Abstract
This paper investigates an understudied generalization of the classical exponential, Rayleigh, and Weibull distributions, known as the power generalized Weibull distribution, particularly in the context of censored data. Characterized by one scale parameter and two shape parameters, the proposed model offers enhanced flexibility for modeling diverse lifetime data patterns and hazard rate behaviors. Notably, its hazard rate function can exhibit five distinct shapes, including upside-down bathtub and bathtub shapes. The study focuses on classical and Bayesian estimation frameworks for the model parameters and associated reliability metrics under a unified hybrid censoring scheme. Methodologies include both point estimation (maximum likelihood and posterior mean estimators) and interval estimation (approximate confidence intervals and Bayesian credible intervals). To evaluate the performance of these estimators, a comprehensive simulation study is conducted under varied experimental conditions. Furthermore, two empirical applications on real-world cancer datasets underscore the efficacy of the proposed estimation methods and the practical viability and flexibility of the explored model compared to eleven other existing lifespan models.
Keywords:
power generalized Weibull; unified hybrid censoring; reliability; classical and Bayes estimations; cancer data modeling MSC:
62F10; 62F15; 62N01; 62N02; 62N05
1. Introduction
Recent advancements in statistical modeling have resulted in the development of numerous flexible lifetime probability distributions designed to analyze lifetime data. Despite their adaptability, many proposed distributions exhibit complex structures due to a substantial number of parameters, often exceeding three, which complicates theoretical investigations and practical applications, particularly in contexts involving censored lifetime data. This complexity has led researchers to prefer classical models, such as the exponential or Weibull distributions, for real-world investigations. Notably, the power generalized Weibull (PGW) distribution, introduced by Bagdonavičius and Nikulin [1] within the framework of accelerated failure time models, remains an unexplored yet highly relevant option in this field. The PGW distribution features one scale parameter and two shape parameters, characterized by its cumulative distribution function (CDF) as
Under the parameter transformations and , the CDF specified in (1) can be reparameterized and rewritten in the form
where is the unknown parameter vector with scale parameter and shape parameters and , and . Dimitrakopoulou et al. [2] developed a novel lifetime model sharing the same CDF in (2), motivated by applications in competing risk strategies. The probability density function (PDF) corresponding to the CDF in (2) can be expressed as
The PGW distribution generalizes multiple classical models; it reduces to the exponential distribution when , the Weibull distribution for , the Rayleigh distribution when and , and the Nadarajah–Haghighi distribution introduced by Nadarajah and Haghighi [3] when . Several studies have explored the PGW distribution. Nikulin and Haghighi [4] assessd the PGW distribution’s validity, demonstrating its utility with censored survival data from cancer patients. Nikulin and Haghighi [5] analyzed the shape properties of the PGW distribution including the PDF and hazard rate function (HRF). Kumar and Dey [6] derived recurrence relations for single and product moments of PGW order statistics. Additionally, El-Din et al. [7] investigated progressive stress accelerated life testing for the PGW distribution using progressively Type-II censored samples (see Pandey and Kumari [8], Sabry et al. [9], and Dey et al. [10] for more detail). The reliability function (RF) and HRF, at a distinct time t, of the PGW distribution can be expressed as
and
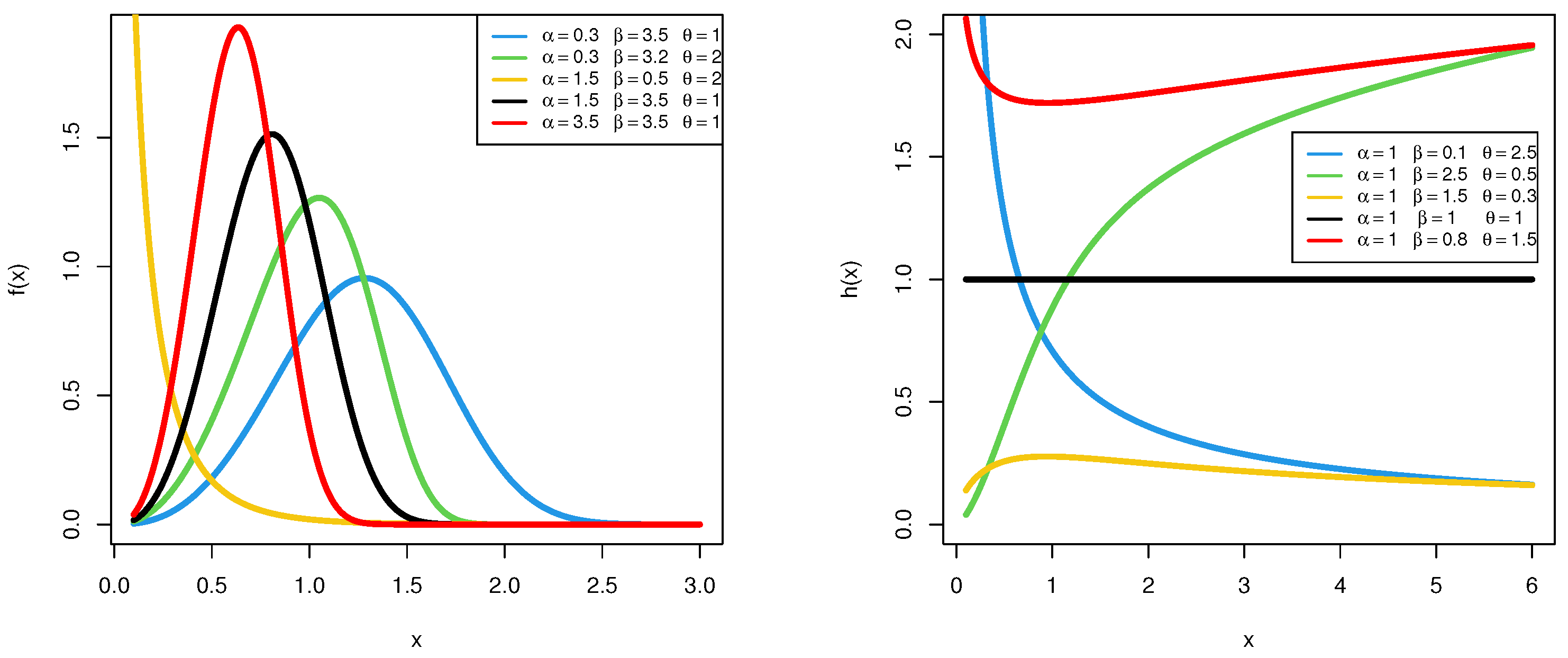
Depending on the values of the two shape parameters and , as shown in Figure 1, the HRF of the PGW distribution can exhibit five distinct forms: constant, monotonically increasing, monotonically decreasing, upside-down bathtub, or bathtub-shaped. Additionally, the PGW density shapes are also depicted in Figure 1. This adaptability positions the PGW model as a highly versatile tool for lifetime data analysis, particularly for datasets that may not be adequately captured by traditional distributions such as the Weibull, gamma, or Nadarajah–Haghighi.
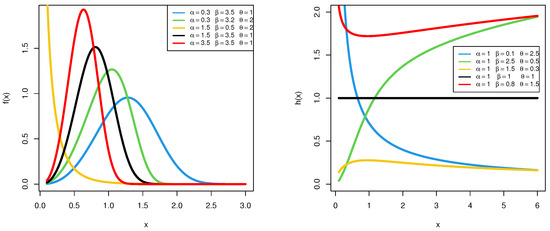
Figure 1.
The PDF (left) and HRF (right) shapes of the PGW distribution.
In reliability and survival analysis, censored data is a common scenario, wherein researchers frequently make observations about only a part of the sample under consideration. The statistical literature extensively documents various censoring mechanisms, including Type-I, Type-II, and hybrid censoring (HC) plans; the latter is a combination of Type-I and Type-II censoring plans. Consider a random sample of size n subjected to a life-testing experiment, where the observed failure times are ordered as . Epstein [11] introduced the HC framework, in which the experiment terminates at a randomly determined endpoint , where and are predefined before the experiment. Childs et al. [12] referred to this censoring approach as the Type-I HC scheme, emphasizing that the experiment duration cannot exceed T. Additionally, they proposed the Type-II HC scheme, which ensures a minimum of m failures by setting . While Type-II HC guarantees at least m failures, it poses the risk of excessively long testing durations. To enhance these frameworks, Chandrasekar et al. [13] introduced generalized Type-I and Type-II HC schemes. However, limitations remained: the generalized Type-I HC scheme did not guarantee a minimum number of failures, while the generalized Type-II HC scheme may result in very few or no observed failures. To address these challenges, Balakrishnan et al. [14] proposed a unified HC (UHC) methodology, which integrates the strengths of generalized HC schemes to provide a more balanced and effective censoring strategy.
Consider a life-testing experiment with n identical units. Prior to the experiment, fix with and thresholds . Balakrishnan et al. [14] defined the UHC plan as follows:
- If , terminate the test at .
- If , stop the test at .
- If , conclude the experiment at .
Let denote the termination time; r refers to the number of observed data, and and denote the number of failures observed before and , respectively. Then, we can observe one of the different six cases as displayed in Table 1.

