Fascination with Fluctuation: Luria and Delbrück’s Legacy
Abstract
1. Introduction
2. Luria–Delbrück’s Work as a Catalyst for Engagement
2.1. The Story
But I soon started wondering, how do phage-resistant bacteria originate? Are they produced by direct action of the phage on a few bacterial cells, about one in a billion? Or do they originate spontaneously by mutations…?
Despite the strength of public opinion and Sir Cyril’s authority denying genes to bacteria, I favored gene mutation origin for my phage-resistant cultures—an arrogant David pitted against the Goliath of physical chemistry. My reasons were several. My interests were in genetics, and I could not conceive of an organism without them. And where are genes there are mutations. Also, the extreme stability of the resistant bacteria spoke for a mutational mechanism. And, finally, I could not really understand Sir Cyril’s mathematics.Luria continues:I struggled with the problem [are mutations directed or spontaneous] for several months, mostly in my own thoughts, and also tried a variety of experiments, none of which worked. The answer finally came to me in February 1943 in the improbable setting of a faculty dance at Indiana University, a few weeks after I had moved there as an instructor…I am not a passionate dancer, but the dances had other attractions for a young bachelor. I was certainly glad to have gone to this one, but not for romantic reasons.
During the pause in the music I found myself standing near a slot machine, watching a colleague putting dimes into it. Though losing most of the time, he occasionally got a return…. I was teasing him about his inevitable losses, when he suddenly hit the jackpot…, gave me a dirty look, and walked away. Right then, I began giving some thought to the actual numerology of slot machines; in so doing it dawned on me that slot machines and bacterial mutations have something to teach each other.
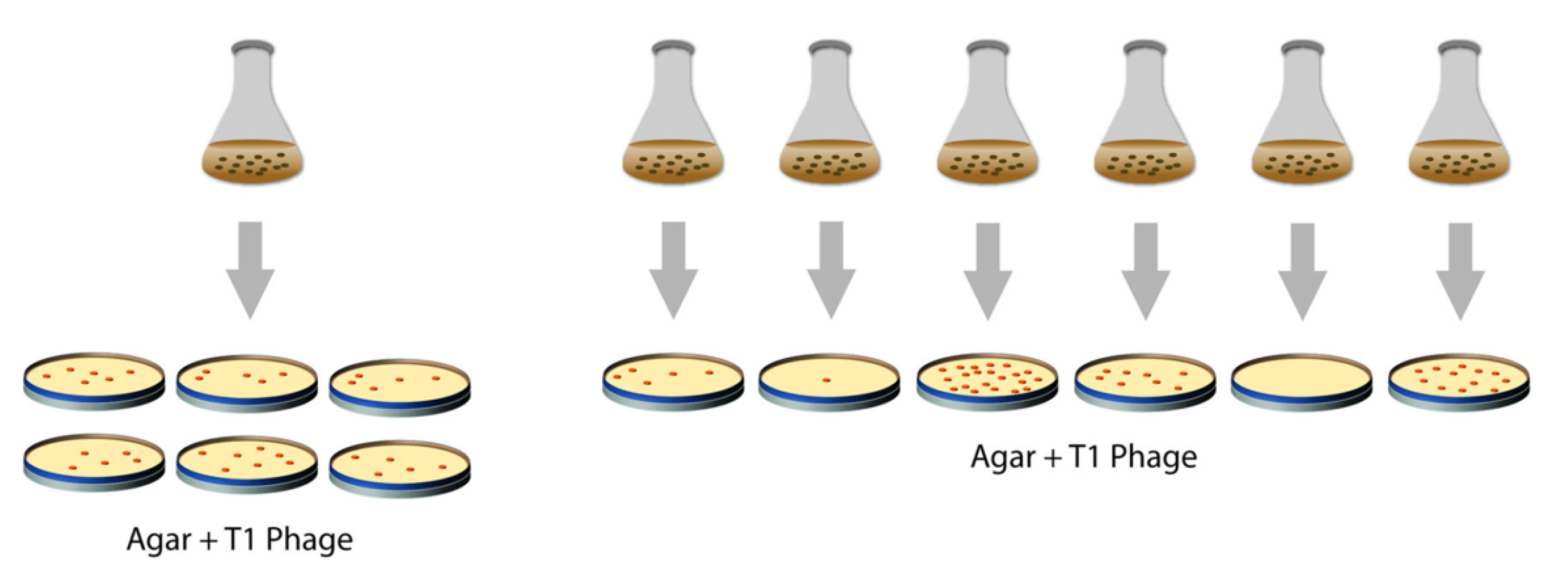
2.2. The Experiment
2.3. The Mathematics Curriculum
3. Mathematical Models
3.1. The Luria–Delbrück Model with Discrete Time
- When a cell divides, a mutation may occur in one of the daughter cells with a small probability p ();
- At any time t, the number of mutated cells in the population is negligible in comparison with . This is justified since the mutation probability p is small;
- Mutations are independent, and both sensitive and mutant cells divide every generation;
- Backward mutations (from a resistant cell to a sensitive one) are negligible.
