossRef]
- Ogren, J.I.; Mamaev, S.; Russano, D.; Li, H.; Spudich, J.L.; Rothschild, K.J. Retinal chromophore structure and schiff base interactions in red-shifted channelrhodopsin-1 from chlamydomonas augustae. Biochemie 2014, 53, 3961–3970. [Google Scholar] [CrossRef]
- Govorunova, E.G.; Spudich, E.N.; Lane, C.E.; Sineshchekov, O.A.; Spudich, J.L. New channelrhodopsin with a red-shifted spectrum and rapid kinetics from mesostigma viride. mBio 2011, 2, e00115-11. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Prigge, M.; Beyrière, F.; Tsunoda, S.P.; Mattis, J.; Yizhar, O.; Hegemann, P.; Deisseroth, K. Red-shifted optogenetic excitation: A tool for fast neural control derived from Volvox carteri. Nat. Neurosci. 2008, 11, 631–633. [Google Scholar] [CrossRef] [PubMed]
- Oda, K.; Vierock, J.; Oishi, S.; Rodriguez-Rozada, S.; Taniguchi, R.; Yamashita, K.; Wiegert, J.S.; Nishizawa, T.; Hegemann, P.; Nureki, O. Crystal structure of the red light-activated channelrhodopsin Chrimson. Nat. Commun. 2018, 9, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Govorunova, E.G.; Sineshchekov, O.A.; Li, H.; Wang, Y.; Brown, L.S.; Spudich, J.L. RubyACRs, nonalgal anion channelrhodopsins with highly red-shifted absorption. Proc. Natl. Acad. Sci. USA 2020, 117, 22833–22840. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.Y.; Knutsen, P.M.; Muller, A.; Kleinfeld, D.; Tsien, R.Y. ReaChR: A red-shifted variant of channelrhodopsin enables deep transcranial optogenetic excitation. Nat. Neurosci. 2013, 16, 1499–1508. [Google Scholar] [CrossRef]
- Inoue, K.; Marín, M.D.C.; Tomida, S.; Nakamura, R.; Nakajima, Y.; Olivucci, M.; Kandori, H. Red-shifting mutation of light-driven sodium-pump rhodopsin. Nat. Commun. 2019, 10, 1–11. [Google Scholar] [CrossRef]
- Ganapathy, S.; Kratz, S.; Chen, Q.; Hellingwerf, K.J.; De Groot, H.J.M.; Rothschild, K.J.; DeGrip, W.J. Redshifted and near-infrared active analog pigments based upon archaerhodopsin-3. Photochem. Photobiol. 2019, 95, 959–968. [Google Scholar] [CrossRef]
- Brown, L.S. Fungal rhodopsins and opsin-related proteins: Eukaryotic homologues of bacteriorhodopsin with unknown functions. Photochem. Photobiol. Sci. 2004, 3, 555–565. [Google Scholar] [CrossRef]
- Ruiz-González, M.X.; Marín, I. New insights into the evolutionary history of type 1 rhodopsins. J. Mol. Evol. 2004, 58, 348–358. [Google Scholar] [CrossRef]
- Pasquale, S.M.; Goodenough, U.W. Cyclic AMP functions as a primary sexual signal in gametes of Chlamydomonas reinhardtii. J. Cell Biol. 1987, 105, 2279–2292. [Google Scholar] [CrossRef]