Table 1.
Unified hybrid censoring scheme cases.
Let be an observed UHC sample, with for simplicity, then the joint likelihood function can be written, ignoring the constant term, as
where
The UHC methodology has been utilized in many studies such as Panahi and Sayyareh [15], Jeon and Kang [16], Dutta and Kayal [17], Dutta et al. [18], Hasaballah et al. [19], Shukla et al. [20], and Alotaibi et al. [21].
Despite the notable versatility of the PGW distribution in modeling diverse datasets, owing to its five distinct HRF shapes and its relatively simple PDF and CDF structures when compared with other Weibull generalizations, limited scholarly focus has been directed, to the best of our knowledge, toward parameter estimation under censored sampling frameworks. Furthermore, we have not identified any studies addressing the estimation of its reliability metrics, such as estimating the RF and HRF, which is very crucial in reliability and survival studies. Motivated by these gaps, this study aims to develop estimators for the PGW distribution parameters and its associated reliability metrics (RF and HRF) under data collected via the UHC methodology. We first investigate the frequentist estimation framework using the maximum likelihood method. This involves deriving point estimators for the parameters as well as constructing approximate confidence intervals (ACIs). Notably, the delta method is employed to approximate the sampling variances of the RF and HRF estimators. Subsequently, we implement a Bayesian estimation approach, using the squared error loss function and Markov chain Monte Carlo (MCMC) techniques to compute posterior estimates. Bayesian credible intervals (BCIs) are also derived from the posterior distributions. To evaluate the performance of these methodologies, a comprehensive simulation study is conducted, varying sample sizes, the desired number of observed data, and thresholds. Finally, two real-world applications are analyzed to illustrate the practical utility of the proposed methods.
The remainder of this study is organized as follows: Section 2 examines the maximum likelihood estimation approach for the model parameters and associated reliability metrics. Section 3 details Bayesian estimation frameworks utilizing the MCMC methodologies. The simulation design is discussed in Section 4, accompanied by the simulation results. Section 5 analyzes two empirical datasets to demonstrate practical applications. Finally, Section 6 concludes the study with key findings and implications.
2. Classical Likelihood Estimation
This section addresses three key objectives: first, deriving classical point estimators for the parameters of the PGW distribution; second, constructing the ACIs for these unknown parameters; and third, obtaining point estimators along with ACIs for the RF and HRF.
2.1. Point Estimation of the Model Parameters
Given a UHC sample , the joint likelihood function can be formulated using the CDF in (2), the PDF in (3) and the likelihood structure from (6) as follows:
The log-likelihood function of (8) is as follows:
The maximum likelihood estimators (MLEs) of the parameters are derived by solving the system of likelihood equations, which are formulated through partial differentiation of the log-likelihood function with respect to each parameter and setting the resulting expressions equal to zero.
and
where , and .
Given the intricate nature of the likelihood equations presented in (9)–(11), obtaining a closed-form analytical solution is not feasible. Instead, iterative techniques such as the Newton–Raphson method must be employed to solve these equations simultaneously, thereby facilitating obtaining the MLEs for the unknown parameters, denoted .
2.2. ACIs for the Model Parameters
Confidence regions for the model parameters rely on deriving the asymptotic variance–covariance matrix. While classical asymptotic theory derives this matrix via the expected Fisher information matrix:
The analytical complexity of computing these expectations for the PGW model motivates the use of the observed Fisher information matrix:
The variance–covariance matrix is then approximated as:
where the main diagonal elements represent the estimated variances of the MLEs for the parameters and , respectively. The second and mixed derivatives are given as follows:
and
where , , , , , , and .
Under the asymptotic properties of the MLEs, it is known that , as n tends to infinity. For practical inference, the ACIs for individual parameters are constructed as
where is the MLE of and denotes the upper quantile of the standard normal distribution.
2.3. Point Estimates and ACIs for the RF and HRF
By employing the invariance principle of maximum likelihood estimation, we can obtain the RF estimator and HRF estimator at any time . These estimators can be derived by substituting the MLEs into the RF and HRF expressions provided in (4) and (5), respectively, as
and
To construct the ACIs for the RF and HRF , we approximate the variances of their MLEs using the delta method. This involves obtaining the partial derivatives of these functions with respect to various parameters. Let and . Evaluated at , these gradients are
and
The approximate variances are then
where is the estimated variance–covariance matrix obtained in (12). The ACIs are:
3. Bayesian Estimation
In recent years, Bayesian inference has become a robust analytical tool across various disciplines. Its ability to integrate prior knowledge with observed data is particularly advantageous in reliability studies, where traditional methods often struggle in the presence of limited data. This section develops Bayesian estimators and associated BCIs for the parameters, RF, and HRF of the PGW distribution. Although alternative loss functions may be applicable, we focus in this analysis on the use of the squared error loss function due to its analytical symmetry and broad applicability.
3.1. Prior, Posterior, and Bayes Estimator
In the absence of conjugate prior distributions for , , and , and due to the computational challenges posed by Jeffreys priors as a result of complex variance–covariance structures, gamma distributions are chosen as priors for these parameters. This decision is driven by three key considerations: (1) the gamma distribution naturally accommodates the non-negative domain of the unknown parameters of the PGW distribution; (2) it offers improved computational efficiency; (3) it simplifies analytical handling in models involving multiple parameters. By assuming the independence between the three unknown parameters and , , and , the joint prior distribution of can be expressed as:
where the hyper-parameters . In Bayesian statistics, the posterior distribution of the parameters given by the data is obtained through integrating the likelihood function of the data given the parameters and the joint prior distribution of the parameters. Using (8) and (13), the joint posterior distribution can be formulated as
where the normalized constant A is given by .
Under squared error loss, the Bayes estimator of the function , which represents any parametric function of , is obtained as the posterior mean. Using (14), the Bayes estimator of , denoted , is then expressed as
The Bayes estimator formulated in (15) involves analytically intractable integrals. To address this computational challenge, we employ MCMC techniques to simulate samples from the posterior distribution defined in (14). These generated samples ease the numerical approximation of Bayes estimates through posterior expectations and support the construction of the BCIs.
3.2. Conditional Distributions and MCMC Paradigm
A key aspect of implementing the MCMC is the derivation of the full conditional posterior distributions for each parameter. Based on the joint posterior in (14), the conditional distributions for the parameters , , and , are as follows:
and
The conditional distributions presented in (16)–(18) lack standard forms, precluding direct sampling methods. To address this challenge, we employ the Metropolis–Hastings (MH) algorithm, an MCMC technique that systematically samples from these non-conjugate posterior distributions. The MH algorithm utilizes a normal proposal distribution, which is initialized based on the MLEs, to generate candidate samples. It subsequently evaluates acceptance probabilities to ensure convergence towards the target posterior distribution. The full MCMC sampling procedure is outlined below.
- Set initial values of and based on the related MLEs.
- For , carry out the next MH steps:
- (a)
- Generate .
- (b)
- Calculate the acceptance probability:
- (c)
- Accept/reject: .
- At iteration j, obtain the RF and HRF as
- Remove the first samples as a burn-in period. Retain the subsequent sequence for Bayesian analysis:
The Bayesian inference results for the parameter (with an analogous approach applicable to other parameters) are derived from the MCMC samples. This includes both the Bayes point estimate and BCI. Under the squared error loss function, the Bayes estimate of is calculated as the posterior mean:
where is the total number of MCMC iterations, and is the burn-in period. The BCI for is constructed using the empirical quantiles of the post-burn-in samples:
where and are the and quantiles of .
4. Monte Carlo Comparisons
Comprehensive Monte Carlo simulations are carried out in this part to evaluate the performance of the proposed methods: maximum likelihood and Bayesian estimations and their interval approaches extensions. To achieve this, by repeating the UHC mechanism 1000 times based on several choices of n, and (for ), we take two levels of PGW populations, namely Pop-1:PGW(0.2, 0.5, 0.8) and Pop-2:PGW(0.4, 0.8, 1.2). For a fixed time point , the actual values of and are set to be 0.9521 and 0.2376 (from Pop-1) and set to be 0.9289 and 0.5707 (from Pop-2), respectively.
Following Kundu [22], the prior parameters are selected such that the resulting prior means align with the true parameter values. For the Bayesian estimations of , , , , or , there are two informative gamma prior setups for namely,
- For Pop-1:PGW(0.2, 0.5, 0.8):
- -
- Prior A[PA]: and ;
- -
- Prior B[PB]: and .
- For Pop-2:PGW(0.4, 0.8, 1.2):
- -
- Prior A[PA]: and ;
- -
- Prior B[PB]: and .
Once 1000 UCS samples are gathered, the MLEs, along with their 95% ACI estimates of , , , , and , are obtained using the ‘’ package proposed by Henningsen and Toomet [23]. Taking and (burn-in), after installing the ‘’ package (proposed by Plummer et al. [24]) in R software 4.2.2, the Bayes point estimations and associated 95% BCI estimations of , , , , or are computed.
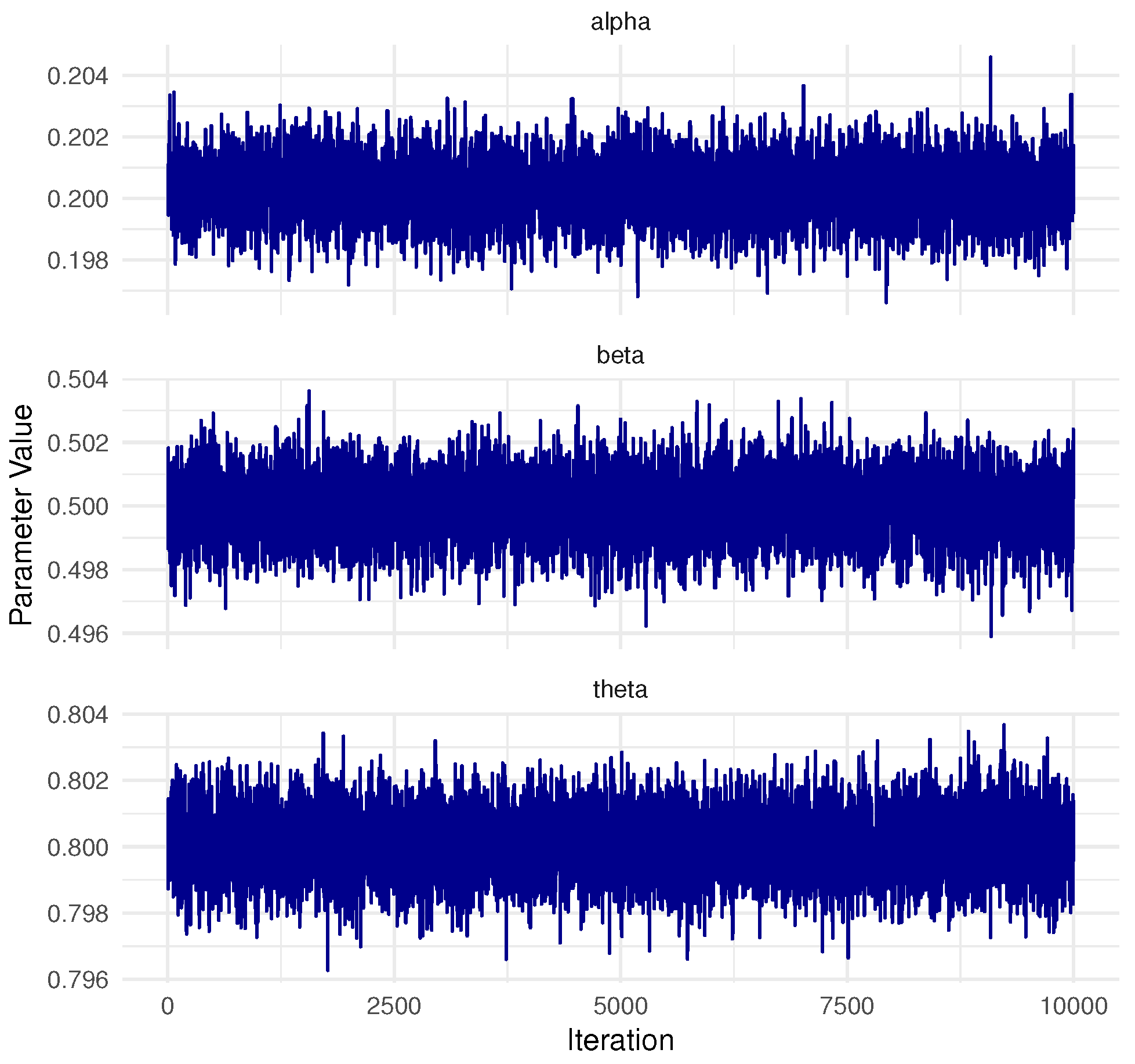
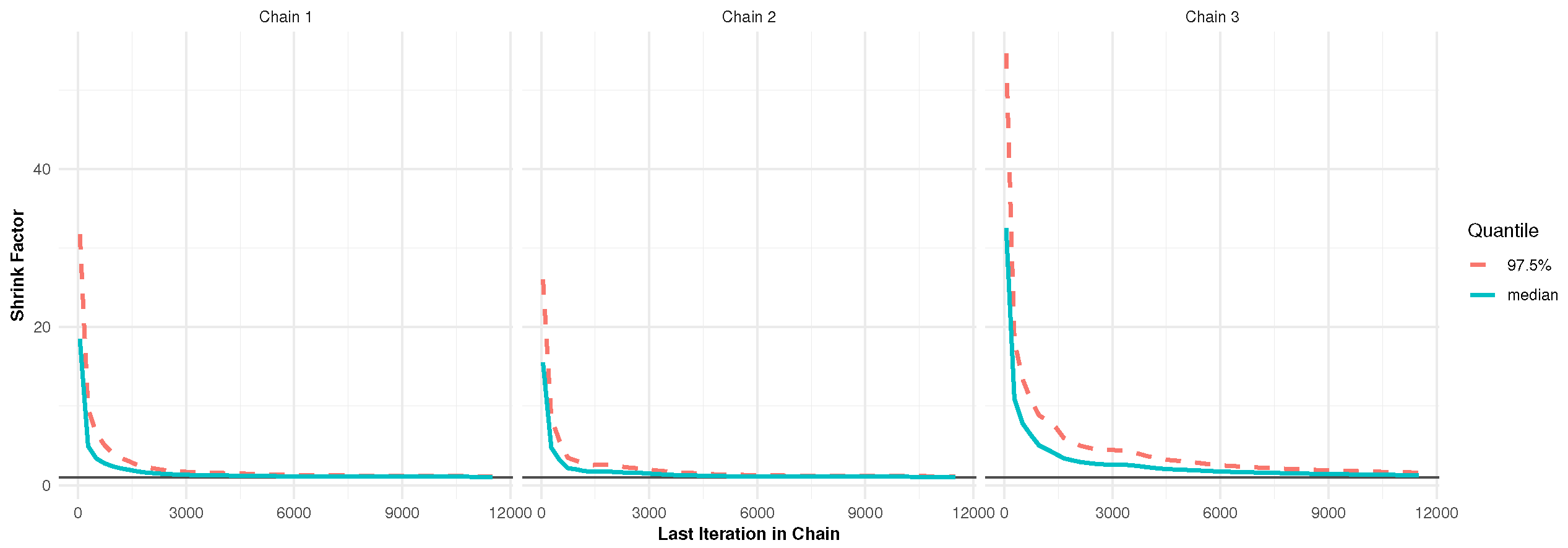
In Bayesian MCMC procedures, obtaining a representative sample from the target posterior distribution hinges on verifying the convergence of the Markov chains, which is a central concern in the analysis. For the dataset from Pop-1 with PA, specified by and , convergence was assessed using two diagnostic tools: trace plots and the Brooks–Gelman–Rubin (BGR) statistic, as illustrated in Figure 2 and Figure 3. The trace plots (in Figure 2) demonstrate that the simulated chains for parameters , , , , and exhibit proper mixing behavior, indicating stability over iterations. Figure 3, presenting the BGR diagnostic, reveals consistency between within-chain and between-chain variances, affirming convergence and the effectiveness of the selected burn-in period in eliminating initial value bias. These results collectively support the validity and robustness of the posterior estimates for the model parameters.
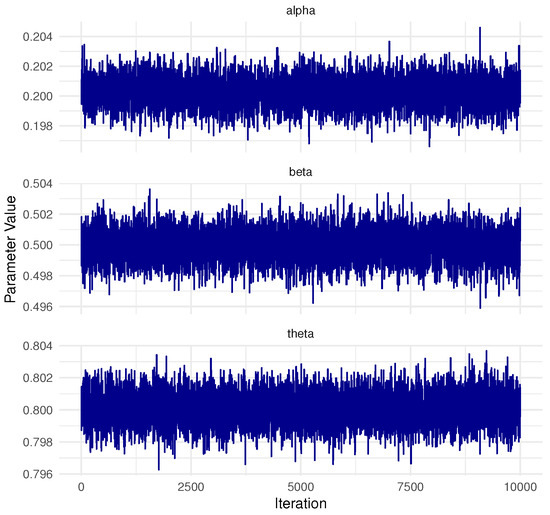
Figure 2.
Trace plots for 10,000 MCMC iterations of , , , , and in Monte Carlo simulation.
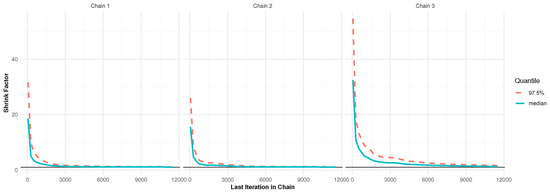
Figure 3.
The BGR plot for 12,000 MCMC iterations in Monte Carlo simulation.
In calculation setups, the average estimates (Av.Es) of (for short) are given by
respectively, where is the offered estimate of at the ith simulated sample.
Comparison of the point estimates are examined in terms of their mean squared errors (MSEs) and mean absolute biases (MABs), as follows:
and
Moreover, the interval estimates are assessed by their average interval lengths (AILs) and the coverage probabilities (CPs), respectively, as
and
where is the indicator function, and and denote the lower and upper bounds, respectively, of the ACI (or BCI) interval of .
In Table 2, Table 3, Table 4, Table 5 and Table 6, the Av.Es, MSEs, and MABs are presented in the first, second, and third columns, respectively. Similarly, Table 7, Table 8, Table 9, Table 10 and Table 11 report the AILs and CPs in the first and second columns, respectively.