3.1.1. Case 1: Directed Mutations
3.1.2. Case 2: Spontaneous Mutations
3.2. The Luria–Delbrück Model with Continuous Time
- 1’
- The mutation rate is , meaning that a mutation of a sensitive cell may occur at any time with probability ();
- 2’
- At any time t, the number of mutated cells in the population is negligible in comparison with . This is justified since the mutation probability is small;
- 3’
- Sensitive cell numbers grow exponentially (and deterministically) at a rate ; that is
- 4’
- Each mutant cell can split at time into two mutant cells with probability , independent of other cells. This probability is the same for all mutants and does not depend on the cell’s age. With this assumption, the number of mutant cells Z will always have an integer value. The splitting rate is the same as the growth rate of sensitive cells.
- 5’
- Backward mutations (from a resistant cell to a sensitive one) are negligible.
3.2.1. Case 1: Directed Mutations
3.2.2. Case 2: Spontaneous Mutations
- (a)
- At time t, the culture had mutants, and one of them divides at time . The probability of this event is
- (b)
- At time t, the culture had mutants and a mutation occurred at time . The probability of this event is
- (c)
- At time t, there were r mutants, and neither a division nor a mutation occurred at time . The probability of this event is
3.3. Determining the Mutation Rate
3.3.1. Method 1—Using a Poisson Approximation (The -Method)
3.3.2. Method 2—Using a Likely Average
This situation is similar to the operation of a (fair) slot machine, where the average return from a limited number of plays is probably considerably less than the input, and improbably, when the jackpot is hit, the return is much bigger than the input [1].
3.3.3. Method 3—The Drake Equation
- Note that if we choose and assume, as we assumed before, and , Equation (42) yields
- Alternatively, to avoid the same type of problems as we discussed when describing Method 2 above, may be chosen so that is the size of the culture when the first mutant appears. At that time, , and is very small and can be ignored. Replacing this value for in Equation (41) and using that , gives the following equation for the mutation rate:
4. Suggestions for Classroom Use
4.1. Possible Uses of the Discrete-Time Model in Mathematics Courses
4.2. Possible Uses of the Continuous-Time Model in Mathematics Courses
- Most students in the sciences are keenly aware of the ongoing political debates about evolution and how school boards nowadays may mandate that teaching evolution in the schools should necessarily be paired with “-parallel” theories, e.g., Lamarckism and intelligent design. To see how, at least in the bacterial world, it can be established that the directed mutations hypothesis is not supported by experimental data provides a high-impact example of the value of hypothesis-driven research.
- The fluctuation test allows students to see a statistical approach not based on comparing group proportions or means (which is almost exclusively what most introductory statistics courses do) and focusing instead on their variances. A discussion asking why averages do not allow for distinguishing between the hypotheses of spontaneous mutations and directed mutations provides an excellent opportunity to build a better understanding of how randomness is quantifiable.
- Most standard courses in mathematical modeling place emphasis on describing the time evolution of systems comprising interacting components through difference or differential equations; that is, using a deterministic approach. In rare cases when some stochasticity is added to the models, it is primarily in assuming that the values of the model parameters are drawn from an underlying distribution of interest (e.g., in the context of looking into a solution’s stability without performing full mathematical analyses). The fluctuation test uses a mathematical model very different from those they may have seen in such courses and, thus, presents an opportunity to broaden the scope of modeling approaches students are exposed to.
- There are many links relating the theory above to standard material taught in other courses. As an example, students in a probability class are introduced to probabilitygenerating functions, but justifying the term-by-term differentiation of the infinite series from Equation (28) is something that they will encounter in Calculus or Real Analysis courses. Verifying that the limit, as , of the solution in Equation (32) agrees with the condition is another good calculus problem. The use of “little o” notation in Equation (24) is an opportunity to talk about rates of growth at infinity.
- Changing the summation index in Equations (19) and (20) is something that comes up often in Differential Equations courses in the context of finding solutions in the form of infinite series—something students generally struggle with. In a Differential Equations course, it would also be of interest to ask students to obtain Equation (21) in an alternative way, without using assumption 4’ and the probabilities . Instead, they may assume that the population of mutant cells grows exponentially at a rate , just as the population of sensitive cells does (assumption 3’). To do that, they should notice that the rate of growth of has two components: (1) a contribution from new mutations in time : , and (2) a contribution from the growth of resistant mutants in time : . Thus, the rate of increase of the average number of resistant mutant bacteria isproviding an intuitive interpretation of Equation (21).
- Once students feel comfortable with the theory outlined above, there are many possible directions for student research projects. They can follow, e.g., the work of Ma [38] to analyze the distribution of mutant cells using discrete convolution powers or read Pakes’s [39] and Kemp’s [40] remarks on the Luria–Delbrück’s distribution to discover more of its mathematical properties, including asymptotic evaluations of the probabilities . The short communication by Goldie [41] suggests an alternative method for finding the PGF and the asymptotics of the probabilities by representing the number of mutant cells Z as a Poisson compound. Examining additional methods for estimating mutation rates, using [37] as a starting point is another possibility.