- Boonyareth, M.; Saranak, J.; Pinthong, D.; Sanvarinda, Y.; Foster, K.W. Roles of cyclic AMP in regulation of phototaxis in Chlamydomonas reinhardtii. Biologia 2009, 64, 1058–1065. [Google Scholar] [CrossRef]
- Avelar, G.M.; Schumacher, R.I.; Zaini, P.A.; Leonard, G.; Richards, T.A.; Gomes, S.L. A rhodopsin-guanylyl cyclase gene fusion functions in visual perception in a fungus. Curr. Biol. 2014, 24, 1234–1240. [Google Scholar] [CrossRef] [PubMed]
- Buensuceso, R.N.C.; Nguyen, Y.; Zhang, K.; Daniel-Ivad, M.; Sugiman-Marangos, S.N.; Fleetwood, A.D.; Zhulin, I.B.; Junop, M.S.; Howell, P.L.; Burrows, L.L. The conserved tetracopeptide repeat-containing C-terminal domain of Pseudomonas aeruginosa FimV is required for its cyclic AMP-dependent and -independent Functions. J. Bacteriol. 2016, 198, 2263–2274. [Google Scholar] [CrossRef] [PubMed]
- Buensuceso, R.N.C.; Daniel-Ivad, M.; Kilmury, S.L.N.; Leighton, T.L.; Harvey, H.; Howell, P.L.; Burrows, L.L. Cyclic AMP-independent control of twitching motility in pseudomonas aeruginosa. J. Bacteriol. 2017, 199, 00188-17. [Google Scholar] [CrossRef] [PubMed]
- Berry, A.; Han, K.; Trouillon, J.; Robert-Genthon, M.; Ragno, M.; Lory, S.; Attree, I.; Elsen, S. cAMP and Vfr control exolysin expression and cytotoxicity of pseudomonas aeruginosa taxonomic outliers. J. Bacteriol. 2018, 200, e00135-18. [Google Scholar] [CrossRef]
- Wang, K.; Duan, C.; Zou, X.; Song, Y.; Li, W.; Xiao, L.; Peng, J.; Yao, L.; Long, Q.; Liu, L. Increased mediator complex subunit 15 expression is associated with poor prognosis in hepatocellular carcinoma. Oncol. Lett. 2018, 15, 4303–4313. [Google Scholar] [CrossRef]
- Yang, F.; Vought, B.W.; Satterlee, J.S.; Walker, A.K.; Sun, Z.-Y.J.; Watts, J.L.; Debeaumont, R.; Saito, R.M.; Hyberts, S.G.; Yang, S.; et al. An ARC/Mediator subunit required for SREBP control of cholesterol and lipid homeostasis. Nat. Cell Biol. 2006, 442, 700–704. [Google Scholar] [CrossRef]
- Thakur, J.K.; Arthanari, H.; Yang, F.; Chau, K.H.; Wagner, G.; Näär, A.M. Mediator subunit Gal11p/MED15 is required for fatty acid-dependent gene activation by yeast transcription factor Oaf1p. J. Biol. Chem. 2008, 284, 4422–4428. [Google Scholar] [CrossRef]
- Syring, I.; Weiten, R.; Schmidt, D.; Steiner, S.; Kristiansen, G. The knockdown of the Mediator complex subunit MED15 restrains urothelial bladder cancer cells’ malignancy. Oncol. Lett. 2018, 16, 3013–3021. [Google Scholar] [CrossRef]
- Zhao, M.; Yang, X.; Fu, Y.; Wang, H.; Ning, Y.; Yan, J.; Chen, Y.-G.; Wang, G. Mediator MED15 modulates transforming growth factor beta (TGFβ)/Smad signaling and breast cancer cell metastasis. J. Mol. Cell Biol. 2013, 5, 57–60. [Google Scholar] [CrossRef]