Table 2.
The point estimation results of .

Table 3.
The point estimation results of .

Table 4.
The point estimation results of .

Table 5.
The point estimation results of .

Table 6.
The point estimation results of .

Table 7.
The 95% interval estimation results of .

Table 8.
The 95% interval estimation results of .

Table 9.
The 95% interval estimation results of .

Table 10.
The 95% interval estimation results of .

Table 11.
The 95% interval estimation results of .
From Table 2, Table 3, Table 4, Table 5, Table 6, Table 7, Table 8, Table 9, Table 10 and Table 11, in terms of the lowest MSEs, MABs, and AILs as well as the highest CPs, we report the following comments:
- In general, all proposed estimators for the PGW parameters , , , , and show satisfactory performance, exhibiting low MSE, MAB, and AIL values, along with high CP values.
- As expected, Bayesian MCMC estimates outperform classical estimates across all parameters. This is due to the Bayesian approach incorporating prior information along with censored data, whereas likelihood methods rely solely on the observed data.
- Bayes point estimates (or BCI estimates) derived from PB consistently outperform those from PA in terms of lower MSEs, MABs, and AILs and higher CPs. This is attributed to the smaller prior variance in PB.
- Regarding interval estimation, the BCI estimates of , , , , or behave surpass those derived from the ACI method in all parameters.
- As n increases, all proposed estimators benefit from reduced MSE, MAB, and AIL values, while CP values improve. A similar trend is observed when increase together.
- When grow, the accuracy of all inferential computations of , , , , or generally tends to be better.
- When , , and grow, it is noted that
- -
- the MSE and MAB results of and decrease, while those of , , and increase;
- -
- the AIL results of , , and decrease, while those of and increase;
- -
- the CP results of and decrease, while those of , and increase.
- As a summary, the Bayes setup using an MCMC-based model is recommended for estimating the distribution parameters and/or reliability features involved in the PGW lifespan model in the presence of unified hybrid censored data.
5. Cancer Data Analysis
In this section, we analyze two real datasets from clinical trials to show how the suggested inference techniques can be used in practice and how the proposed PGW model performs better than others; one dataset shows the remission times of bladder cancer patients, and the other shows the survival rates of head and neck cancer patients.
5.1. Bladder Cancer
Bladder cancer is a disease where abnormal cells grow uncontrollably in the bladder’s lining, potentially spreading to other layers or organs. It often causes symptoms like blood in the urine, frequent urination, or pain while urinating. We now shall illustrate the analysis of a genuine real-world dataset that represents the remission times (in months) for 128 bladder cancer patients; see Table 12. In clinical trials, the remission time in bladder cancer is the interval during which a patient remains free of detectable cancer after successful treatment. It can range from months to years, influenced by factors such as tumor grade, treatment response, and patient health, with ongoing surveillance essential to catch potential recurrence early. This data is available in Lee and Wang [25] and rediscussed by Alotaibi et al. [26] and Alotaibi et al. [27].

Table 12.
Remission times for bladder cancer patients.
Using eleven lifetime models (reported in Table 13) that provide different failure rates as competitors, we now analyze the PGW distribution from the whole bladder cancer data. To establish this, to choose the best lifetime model, eight model selection metrics are considered: (1) negative log-likelihood (N-LL); (2) Akaike information (A.I.); (3) Bayesian information (B.I.); (4) consistent Akaike information (CA.I.); (5) Hannan–Quinn information (HQ.I.); (6) Anderson–Darling (A–D); (7) Cramér–von Mises (CvM); and (8) Kolmogorov–Smirnov (K–S) with its p-value.

Table 13.
Eleven competitive models of the PGW distribution.
The fitted metrics (1)–(6) are computed using the ‘’ package (by Marinho et al. [38]); refer to Table 14. Furthermore, each model parameter’s MLE (together with its standard error (Std.Err)) is fitted and presented in Table 14. Since the most suitable choice should match the lowest value of all the provided metrics (1)–(8), except the greatest P-value, the findings in Table 14 demonstrate that the PGW model is superior to the others.

Table 14.
Fitting results of the PGW and its competitors from bladder cancer data.
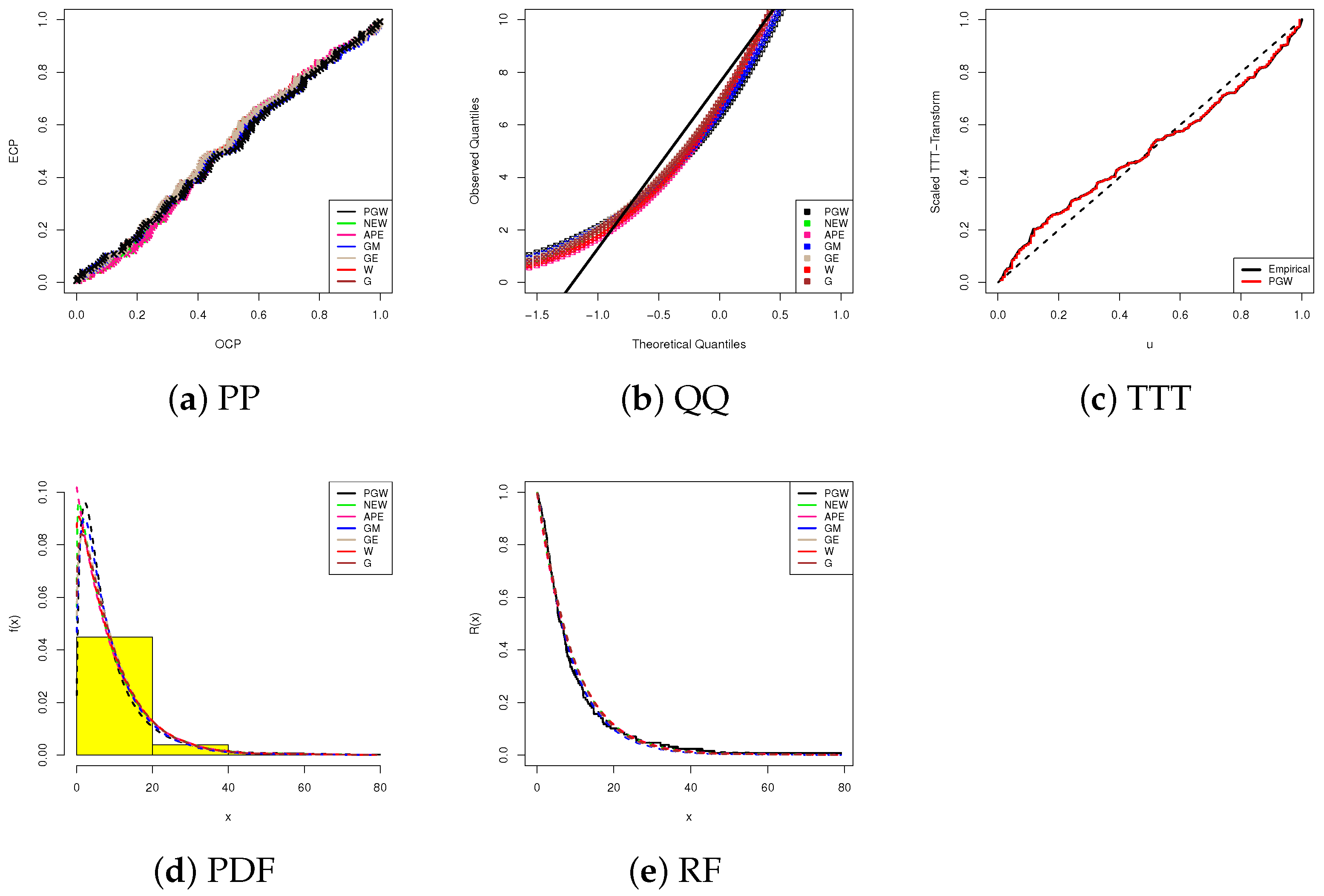
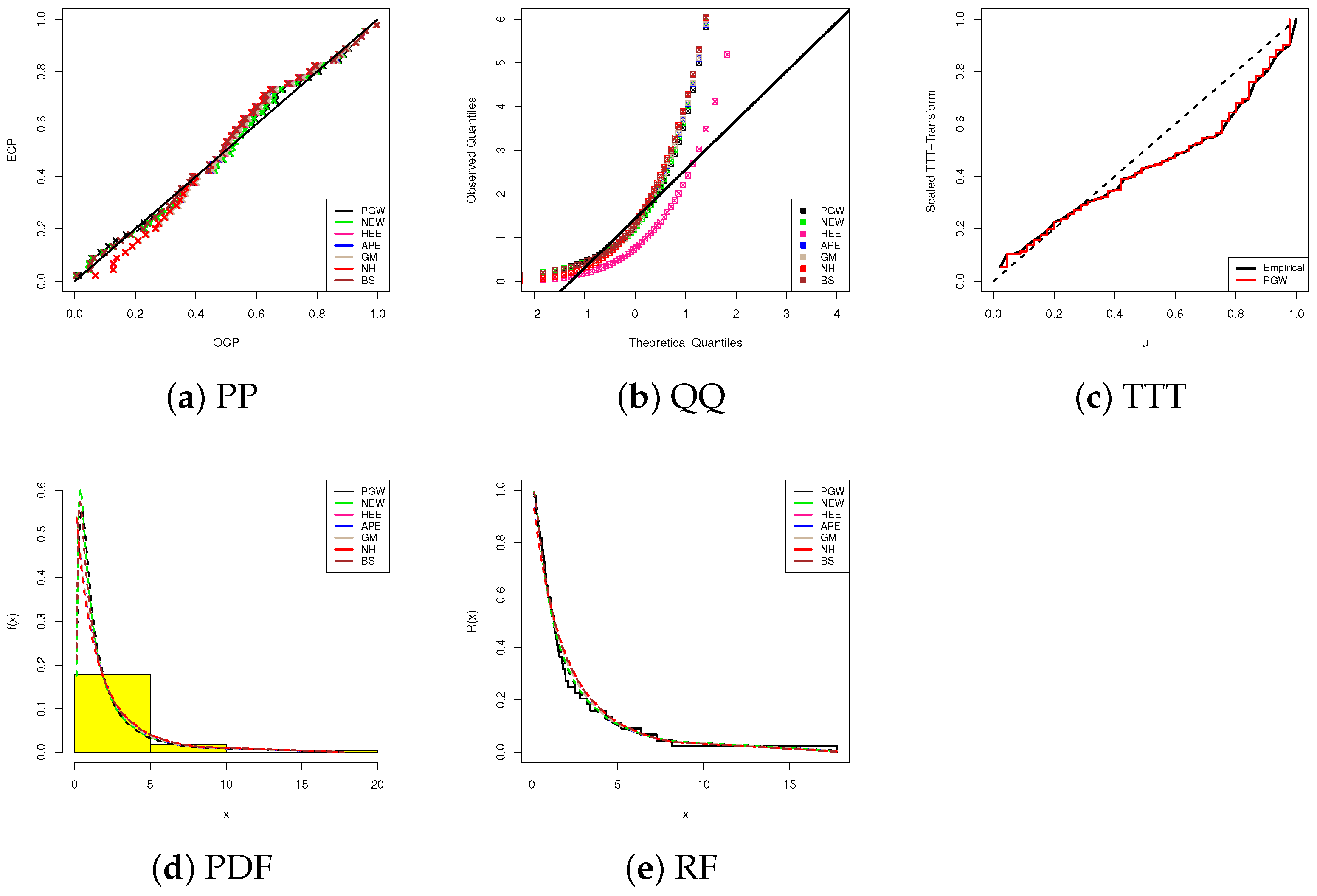
Using data visualizations, in Figure 4, we display our fitting results based on five visual tools, namely, (1) probability-probability (PP), (2) quantile-quantile (QQ), (3) scale TTT-transform (4), (iv) fitted PDF lines with bladder cancer histograms, and (5) fitted RF lines for the highest competitive models for the PGW distribution that provide the largest p-value. All subplots in Figure 4a–d support our conclusion that the proposed PGW lifetime model is the best choice when bladder cancer data is analyzed. Figure 4e reveals that the analyzed bladder cancer data provides an inverted bathtub failure rate and confirms one of the features of PGW failure rates discussed in Section 1.
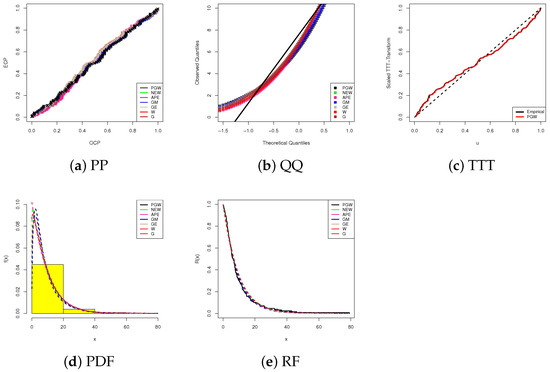
Figure 4.
Fitting diagrams of the PGW and its competitors from bladder cancer data.
From the entire bladder cancer dataset (reported in Table 12), using several levels of and (for ), different UHC samples are collected; see Table 15. For each sample (), we obtain the MLEs and MCMC estimators (along with their Std.Errs) as well as the 95% ACI/BCI estimators (along with their widths) of , , , , and (at ); see Table 16. Because the prior information about the PGW parameters , , and from the entire bladder cancer data is not available, improper gamma priors are used because they serve as noninformative or weakly informative priors, allowing the data to predominantly drive the posterior inference without imposing strong prior beliefs. Making use of gamma improper priors (by setting ) of , , and , all Bayes examinations are implemented when and . The findings in Table 16 exhibit that Bayesian MCMC estimates (in addition to the associated 95% BCI estimates) behave superiorly and precisely compared to those developed by likelihood estimates (in addition to associated 95% ACI estimates) in terms of lower standard errors and shorter interval widths.