- If one is interested in more recent mathematical developments, the paper by Zheng in Chance [42], written for a general audience, can be used as a continuation of our Section 2.1. This pleasant read addresses the time span between the publications of the Luria–Delbrück paper [1] and that by Lea and Coulson [11], discussing unpublished efforts and anecdotes. This is followed by descriptions of more recent work on the topic. The review paper by the same author [12] provides a rigorous review of the mathematical literature until 1999, which will be of interest to those who pursue research in the field.
- A fine distinction of interest based on the assumptions under which the models are developed can be studied by the work of Luria–Delbrück [1], Lea and Coulson [11] and M.S. Bartlett. As mentioned earlier, Luria and Delbrück assumed deterministic growth of the sensitive and the mutant cells, while Lea and Coulson used a deterministic growth for the sensitive cells but assumed the growth of mutants followed a Yule branching process. Bartlett on the other hand, treats the growth of both sensitive cells and mutant cells as Yule processes. See [12,43] for the interesting history behind Bartlett’s work and references to it.
- Finally, it is important to help students realize that mathematical models for biology and medicine rely on assumptions that should be considered and weighed with great care. Due to those assumptions, mathematical models generally present useful but incomplete descriptions of cellular and molecular mechanisms. Students should be aware of the risks of drawing dubious conclusions based on simplifications that are more about mathematical tractability than about biological reality. There are certain mutations that are extremely hard to predict but can have significant biological consequences (see e.g., [44,45]). The accurate modeling of such processes may require different approaches or new mathematical treatments, thus advancing research in both mathematics and biology.
5. Educational Simulations, Mathematical Manipulatives, and Wet Laboratories
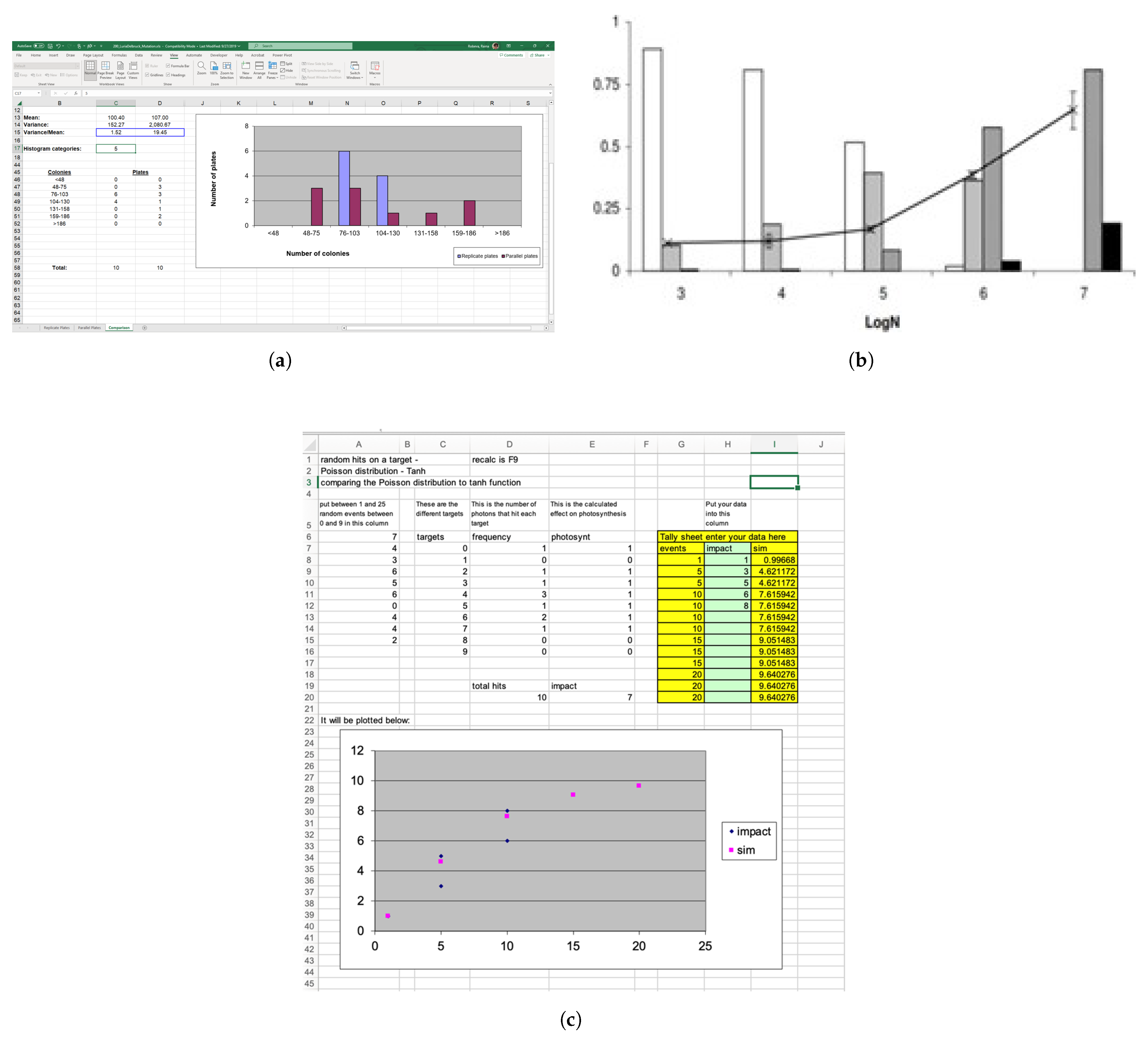
5.1. Simulations
5.2. Mathematical Manipulatives
Teachers need to learn how to encourage student exploration, related discussion, and reflection about the prospective math concept they teach. They need to be comfortable with students’ exploration of the math concepts and possibly wandering off the ‘correct’ track or even to represent quantities in real-world contexts, challenging the teachers’ own mathematical viewpoint. Teachers cannot assume that when students use manipulatives they will automatically draw the correct conclusions from them … Teachers need to keep in mind that the student does not already possess this knowledge and still needs to make the correct connections between the manipulative and the underlying math concept. While math manipulatives are a valuable tool in the instruction of mathematics, teachers need to bridge the manipulatives to the representational and then abstract understanding in mathematics so that students internalize their understanding. Just using manipulatives by themselves without this may not have great value.
5.3. Wet Laboratories