- Schlieker, C.; Korbel, G.A.; Kattenhorn, L.M.; Ploegh, H.L. A Deubiquitinating activity is conserved in the large tegument protein of the herpesviridae. J. Virol. 2005, 79, 15582–15585. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Wang, K.; Li, J.; Zheng, C. Herpes simplex virus 1 ubiquitin-specific protease UL36 inhibits beta interferon production by deubiquitinating TRAF3. J. Virol. 2013, 87, 11851–11860. [Google Scholar] [CrossRef] [PubMed]
- Lander, G.C.; Estrin, E.; Matyskiela, M.E.; Bashore, C.; Nogales, E.; Martin, A. Complete subunit architecture of the proteasome regulatory particle. Nat. Cell Biol. 2012, 482, 186–191. [Google Scholar] [CrossRef] [PubMed]
- Jossé, L.; Harley, M.E.; Pires, I.M.S.; Hughes, D.A. Fission yeast Dss1 associates with the proteasome and is required for efficient ubiquitin-dependent proteolysis. Biochem. J. 2005, 393, 303–309. [Google Scholar] [CrossRef]
- Wang, Q.; Young, P.; Walters, K.J. Structure of S5a bound to monoubiquitin provides a model for polyubiquitin recognition. J. Mol. Biol. 2005, 348, 727–739. [Google Scholar] [CrossRef]
- Walters, K.J.; Goh, A.M.; Wang, Q.; Wagner, G.; Howley, P.M. Ubiquitin family proteins and their relationship to the proteasome: A structural perspective. Biochim. Biophys. Acta Bioenerg. 2004, 1695, 73–87. [Google Scholar] [CrossRef]
- Venkitaraman, A.R. Cancer susceptibility and the functions of BRCA1 and BRCA2. Cell 2002, 108, 171–182. [Google Scholar] [CrossRef]
- Marston, N.J.; Richards, W.J.; Hughes, D.; Bertwistle, D.; Marshall, C.J.; Ashworth, A. Interaction between the product of the breast cancer susceptibility gene BRCA2 and DSS1, a protein functionally conserved from yeast to mammals. Mol. Cell. Biol. 1999, 19, 4633–4642. [Google Scholar] [CrossRef]
- Boöttcher, S.; Klupp, B.G.; Granzow, H.; Fuchs, W.; Michael, K.; Mettenleiter, T.C. Identification of a 709-amino-acid internal nonessential region within the essential conserved tegument protein (p)UL36 of pseudorabies virus. J. Virol. 2006, 80, 9910–9915. [Google Scholar] [CrossRef]
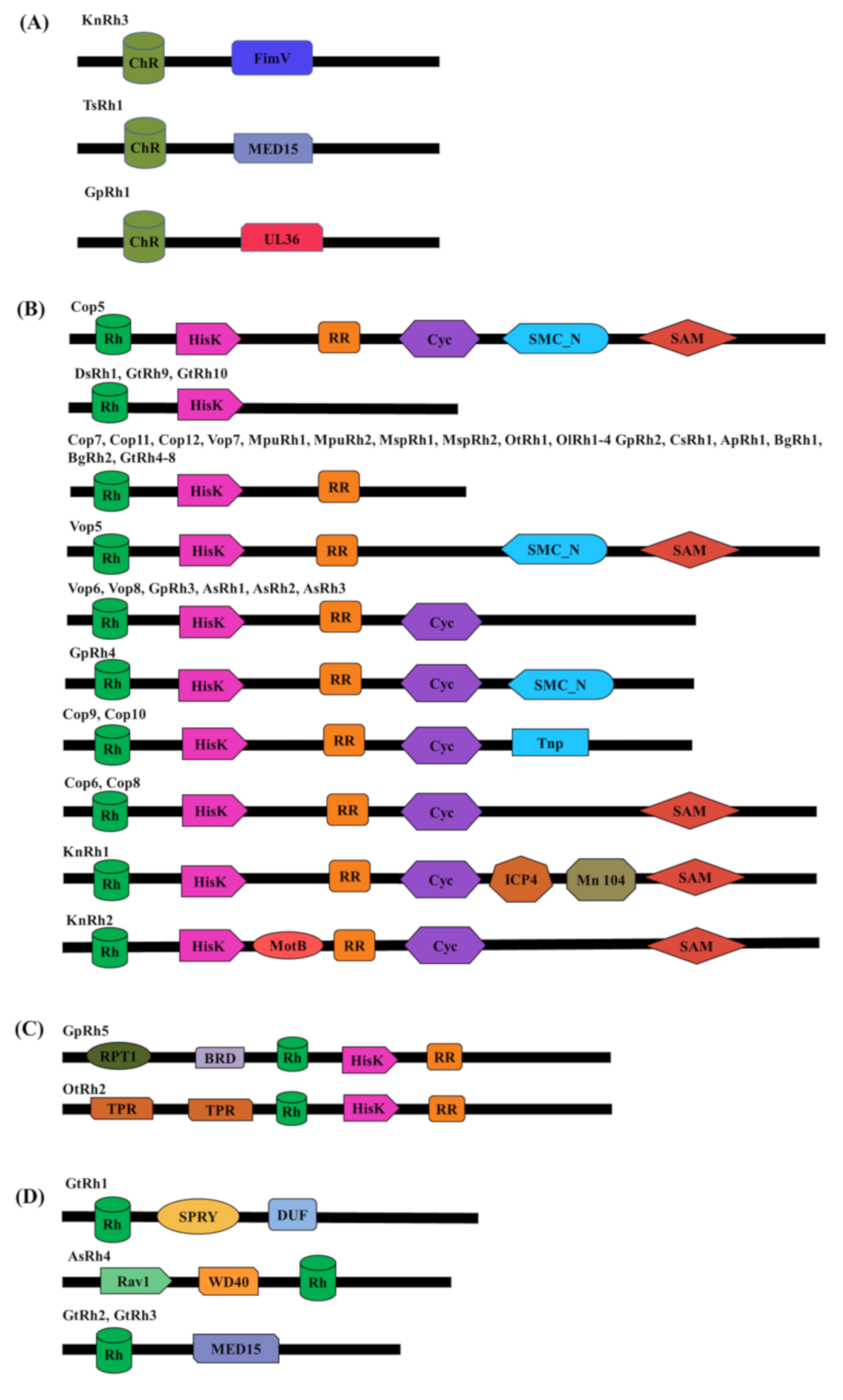

| Modular Domain | Channelrhodopsin | Functional Role and Optogenetic Potential |
|---|---|---|
| FimV (Peptidoglycan binding protein) | KnRh3 | In bacteria: Controls bacterial pathogenesis by indirectly activating adenylyl cyclase and hence cAMP level. |
| MED15 (Subunit of mediator complex) | TsRh1 | In mammals: Regulates cholesterol and lipid homeostasis. Promotes cancerous growth and is used as a biomarker for malignancies. |
| UL36 (Large tegument protein) | GpRh1 | Regulates viral entry to the cells. |
| Function of the Residue | Proton Acceptor | Proton Donor | DC Gate | Stabilizes Proton Acceptor | Retinal Attachment |