Table 15.
Several UHC samples from bladder cancer data.

Table 16.
Estimates of , , , , and from bladder cancer data.
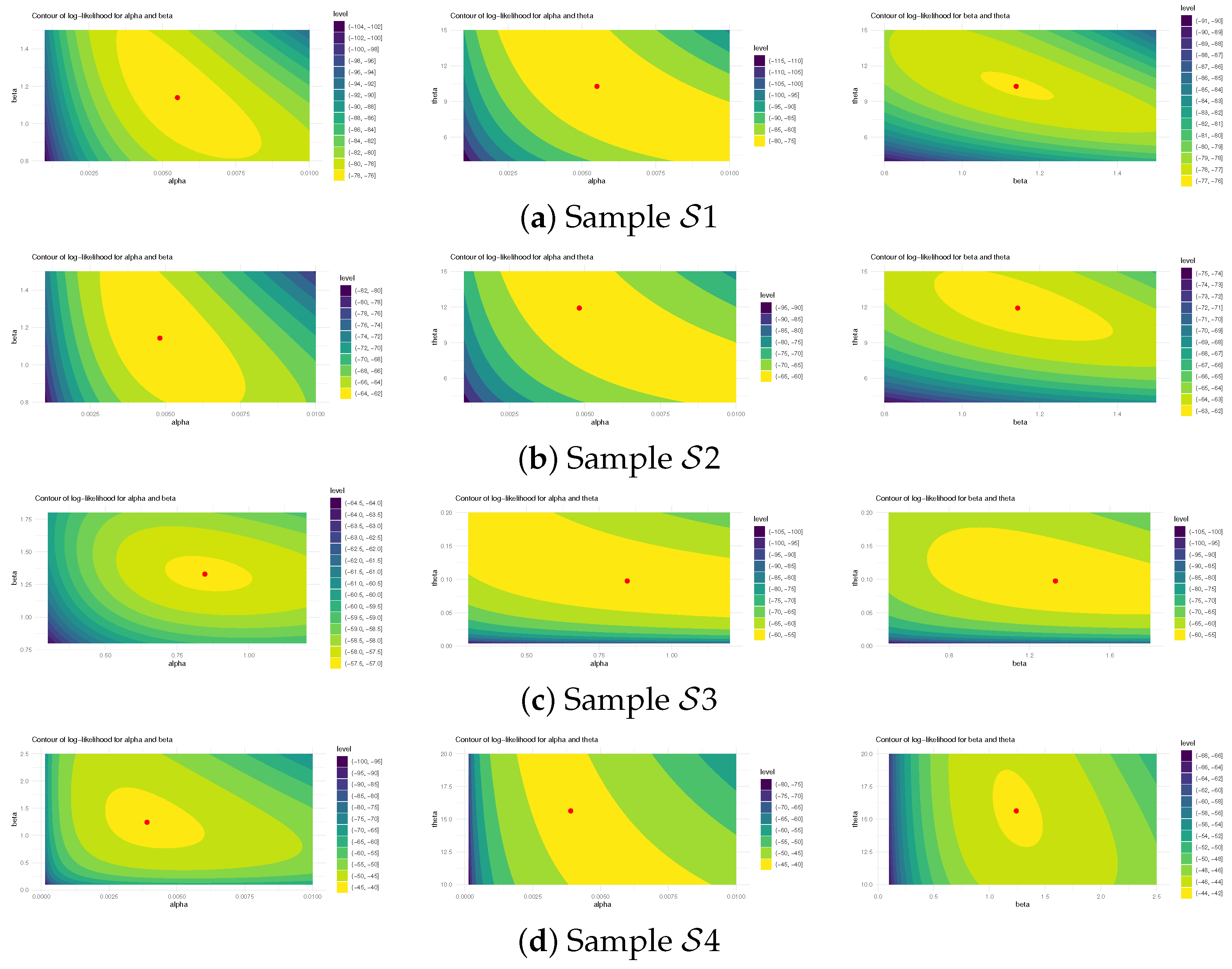
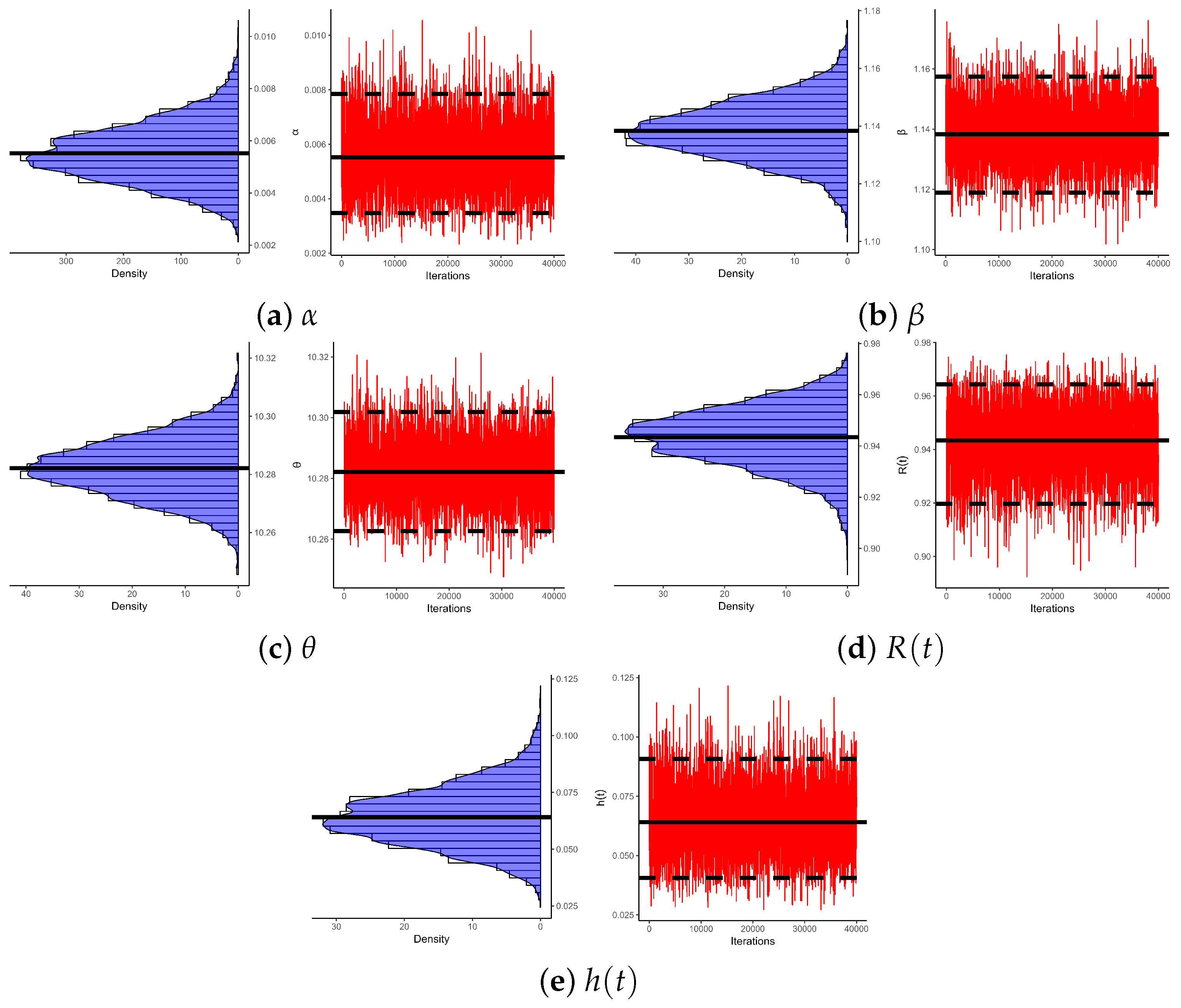
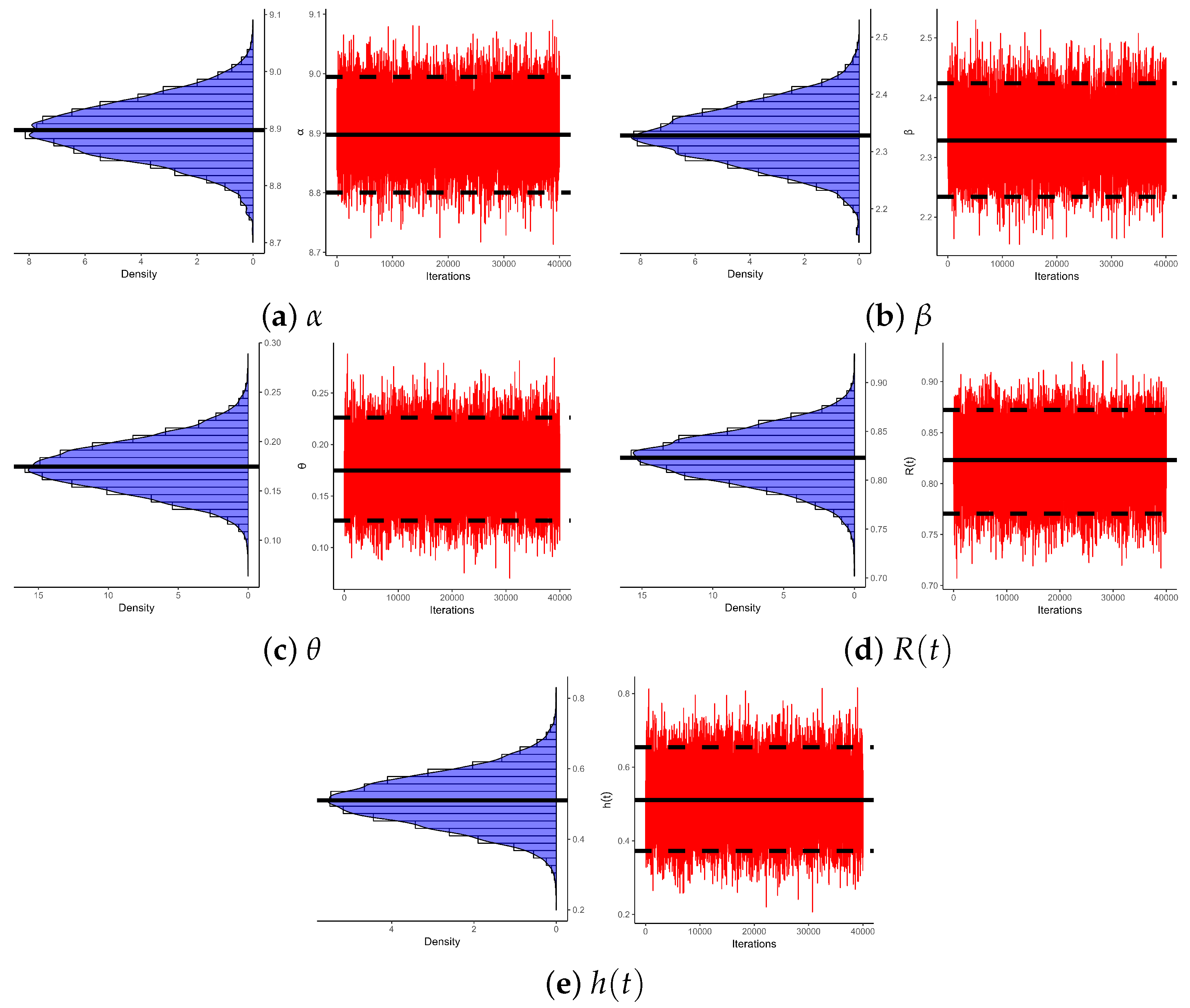
Figure 5 illustrates log-likelihood contours (from ) to assess the existence and uniqueness of likelihood estimates for , , and . It demonstrates that the offered most frequent estimates of , , or calculated from bladder cancer samples exist and are unique. These graphical findings are consistent with the numerical outcomes in Table 16, making them suitable as beginning values for subsequent computations. Additionally, using 1 (as a representative case), Figure 6 provides Gaussian kernel density estimates and trace plots for , , , , and . The MCMC chains (40,000 samples) exhibit decent mixing and confirm that the burn-in size () is sufficient to eliminate initial transients and ensure proper mixing. Figure 6 also indicates that the marginal estimates of and are positively skewed, those of are negatively skewed, while those of and are essentially symmetric.
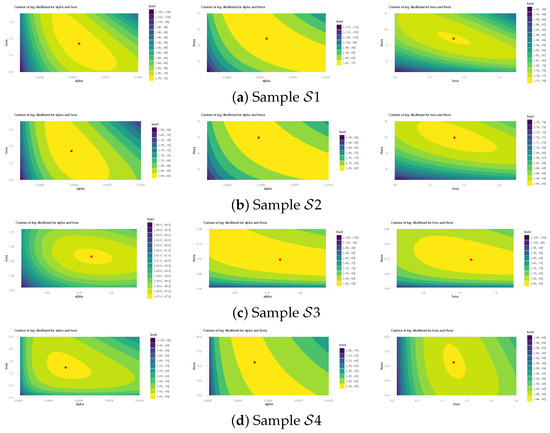
Figure 5.
The log-likelihood contours for , , and from bladder cancer data.
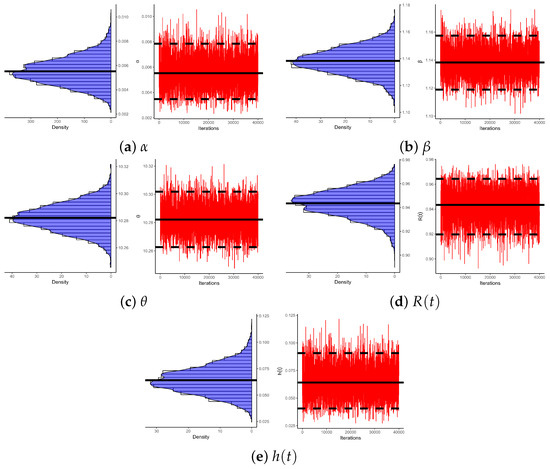
Figure 6.
Density/trace plots of , , , , and from bladder cancer data.
5.2. Head and Neck Cancer
Head and neck cancer patients typically receive treatment that combines radiation and chemotherapy to improve outcomes and target cancer more effectively. This technique helps shrink tumors, reduce recurrence, and enhance the effectiveness of each treatment. This example examines the survival rates (in days) of 44 head and neck (HN) cancer patients who had a combination of chemotherapy and radiation; see Efron [39]. Later, the HN cancer dataset was analyzed by Elshahhat and Rastogi [40], Elshahhat and Nassar [41] and Alotaibi et al. [21], among others. For computational purposes, each survival rate included in the proposed HN cancer data is divided by one hundred and listed. The newly transformed HN cancer dataset is provided in Table 17.

Table 17.
Survival rates of HN cancer patients.
Using the whole HN cancer dataset and the same fitting scenarios mentioned in Section 5.1, we will demonstrate the PGW distribution’s superiority and adaptability over the nine lifespan models listed in Table 13. Table 18 reports the fitted values of N–LL, A.I, B.I, CA.I, HQ.I, A–D, CvM, and KS (p-value) in addition to the MLEs and associated standard errors. With the exception of the highest P-value, the PGW model correlates to the lowest level of all the metrics provided, making it an excellent choice when compared to other comparable models, according to Table 18. Figure 7 supports the findings reported in Table 18, indicating that the PGW lifetime model is the best model when all the HN cancer survival times are analyzed.