It is important to provide students in biology and mathematics with opportunities to interact and collaborate with one another. However, it can be a challenge to develop integrated courses that are accessible and useful to both sets of students. In this paper, we describe the development, implementation, and assessment of a team-taught course that was developed to provide undergraduate students with a truly integrated experience by incorporating a wet laboratory into a mathematics course …The wet laboratory in Math 236 enabled students to undertake experimentation, data collection, mathematical modeling, and statistical computation, all essential to developing models of biological processes. Each lab addresses core concepts in the categories outlined in BIO 2010 [2]: calculus, linear algebra, dynamical systems, computation, data structures, rate of change, modeling, equilibria and stability, structure, and interactions. Although each lab touches on each of these areas, the tools become more sophisticated as the semester progresses.
6. Why Measuring Mutation Rates Matters in Everyday Life
6.1. Radiation: Hiroshima/Nagasaki/Fukushima/Chernobyl/Three Mile Island
The disaster at the Chernobyl nuclear power plant in 1986 released 80 petabecquerels of radioactive cesium, strontium, plutonium, and other radioactive isotopes into the atmosphere, polluting 200,000 of land in Europe. As we discuss here, several studies have since shown associations between high and low levels of radiation and the abundance, distribution, life history, and mutation rates of plants and animals. However, this research is the consequence of investment by a few individuals rather than a concerted research effort by the international community, despite the fact that the effects of the disaster are continent-wide. A coordinated international research effort is therefore needed to further investigate the effects of the disaster, the knowledge that could be beneficial if there are further nuclear accidents, including the threat of a ‘dirty bomb’.
6.2. Cancer Chemotherapy—Evolution of Resistance
6.3. Antibiotic Resistance
Antimicrobial-resistant infections kill 700,000 patients every year …By 2050, they are projected to cause 10 million deaths per year at a cumulative global cost of $100 trillion. Professional societies and international health agencies, including the United Nations, have declared escalating antimicrobial resistance as one of the gravest and most urgent threats to global public health and issued calls for action.
6.4. Environmental Screening for Mutagens and Teratogens: Ames Test
The initial uses of the Salmonella assay led to the startling (at the time) recognition that our environment is replete with mutagens, including fungal toxins, combustion emissions, industrial chemicals, and drugs. The Salmonella assay was essential to this effort, providing the means by which researchers discovered for the first time that much of our environment had mutagenic activity, including cigarette smoke…
The rapidly increasing number of new production chemicals coupled with the stringent implementation of global chemical management programs necessitates a paradigm shift towards broader uses of low-cost and high-throughput ecotoxicity testing strategies as well as a deeper understanding of cellular and sub-cellular mechanisms of ecotoxicity that can be used in effective risk assessment. The latter will require the automated acquisition of biological data, new capabilities for big data analysis as well as computational simulations capable of translating new data into in vivo relevance.