|---|---|---|---|---|---|
| No. corresponds to ChR2 | 253 | 156 | 128 | 120 | 257 |
| ChR2 | D253 | D156 | C128 | R120 | K257 |
| KnRh3 | D250 | D154 | C126 | R118 | K254 |
| TsRh1 | D236 | D139 | C111 | R103 | K240 |
| GpRh1 | D213 | D116 | C88 | R80 | K217 |
| Modular Domain | Modular Rhodopsins | Cellular Role and Optogenetic Potential |
|---|---|---|
| HisK | DsRh1, GtRh4-10, Cop5-12, Vop5-8, AsRh1-3, GpRh2-5, KnRh1 & 2, OtRh1&2, OlRh1-4, MpuRh1&2, Msp1&2, CsRh1, ApRh1, BgRh1&2 | Part of two-component signaling; regulates gene expression |
| HisK-RR (Histidine kinase-response regulator) Two-component signaling system | GtRh4-8, Cop5-12, Vop5-8, AsRh1-3, GpRh2-5, KnRh1 & 2, OtRh1&2, OlRh1-4, MpuRh1&2, Msp1&2, CsRh1, ApRh1, BgRh1&2 | Regulates gene expression and various other cell processes via output domain like helix-turn-helix (HTH), RNA, enzyme, or ligand-binding domain. |
| Cyc (Cyclase) | Cop5, 6, 8, 9 &10, Vop6&8, AsRh1-3, GpRh3&4, KnRh1 & 2 | Regulates the level of secondary messengers: cAMP and cGMP. |
| SMC_N (Structural Maintenance of chromosome _N terminal) | Cop5, Vop5, GpRh4 | Stabilizes the chromosome, helps in its proper segregation during cell division and DNA repair. |
| Tnp (Transposase) | Cop9 & 10 | Recognizes the transposable elements in DNA and catalyzes their movement to another DNA. |
| SAM (Sterile alpha motif) | Cop5-8, Vop5, KnRh1 & 2 | Mediate protein-protein interactions, RNA and lipid binding; regulates transcription factor |
| ICP4 (Infected-cell polypeptide 4) | KnRh1 | Major transcription factor of herpes simplex virus type1 (HSV-1) |
| Mn104 (Microneme/rhoptry) | KnRh1 | Helps in invading host cell by apicomplexan parasites; N-terminal region proposed to serve as a signal peptide for ER |
| MotB (Flagellar motor protein) | KnRh2 | MotB acts as a stator in the proton pump. |
| RPT1 (Regulatory Particle Triple ATPase) | GpRh5 | Forms a part of 26S proteasomal complex |
| BRD (Bromodomain) | GpRh5 | Modulate gene expression by associating with acetylated lysine on histone |
| TPR (Tetracopeptide repeat) | OtRh2 | Regulates virulence in bacteria; translocation of receptors to their respective organelles in different systems |
| SPRY [Spore lysis A (Spl A) in Dictyostelium discoideum and mammalian Ryanodine receptor (RYR)] | GtRh1 | Substrate binding for ubiquitination in ubiquitin ligase family proteins; involved in the various immune response |
| DUF (Domain of unknown function) | GtRh1 | Mediate protein-protein interaction and transcription repression; ATP dependent protein kinase; enzymatic part of dicer; virulence and pathogenesis. |