Table 18.
Fitting results of the PGW and its competitors from HN cancer data.
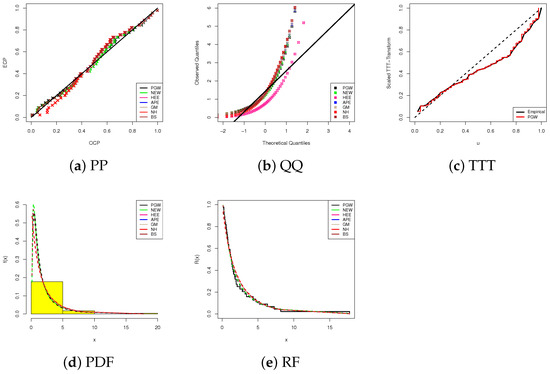
Figure 7.
Fitting diagrams of the PGW and its competitors from HN cancer data.
From Table 12, taking various choices of and for , four UHC samples from the full HN cancer data are generated; see Table 19. Using the MCMC technique, we recreated 50,000 Markovian variations, with the first 10,000 serving as burn-in variations. The MCMC sampler’s beginning values of , , and are set to fitted MLE values. Because there is no known previous knowledge, we put for . Table 20 displays maximum likelihood and Bayes’ estimates (with standard errors), as well as 95% asymptotic/credible interval limits (with widths) for , , , , and (at ). It is noted from Table 20 that the Bayesian point (or credible) estimates of , , , , or obtained using the Bayes’ technique perform better than those acquired using the likelihood approach, as demonstrated by the lowest standard error and interval width values.

Table 19.
Several UHC samples from HN cancer data.