7. Conclusions
- (1)
- (2)
- CUREs: Course-based Undergraduate Research Experiences [135], where the students are engaged in a research problem that they can investigate with powerful tools;
- (3)
- USE Cit Sci: Undergraduate Student Experiences with Citizen Science [136];
- (4)
- ICBL: Investigative Case Based Learning [137]: the National Center for Case Study Teaching in Science at the University of Buffalo has recently transferred its longheld repository of vetted cases to the National Science Teaching Organization’s site: https://www.nsta.org/case-studies accessed on 15 January 2023;
- (5)
- Problem-based Learning [138]; we maintain a clearinghouse of vetted problems at ITUE (the Institute for Transforming University Education—https://itue.udel.edu/ accessed on 15 January 2023;
- (6)
- Problem Spaces (Donovan: https://bioquest.org/bedrock/problem_spaces/) accessed on 15 January 2023;
- (7)
- Question Formulation Technique and Problem-Posing [139]; The Right Question Institute: https://rightquestion.org accessed on 15 January 2023; BioQUEST: https://bioquest.org/ accessed on 15 January 2023 [140,141,142,143];
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Luria, S.E.; Delbrück, M. Mutations of bacteria from virus sensitivity to virus resistance. Genetics 1943, 28, 491. [Google Scholar] [CrossRef] [PubMed]
- National Research Council. BIO2010: Transforming Undergraduate Education for Future Research Biologists; National Academies Press: Washington, DC, USA, 2003. [Google Scholar]
- National Research Council. A New Biology for the 21st Century; National Academies Press: Washington, DC, USA, 2009. [Google Scholar]
- Jungck, J.R.; Robeva, R.; Gross, L.J. Mathematical biology education: Changes, communities, connections, and challenges. Bull. Math. Biol. 2020, 82, 1–14. [Google Scholar] [CrossRef] [PubMed]
- National Research Council. The Mathematical Sciences in 2025; National Academies Press: Washington, DC, USA, 2013. [Google Scholar]
- AAAS. Vision and Change: A Call to Action—A Summary of Recommendations; AAAS: Washington, DC, USA, 2010. [Google Scholar]
- Sturmfels, B. Can biology lead to new theorems? Annu. Rep. Clay Math. Inst. 2005, 1468, 13–26. [Google Scholar]
- Kac, M.; Rota, G.-C.; Schwartz, J.T. Discrete Thoughts: Essays on Mathematics, Science and Philosophy; Springer: Berlin/Heidelberg, Germany, 2009. [Google Scholar]
- Luria, S.E. A Slot Machine, a Broken Test Tube: An Autobiography; Harper & Row Publishers: New York, NY, USA, 1984. [Google Scholar]
- Henry, M.A.; Shorter, S.; Charkoudian, L.; Heemstra, J.M.; Corwin, L.A. Fail is not a four-letter word: A theoretical framework for exploring undergraduate students’ approaches to academic challenge and responses to failure in STEM learning environments. CBE-Life Sci. Educ. 2019, 18, ar11. [Google Scholar] [CrossRef] [PubMed]
- Lea, D.E.; Coulson, C.A. The distribution of the numbers of mutants in bacterial populations. J. Genet. 1949, 49, 264–285. [Google Scholar] [CrossRef]
- Zheng, Q. Progress of a half century in the study of the Luria–Delbrück distribution. Math. Biosci. 1999, 162, 1–32. [Google Scholar] [CrossRef]
- Cairns, J.; Overbaugh, J.; Miller, S. The origin of mutants. Nature 1988, 335, 142–145. [Google Scholar] [CrossRef]
- Sarkar, S. On the possibility of directed mutations in bacteria: Statistical analyses and reductionist strategies. PSA Proc. Bienn. Meet. Philos. Sci. Assoc. 1990, 1990, 111–124. [Google Scholar] [CrossRef]
- Hall, B.G. Selection-induced mutations occur in yeast. Proc. Natl. Acad. Sci. USA 1992, 89, 4300–4303. [Google Scholar] [CrossRef]
- Heidenreich, E. Adaptive mutation in saccharomyces cerevisiae. Crit. Rev. Biochem. Mol. Biol. 2007, 42, 285–311. [Google Scholar] [CrossRef]
- Holmes, C.M.; Ghafari, M.; Abbas, A.; Saravanan, V.; Nemenman, I. Luria–Delbrück, revisited: The classic experiment does not rule out Lamarckian evolution. Phys. Biol. 2017, 14, 055004. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Q. Mathematical issues arising from the directed mutation controversy. Genetics 2003, 164, 373–379. [Google Scholar] [CrossRef] [PubMed]