| Rav1 (Regulator of V-ATPase of vacuole membrane protein 1) | AsRh4 | Regulates the assembly of V-ATPase (ATP powered H+ pump in vacuole forming organelles) |
| WD40 | AsRh4 | Mediate protein–protein interaction |
| Function of the Residue | Ion Pumping | Proton-Release to Outside | Retinal Attachment | |||||
|---|---|---|---|---|---|---|---|---|
| No. corresponds to BR | 85 | 89 | 90 | 96 | 212 | 194 | 204 | 216 |
| BR | D | T | T | D | D | E | E | K |
| HR | T90 | S94 | T95 | A101 | D217 | E198 | T209 | K221 |
| KR2 (Na+) | N112 | D116 | V117 | Q123 | D251 | L227 | R243 | K255 |
| ASR1 | D75 | T79 | T80 | S86 | P206 | S188 | D198 | K210 |
| SR2 | D75 | T79 | T80 | F86 | D201 | L188 | D193 | K205 |
| RhoGC | E254 | T258 | C259 | L265 | D380 | S364 | A372 | K384 |
| RhoPDE | E164 | T168 | C167 | W175 | D292 | Q276 | G284 | K296 |
| AsRh4 | D2593 | T2597 | T2598 | D2604 | D2718 | G2701 | E2710 | K2722 |
| GtRh1 | F152 | S156 | T157 | I163 | D297 | G280 | K289 | K301 |
| GtRh2/3 | D95 | T99 | C100 | T106 | D248 | T232 | E240 | K252 |
| Cop5 | M113 | T117 | T118 | L124 | D239 | M223 | E231 | K243 |
| Cop6 | Q170 | T174 | T175 | I181 | N294 | V279 | - | K298 |
| Cop7 | Q161 | S165 | T166 | M172 | D287 | W271 | E279 | K291 |
| Cop8 | L67 | T71 | A72 | I78 | D194 | D178 | S186 | K198 |
| Cop9-10 | L141 | T145 | A146 | I152 | D268 | D252 | S260 | K272 |
| Cop11 | C95 | T99 | T100 | L106 | D279 | L263 | E271 | K283 |
| Cop12 | C95 | T99 | T100 | L106 | D221 | L205 | E213 | K225 |
| Vop5 | M157 | T161 | T162 | L168 | D283 | L267 | E275 | K287 |
| Vop6 | Q153 | T157 | T158 | I164 | N278 | L263 | - | K282 |
| Vop7 | Q147 | S151 | T152 | M158 | D272 | W256 | E264 | K276 |
| MspRh1 | E140 | T144 | T145 | I151 | D284 | F268 | Q276 | K288 |
| MspRh2 | E142 | G146 | T147 | L153 | D299 | S283 | L291 | K303 |
| MpuRh1 | E140 | T144 | T145 | I151 | D300 | F284 | Q292 | K304 |
| MpuRh2 | S151 | S155 | T156 | L162 | D328 | A312 | A320 | K332 |
| GtRh4 | Q92 | T96 | T97 | V103 | D225 | S209 | Y217 | K229 |
| GtRh5 | Q222 | T226 | T227 | V233 | D355 | G339 | Y347 | K359 |
| GtRh6 | Q234 | T238 | T239 | V245 | D367 | G351 | Y359 | K371 |
| GtRh7 | Q116 | T120 | T121 | V127 | D249 | S233 | Y241 | K253 |
| GtRh8 | Q226 | T230 | T231 | V237 | D359 | L343 | Y351 | K363 |
| GtRh9 | Q229 | T233 | T234 | I240 | D362 | L346 | Y354 | K366 |
| GtRh10 | Q192 | T196 | T197 | V203 | D325 | L309 | F317 | K329 |
| BgRh1/2 | E173 | T177 | T178 | S184 | D302 | L286 | E294 | K306 |
| OtRh1 | E181 | T185 | T186 | L192 | D314 | M298 | E306 | K318 |
| OtRh2 | E476 | T480 | T481 | L487 | D609 | M593 | E601 | K613 |
| OlRh1 | E204 | T208 | T209 | L215 | D337 | L321 | E329 | K341 |
| OlRh2 | E260 | T264 | T265 | L271 | D393 | L377 | E385 | K397 |