Table 20.
Estimates of , , , , and from HN cancer data.
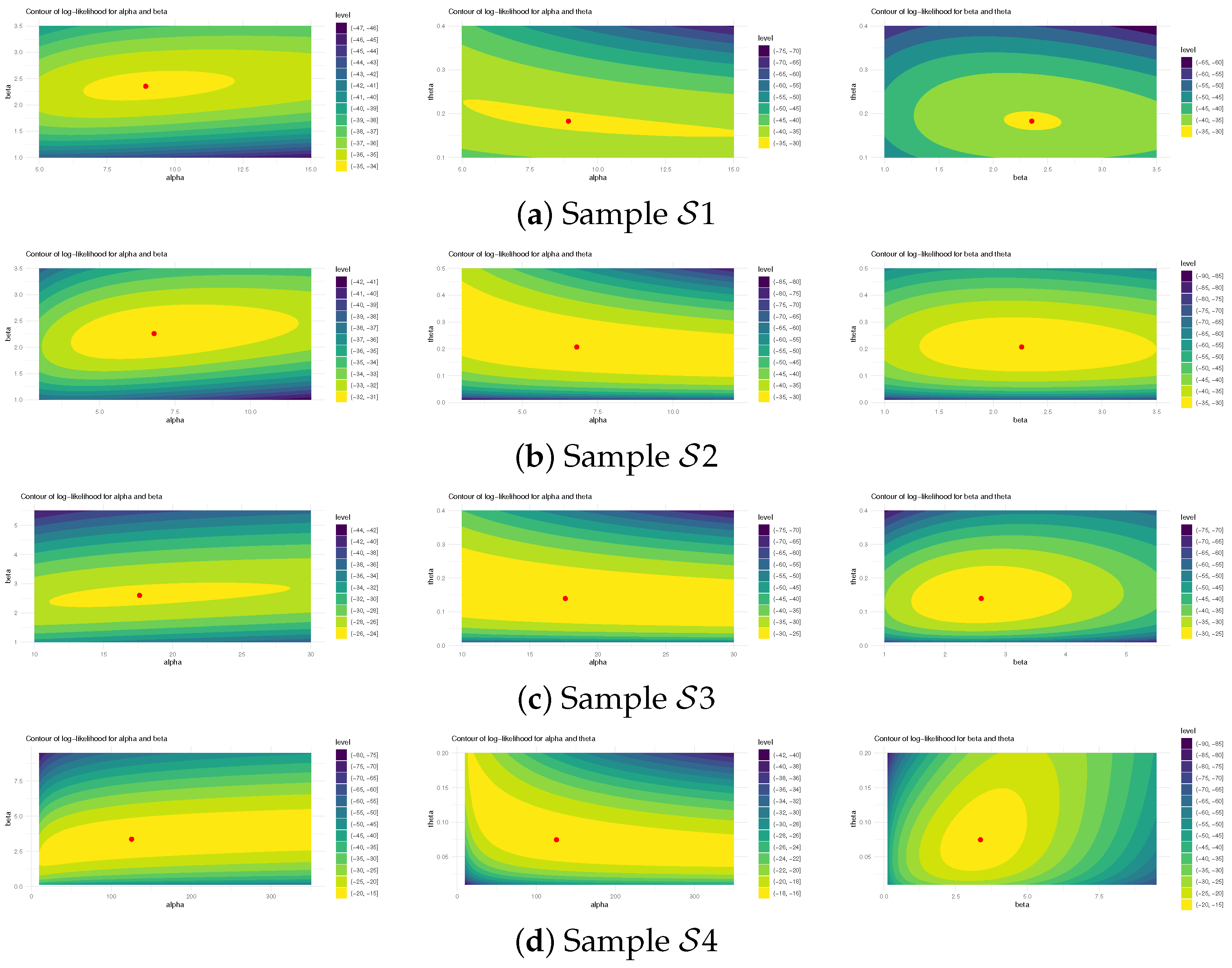
Using (reported in Table 19) to examine the existence and uniqueness of likelihood estimates for , , and , Figure 8 depicts their log-likelihood contours. It indicates that the supplied most frequent estimates of , , or determined from HN cancer samples occurred and are unique. These graphics are aligned with the numerical results in Table 20, making them appropriate as starting values for Bayesian computations. Figure 9 depicts Gaussian kernel density estimates and trace plots for , , , , and , using 1 (as an example). The MCMC chains () show good mixing; demonstrate that the burn-in size () is adequate to remove initial transients and guarantee proper mixing; and reveal that all posterior samples gathered for , , , , or are almost symmetric.
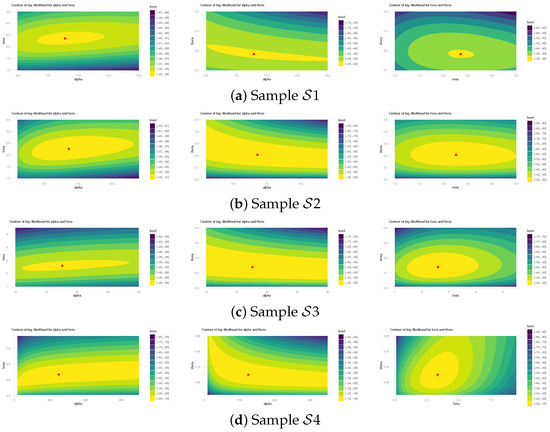
Figure 8.
The log-likelihood contours for , , and from HN cancer data.
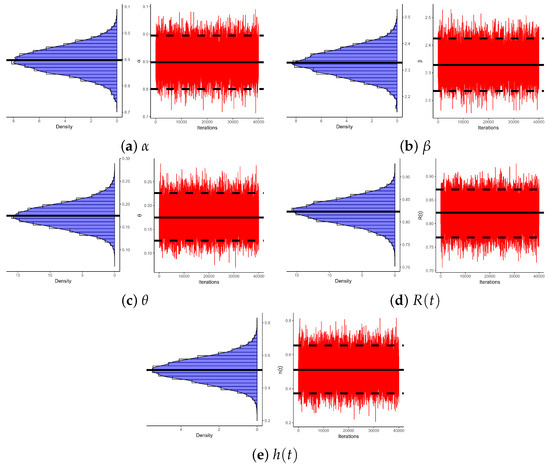
Figure 9.
Density/trace plots of , , , , and from HN cancer data.
6. Conclusions
This study investigated the power generalized Weibull distribution, which functions as a flexible extension of the exponential, Rayleigh, Nadarajah–Haghighi, and Weibull models within a unified hybrid censoring framework. The distribution’s ability to model five distinct hazard rate shapes—namely constant, increasing, decreasing, bathtub, and upside-down bathtub—was employed to analyze censored lifetime data. Classical estimation techniques, including maximum likelihood estimation accompanied by delta method-based variance approximation, were developed for parameter estimation and reliability metrics. Bayesian approaches utilizing gamma prior distributions and Markov chain Monte Carlo sampling methods were applied to derive posterior estimates and credible intervals. Simulations revealed that Bayesian methods, particularly those employing informative priors, attained greater precision in small sample sizes, characterized by lower mean squared errors and narrower interval widths compared to classical methods. Two examples of cancer clinical trials are examined to show how the proposed approaches can be used in practical circumstances and to demonstrate the model’s superiority over alternative distributions, as confirmed by goodness-of-fit tests. To sum up, based on both clinical scenarios, one can conclude that the power generalized Weibull lifespan model is more suited to assessing various sets of real-world data from the medical area than eleven common statistical models with varying failure rates. Furthermore, it proves to be effective in computing reliability measures when the data are collected under the proposed censoring scheme. A noted limitation is the computational complexity associated with Markov chain Monte Carlo sampling techniques for high-dimensional parameter spaces. Future research should explore more complex censoring designs in the presence of power generalized Weibull distribution, such as adaptive progressive censoring or unified progressive hybrid censoring methodologies.
Author Contributions
Methodology, M.N., R.A. and A.E.; Funding acquisition, R.A.; Software, A.E.; Supervision, M.N. and R.A.; Writing—original draft, M.N. and R.A.; Writing—review andediting, M.N. and A.E. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Princess Nourah bint Abdulrahman University researchers supporting project number (PNURSP2025R50), Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia.
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Bagdonavi čius, V.; Nikulin, M. Accelerated Life Models: Modeling and Statistical Analysis; Chapman and Hall/CRC: New York, NY, USA, 2001. [Google Scholar]
- Dimitrakopoulou, T.; Adamidis, K.; Loukas, S. A lifetime distribution with an upside-down bathtub-shaped hazard function. IEEE Trans. Reliab. 2007, 56, 308–311. [Google Scholar] [CrossRef]
- Nadarajah, S.; Haghighi, F. An extension of the exponential distribution. Statistics 2011, 45, 543–558. [Google Scholar] [CrossRef]
- Nikulin, M.; Haghighi, F. A Chi-Squared Test for the Generalized Power Weibull Family for the Head-and-Neck Cancer Censored Data. J. Math. Sci. 2006, 133, 1333–1341. [Google Scholar] [CrossRef]
- Nikulin, M.; Haghighi. On the power generalized Weibull family: Model for cancer censored data. Metron 2009, 67, 75–86. [Google Scholar]
- Kumar, D.; Dey, S. Power generalized Weibull distribution based on order statistics. J. Stat. Res. 2017, 51, 61–78. [Google Scholar] [CrossRef]
- El-Din, M.M.; Abd El-Raheem, A.M.; Abd El-Azeem, S.O. On progressive-stress accelerated life testing for power generalized Weibull distribution under progressive type-II censoring. J. Stat. Appl. Probab. Lett. 2018, 5, 131–143. [Google Scholar] [CrossRef]
- Pandey, R.; Kumari, N. Bayesian analysis of power generalized Weibull distribution. Int. J. Appl. Comput. Math. 2018, 4, 1–16. [Google Scholar] [CrossRef]
- Sabry, M.A.; Muhammed, H.Z.; Nabih, A.; Shaaban, M. Parameter estimation for the power generalized Weibull distribution based on one-and two-stage ranked set sampling designs. J. Stat. Appl. Probab. 2019, 8, 113–128. [Google Scholar]
- Dey, S.; Nassar, M.; Ali, S.; Kumar, D.; Raheem, E. Comparison of Estimation Methods of the Power Generalized Weibull Distribution. Statistica 2022, 82, 339–372. [Google Scholar]
- Epstein, B. Truncated life tests in the exponential case. Ann. Math. Stat. 1954, 25, 555–564. [Google Scholar] [CrossRef]