- Zheng; Q On a logical difficulty in the directed mutation debate. Genet. Res. 2009, 91, 5–7. [CrossRef] [PubMed]
- Lang, G.I.; Murray, A.W. Estimating the per-base-pair mutation rate in the yeast Saccharomyces cerevisiae. Genetics 2008, 178, 67–82. [Google Scholar] [CrossRef] [PubMed]
- Ford, C.B.; Shah, R.R.; Maeda, M.K.; Gagneux, S.; Murray, M.B.; Cohen, T.; Johnston, J.C.; Gardy, J.; Lipsitch, M.; Fortune, S.M. Mycobacterium tuberculosis mutation rate estimates from different lineages predict substantial differences in the emergence of drug-resistant tuberculosis. Nat. Genet. 2013, 45, 784–790. [Google Scholar] [CrossRef] [PubMed]
- Witkin, E.M. Inherited differences in sensitivity to radiation in escherichia coli. Proc. Natl. Acad. Sci. USA 1946, 32, 59–68. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Q. Estimation of rates of non-neutral mutations when bacteria are exposed to subinhibitory levels of antibiotics. Bull. Math. Biol. 2022, 84, 131. [Google Scholar] [CrossRef] [PubMed]
- Datta, M.S.; Kishony, R. A spotlight on bacterial mutations for 75 years. Nature 2018, 563, 633–644. [Google Scholar] [CrossRef] [PubMed]
- Klompe, S.E.; Sternberg, S.H. Harnessing “a billion years of experimentation”: The ongoing exploration and exploitation of CRISPR–Cas immune systems. Cris. J. 2018, 1, 141–158. [Google Scholar] [CrossRef] [PubMed]
- Schlegel, S.; Genevaux, P.; de Gier, J.-W. Isolating Escherichia coli strains for recombinant protein production. Cell. Mol. Life Sci. 2017, 74, 891–908. [Google Scholar] [CrossRef] [PubMed]
- Tintle, N.; Chance, B.L.; Cobb, G.W.; Rossman, A.J.; Roy, S.; Swanson, T.; VanderStoep, J. Introduction to Statistical Investigations; John Wiley & Sons: Hoboken, NJ, USA, 2020. [Google Scholar]
- Baake, E. The Luria-Delbrück experiment: Are mutations spontaneous or directed. Newsl. Euro. Math. Soc. 2009, 69, 17–20. [Google Scholar]
- Zheng, Q. On Haldane’s formulation of Luria and Delbrück’s mutation model. Math. Biosci. 2007, 209, 500–513. [Google Scholar] [CrossRef] [PubMed]
- Armitage, P. The statistical theory of bacterial populations subject to mutation. J. R. Stat. Soc. Ser. (Methodol.) 1952, 14, 1–33. [Google Scholar] [CrossRef]
- Zheng, Q. A new practical guide to the Luria–Delbrück protocol. Mutat. Res. Mol. Mech. Mutagen. 2015, 781, 7–13. [Google Scholar] [CrossRef]
- Lazowski, K. Efficient, robust, and versatile fluctuation data analysis using MLE MUtation rate calculator (mlemur). bioRxiv 2023. [Google Scholar] [CrossRef]
- Drake, J. The Molecular Basis of Mutation; Holden-Day: San Francisco, CA, USA, 1970. [Google Scholar]
- Zheng, Q. WebSalvador: A web tool for the Luria-Delbrük experiment. Microbiol. Resour. Announc. 2021, 10, e00314-21. [Google Scholar] [CrossRef]
- Zheng, Q. rSalvador: An R package for the fluctuation experiment. G3 Genes Genomes Genet. 2017, 7, 3849–3856. [Google Scholar] [CrossRef]
- Zheng, Q. New approaches to mutation rate fold change in Luria–Delbrück fluctuation experiments. Math. Biosci. 2021, 335, 108572. [Google Scholar] [CrossRef]
- Foster, P.L. Methods for determining spontaneous mutation rates. Methods Enzymol. 2006, 409, 195–213. [Google Scholar]
- Ma, W.T.; Sarkar, S. Analysis of the Luria–Delbrück distribution using discrete convolution powers. J. Appl. Probab. 1992, 29, 255–267. [Google Scholar] [CrossRef]
- Pakes, A.G. Remarks on the Luria–Delbrück distribution. J. Appl. Probab. 1993, 30, 991–994. [Google Scholar] [CrossRef]
- Kemp, A.W. Comments on the Luria-Delbrück distribution. J. Appl. Probab. 1994, 31, 822–828. [Google Scholar] [CrossRef]
- Goldie, C.M. Asymptotics of the Luria-Delbrück distribution. J. Appl. Probab. 1995, 32, 840–841. [Google Scholar] [CrossRef]
- Zheng, Q. The Luria-Delbrück distribution: Early statistical thinking about evolution. Chance 2010, 23, 15–18. [Google Scholar] [CrossRef]
- Zheng, Q. On Bartlett’s formulation of the Luria–Delbrück mutation model. Math. Biosci. 2008, 215, 48–54. [Google Scholar] [CrossRef]
- McDermott, D.H.; Gao, J.-L.; Liu, Q.; Siwicki, M.; Martens, C.; Jacobs, P.; Velez, D.; Yim, E.; Bryke, C.R.; Hsu, N.; et al. Chromothriptic cure of whim syndrome. Cell 2015, 160, 686–699. [Google Scholar] [CrossRef]