| OlRh3 | E188 | T192 | T193 | L199 | D321 | L305 | E313 | K325 |
| OlRh4 | E115 | T119 | T120 | L126 | D248 | L232 | E240 | K252 |
| DsRh1 | Q140 | S144 | T145 | M151 | D268 | L252 | E260 | K272 |
| GpRh2 | C91 | T95 | T96 | L102 | D217 | L201 | E209 | K221 |
| GpRh3 | Q85 | T89 | T90 | I96 | N209 | A194 | - | K213 |
| GpRh4 | M67 | T71 | T72 | L78 | D193 | L177 | E185 | K197 |
| GpRh5 | Q1412 | S1416 | T1417 | M1423 | D1537 | L1521 | E1529 | K1541 |
| CsRh1 | M144 | A148 | T149 | T155 | D269 | L253 | E261 | K273 |
| ApRh1 | M67 | A71 | T72 | T78 | D192 | A176 | E184 | K196 |
| AsRh1 | N122 | T126 | T127 | L133 | N248 | L232 | T240 | K252 |
| AsRh2 | N123 | T127 | T128 | L134 | N249 | L233 | S241 | K253 |
| AsRh3 | Q78 | T82 | T83 | V89 | N203 | L187 | C195 | K207 |
| KnRh1 | Q166 | T170 | T171 | M177 | D292 | L276 | E284 | K296 |
| KnRh2 | Q95 | T99 | T100 | L106 | E221 | T205 | E213 | K225 |
| Rhodopsin | 105th Position/ Corresponding Amino Acid | Polar/Non-Polar aa | Green/Blue Shifted |
|---|---|---|---|
| Green PR | Leucine | Non-Polar | Green |
| Blue PR | Glutamine | Polar | Blue |
| KnRh3, TsRh1 and GpRh3 | Isoleucine | Non-polar | Green |
| Cop8-12, GpRh2, ApRh1, AsRh2 | Isoleucine | Non-polar | Green |
| MspRh1, MpuRh1, AsRh3-4, OtRh1-2, OlRh1-4, DsRh1, GtRh2,3 | Leucine | Non-polar | Green |
| Cop5-7, Vop5-7, GpRh3-5, GtRh4-10, AsRh1, MspRh2, MpuRh2, CsRh1, BgRh1-2, KnRh1-2 | Methionine | Non-polar | Green |
| GtRh1 | Aspartate | Acidic | unknown |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Awasthi, M.; Sushmita, K.; Kaushik, M.S.; Ranjan, P.; Kateriya, S. Novel Modular Rhodopsins from Green Algae Hold Great Potential for Cellular Optogenetic Modulation Across the Biological Model Systems. Life 2020, 10, 259. https://doi.org/10.3390/life10110259
Awasthi M, Sushmita K, Kaushik MS, Ranjan P, Kateriya S. Novel Modular Rhodopsins from Green Algae Hold Great Potential for Cellular Optogenetic Modulation Across the Biological Model Systems. Life. 2020; 10(11):259. https://doi.org/10.3390/life10110259
Chicago/Turabian StyleAwasthi, Mayanka, Kumari Sushmita, Manish Singh Kaushik, Peeyush Ranjan, and Suneel Kateriya. 2020. "Novel Modular Rhodopsins from Green Algae Hold Great Potential for Cellular Optogenetic Modulation Across the Biological Model Systems" Life 10, no. 11: 259. https://doi.org/10.3390/life10110259
APA StyleAwasthi, M., Sushmita, K., Kaushik, M. S., Ranjan, P., & Kateriya, S. (2020). Novel Modular Rhodopsins from Green Algae Hold Great Potential for Cellular Optogenetic Modulation Across the Biological Model Systems. Life, 10(11), 259. https://doi.org/10.3390/life10110259