- Childs, A.; Chandrasekar, B.; Balakrishnan, N.; Kundu, D. Exact likelihood inference based on Type-I and Type-II hybrid censored samples from the exponential distribution. Ann. Inst. Stat. Math. 2003, 55, 319–330. [Google Scholar] [CrossRef]
- Chandrasekar, B.; Childs, A.; Balakrishnan, N. Exact likelihood inference for the exponential distribution under generalized Type-I and Type-II hybrid censoring. Nav. Res. Logist. 2004, 51, 994–1004. [Google Scholar] [CrossRef]
- Balakrishnan, N.; Rasouli, A.; Sanjari Farsipour, N. Exact likelihood inference based on an unified hybrid censored sample from the exponential distribution. J. Stat. Comput. Simul. 2008, 78, 475–488. [Google Scholar] [CrossRef]
- Panahi, H.; Sayyareh, A. Estimation and prediction for a unified hybrid-censored Burr Type XII distribution. J. Stat. Comput. Simul. 2016, 86, 55–73. [Google Scholar] [CrossRef]
- Jeon, Y.E.; Kang, S.B. Estimation of the Rayleigh distribution under unified hybrid censoring. Austrian J. Stat. 2021, 50, 59–73. [Google Scholar] [CrossRef]
- Dutta, S.; Kayal, S. Bayesian and non-Bayesian inference of Weibull lifetime model based on partially observed competing risks data under unified hybrid censoring scheme. Qual. Reliab. Eng. Int. 2022, 38, 3867–3891. [Google Scholar] [CrossRef]
- Dutta, S.; Lio, Y.; Kayal, S. Parametric inferences using dependent competing risks data with partially observed failure causes from MOBK distribution under unified hybrid censoring. J. Stat. Comput. Simul. 2024, 94, 376–399. [Google Scholar] [CrossRef]
- Hasaballah, M.M.; Tashkandy, Y.A.; Balogun, O.S.; Bakr, M.E. Statistical inference of unified hybrid censoring scheme for generalized inverted exponential distribution with application to COVID-19 data. AIP Adv. 2024, 14, 045111. [Google Scholar] [CrossRef]
- Shukla, A.K.; Soni, S.; Kumar, K. Statistical inference and prediction in unified hybrid censored power Lindley distribution. Life Cycle Reliab. Saf. Eng. 2025, 14, 183–204. [Google Scholar] [CrossRef]
- Alotaibi, R.; Nassar, M.; Khan, Z.A.; Alajlan, W.A.; Elshahhat, A. Entropy evaluation in inverse Weibull unified hybrid censored data with application to mechanical components and head-neck cancer patients. AIMS Math. 2025, 10, 1085–1115. [Google Scholar] [CrossRef]
- Kundu, D. Bayesian inference and life testing plan for the Weibull distribution in presence of progressive censoring. Technometrics 2008, 50, 144–154. [Google Scholar] [CrossRef]
- Henningsen, A.; Toomet, O. maxLik: A package for maximum likelihood estimation in R. Comput. Stat. 2011, 26, 443–458. [Google Scholar] [CrossRef]
- Plummer, M.; Best, N.; Cowles, K.; Vines, K. CODA: Convergence diagnosis and output analysis for MCMC. R News 2006, 6, 7–11. [Google Scholar]
- Lee, E.T.; Wang, J.W. Statistical Methods for Survival Data Analysis, 3rd ed.; Wiley: New York, NY, USA, 2003. [Google Scholar]
- Alotaibi, R.; Nassar, M.; Khan, Z.A.; Elshahhat, A. Analysis of Weibull progressively first-failure censored data with beta-binomial removals. AIMS Math. 2024, 9, 24109–24142. [Google Scholar] [CrossRef]
- Alotaibi, R.; Nassar, M.; Elshahhat, A. A new extended Pham distribution for modelling cancer data. J. Radiat. Res. Appl. Sci. 2024, 17, 100961. [Google Scholar] [CrossRef]
- Peng, X.; Yan, Z. Estimation and application for a new extended Weibull distribution. Reliab. Eng. Syst. Saf. 2014, 121, 34–42. [Google Scholar] [CrossRef]
- Pinho, L.G.B.; Cordeiro, G.M.; Nobre, J.S. The Harris extended exponential distribution. Commun. Stat.-Theory Methods 2015, 44, 3486–3502. [Google Scholar] [CrossRef]
- Oguntunde, P.E.; Balogun, O.S.; Okagbue, H.I.; Bishop, S.A. The Weibull-exponential distribution: Its properties and applications. J. Appl. Sci. 2015, 15, 1305–1311. [Google Scholar] [CrossRef]
- Marshall, A.W.; Olkin, I. Life Distributions; Springer: New York, NY, USA, 2007. [Google Scholar]
- Mahdavi, A.; Kundu, D. A new method for generating distributions with an application to exponential distribution. Commun. Stat. Theory Methods 2017, 46, 6543–6557. [Google Scholar] [CrossRef]
- Gupta, R.D.; Kundu, D. Generalized exponential distribution: Different method of estimations. J. Stat. Comput. Simul. 2001, 69, 315–337. [Google Scholar] [CrossRef]
- Birnbaum, Z.W.; Saunders, S.C. A probabilistic interpretation of Miner’s rule. SIAM J. Appl. Math. 1968, 16, 637–652. [Google Scholar] [CrossRef]
- Weibull, W. A statistical distribution function of wide applicability. J. Appl. Mech. 1951, 18, 293–297. [Google Scholar] [CrossRef]
- Johnson, N.; Kotz, S.; Balakrishnan, N. Continuous Univariate Distributions, 2nd ed.; John Wiley & Sons: New York, NY, USA, 1994. [Google Scholar]
- Pham, H. A vtub-shaped hazard rate function with applications to system safety. Int. J. Reliab. Appl. 2002, 3, 1–16. [Google Scholar]
- Marinho, P.R.D.; Silva, R.B.; Bourguignon, M.; Cordeiro, G.M.; Nadarajah, S. AdequacyModel: An R package for probability distributions and general purpose optimization. PLoS ONE 2019, 14, e0221487. [Google Scholar] [CrossRef]
- Efron, B. Logistic regression, survival analysis, and the Kaplan-Meier curve. J. Am. Stat. Assoc. 1988, 83, 414–425. [Google Scholar] [CrossRef]
- Elshahhat, A.; Rastogi, M.K. Bayesian Life Analysis of Generalized Chen’s Population Under Progressive Censoring. Pak. J. Stat. Oper. Res. 2022, 18, 675–702. [Google Scholar] [CrossRef]
- Elshahhat, A.; Nassar, M. Analysis of adaptive Type-II progressively hybrid censoring with binomial removals. J. Stat. Comput. Simul. 2023, 93, 1077–1103. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).