- Tan, J.; Pieper, K.; Piccoli, L.; Abdi, A.; Foglierini, M.; Geiger, R.; Tully, C.M.; Jarrossay, D.; Ndungu, F.M.; Wambua, J.; et al. A lair1 insertion generates broadly reactive antibodies against malaria variant antigens. Nature 2016, 529, 105–109. [Google Scholar] [CrossRef]
- Robson, R.L.; Burns, S. Gain in student understanding of the role of random variation in evolution following teaching intervention based on Luria–Delbruck experiment. J. Microbiol. Biol. Educ. 2011, 12, 3–7. [Google Scholar] [CrossRef]
- Meneely, P.M. Pick your Poisson: An educational primer for Luria and Delbrück’s classic paper. Genetics 2016, 202, 371–375. [Google Scholar] [CrossRef]
- Jungck, J. Mathematical biology education: Modeling makes meaning. Math. Model. Nat. Phenom. 2011, 6, 1–21. [Google Scholar] [CrossRef]
- Jungck, J.R. If life is analog, why be discrete? Middle-out modeling in mathematical biology. In BIOMAT 2011; World Scientific: Singapore, 2012; pp. 376–391. [Google Scholar]
- Rice, K.; Lumley, T. Graphics and statistics for cardiology: Comparing categorical and continuous variables. Heart 2016, 102, 349–355. [Google Scholar] [CrossRef] [PubMed]
- Hampton, R.E. The rhetorical and metaphorical nature of graphics and visual schemata. Rhetor. Soc. Q. 1990, 20, 347–356. [Google Scholar] [CrossRef]
- Carvajal-Rodríguez, A. Teaching the fluctuation test in silico by using Mutate: A program to distinguish between the adaptive and spontaneous mutation hypotheses. Biochem. Mol. Biol. Educ. 2012, 40, 277–283. [Google Scholar] [CrossRef] [PubMed]
- Wimsatt, W.; Shank, J. Modeling; Academic Press: Cambridge, MA, USA, 2006. [Google Scholar]
- Furner, J.M.; Worrell, N.L. The importance of using manipulatives in teaching math today. Transformations 2017, 3, 2. [Google Scholar]
- Ross, J.A.; Hogaboam-Gray, A.; Hannay, L. Predictors of teachers’ confidence in their ability to implement computer-based instruction. J. Educ. Comput. Res. 1999, 21, 75–97. [Google Scholar] [CrossRef]
- Jungck, J.R.; Gaff, H.; Weisstein, A.E. Mathematical manipulative models: In defense of “beanbag biology”. CBE-Life Sci. Educ. 2010, 9, 201–211. [Google Scholar] [CrossRef]
- Buonaccorsi, V.; Skibiel, A. A ‘striking’ demonstration of the Poisson distribution. Teach. Stat. 2005, 27, 8–10. [Google Scholar] [CrossRef]
- Haddix, P.L.; Danderson, C.A. Mutation rate simulation by dice roll: Practice with the Drake equation. J. Microbiol. Biol. Educ. 2018, 19, 19.2.73. [Google Scholar] [CrossRef]
- Cornette, J.L.; Ackerman, R.A. Calculus for the Life Sciences: A Modeling Approach; American Mathematical Society: Providence, RI, USA, 2019; Volume 29. [Google Scholar]
- Sanft, R.; Walter, A. Experimenting with mathematical biology. PRIMUS 2016, 26, 83–103. [Google Scholar] [CrossRef]
- Sanft, R.; Walter, A. Exploring Mathematical Modeling in Biology through Case Studies and Experimental Activities; Academic Press: Cambridge, MA, USA, 2020. [Google Scholar]
- Aikens, M.L. Meeting the needs of a changing landscape: Advances and challenges in undergraduate biology education. Bull. Math. Biol. 2020, 82, 1–20. [Google Scholar] [CrossRef]
- Eaton, C.D.; Highlander, H.C. The case for biocalculus: Design, retention, and student performance. CBE-Life Sci. Educ. 2017, 16, ar25. [Google Scholar] [CrossRef] [PubMed]
- Haddix, P.L.; Paulsen, E.T.; Werner, T.F. Measurement of mutation to antibiotic resistance: Ampicillin resistance in Serratia marcescens. Bioscene 2000, 26, 17–21. [Google Scholar]
- Green, D.S.; Bozzone, D.M. A test of hypotheses about random mutation: Using classic experiments to teach experimental design. Am. Biol. Teach. 2001, 63, 54–58. [Google Scholar] [CrossRef]
- Hester, L.L.; Sarvary, M.A.; Ptak, C.J. Mutation and selection: An exploration of antibiotic resistance in serratia marcescens. Proc. Assoc. Biol. Lab. 2014, 35, 140–183. [Google Scholar]
- Smith, G.P.; Golomb, M.; Billstein, S.K.; Smith, S.M. The Luria-Delbrück fluctuation test as a classroom investigation in Darwinian evolution. Am. Biol. Teach. 2015, 77, 614–619. [Google Scholar] [CrossRef]
- Amir, A.; Balaban, N.Q. Learning from noise: How observing stochasticity may aid microbiology. Trends Microbiol. 2018, 26, 376–385. [Google Scholar] [CrossRef]
- Hutchison, E.A.; Scheffler, A.; Militello, K.T.; Reinhardt, J.; Nedelkovska, H.; Jamburuthugoda, a.V.K. An undergraduate laboratory exploring mutational mechanisms in Escherichia coli based on the Luria-Delbrück experiment. J. Microbiol. Biol. Educ. 2022, 23, e00211-21. [Google Scholar] [CrossRef]
- Møller, A.P.; Mousseau, T.A. Biological consequences of Chernobyl: 20 years on. Trends Ecol. Evol. 2006, 21, 200–207. [Google Scholar] [CrossRef]
- Sacks, B.; Meyerson, G.; Siegel, J.A. Epidemiology without biology: False paradigms, unfounded assumptions, and specious statistics in radiation science (with commentaries by Inge Schmitz-Feuerhake and Christopher Busby and a reply by the authors). Biol. Theory 2016, 11, 69–101. [Google Scholar] [CrossRef]
- Selya, R. Salvador Luria: An Immigrant Biologist in Cold War America; MIT Press: Cambridge, MA, USA, 2022. [Google Scholar]
- Abbott, A. Why did the FBI track Nobel-winning microbiologist Salvador Luria? Nature 2022, 612, 25–26. [Google Scholar] [CrossRef]
- Aktipis, C.A.; Kwan, V.S.; Johnson, K.A.; Neuberg, S.L.; Maley, C.C. Overlooking evolution: A systematic analysis of cancer relapse and therapeutic resistance research. PLoS ONE 2011, 6, e26100. [Google Scholar] [CrossRef] [PubMed]
- Merlo, L.M.; Pepper, J.W.; Reid, B.J.; Maley, C.C. Cancer as an evolutionary and ecological process. Nat. Rev. Cancer 2006, 6, 924–935. [Google Scholar] [CrossRef] [PubMed]
- Dujon, A.M.; Aktipis, A.; Alix-Panabières, C.; Amend, S.R.; Boddy, A.M.; Brown, J.S.; Capp, J.-P.; DeGregori, J.; Ewald, P.; Gatenby, R.; et al. Identifying key questions in the ecology and evolution of cancer. Evol. Appl. 2021, 14, 877–892. [Google Scholar] [CrossRef] [PubMed]
- Enriquez-Navas, P.M.; Wojtkowiak, J.W.; Gatenby, R.A. Application of evolutionary principles to cancer therapy. Cancer Res. 2015, 75, 4675–4680. [Google Scholar] [CrossRef]
- Smith, C.J.; Perfetti, T.A.; Berry, C.; Brash, D.E.; Bus, J.; Calabrese, E.; Clemens, R.A.; Greim, H.; MacGregor, J.T.; Maronpot, R.; et al. Bruce Nathan Ames-Paradigm shifts inside the cancer research revolution. Mutat. Res. Mutat. Res. 2021, 787, 108363. [Google Scholar] [CrossRef]
- Cole, J.; Arlett, C.; Green, M. The fluctuation test as a more sensitive system for determining induced mutation in l5178y mouse lymphoma cells. Mutat. Res. Mol. Mech. Mutagen. 1976, 41, 377–386. [Google Scholar] [CrossRef]
- Frank, S.A. Somatic mosaicism and cancer: Inference based on a conditional Luria–Delbrück distribution. J. Theor. Biol. 2003, 223, 405–412. [Google Scholar] [CrossRef]
- Kendal, W.S.; Frost, P. Pitfalls and practice of Luria-Delbrück fluctuation analysis: A review. Cancer Res. 1988, 48, 1060–1065. [Google Scholar]
- Law, L. Origin of the resistance of leukaemic cells to folic acid antagonists. Nature 1952, 169, 628–629. [Google Scholar] [CrossRef]
- Skipper, H.E. The forty-year-old mutation theory of Lurla and Delbrück and its pertinence to cancer chemotherapy. Adv. Cancer Res. 1983, 40, 331–363. [Google Scholar]
- Tlsty, T.D.; Margolin, B.H.; Lum, K. Differences in the rates of gene amplification in nontumorigenic and tumorigenic cell lines as measured by Luria-Delbrück fluctuation analysis. Proc. Natl. Acad. Sci. USA 1989, 86, 9441–9445. [Google Scholar] [CrossRef] [PubMed]
- Goldie, J.H.; Coldman, A.J. Drug Resistance in Cancer: Mechanisms and Models; Cambridge University Press: Cambridge, UK, 1998. [Google Scholar]
- Durrett, R. Branching process models of cancer. In Branching Process Models of Cancer; Springer: Berlin/Heidelberg, Germany, 2015; pp. 1–63. [Google Scholar]
- Pinedo, H.M.; Giaccone, G. Drug Resistance in the Treatment of Cancer; Cambridge University Press: Cambridge, UK, 1998. [Google Scholar]
- Teicher, B.A. Antiangiogenic Agents in Cancer Therapy; Springer: Berlin/Heidelberg, Germany, 1998. [Google Scholar]
- Zhu, X.; Li, S.; Xu, B.; Luo, H. Cancer evolution: A means by which tumors evade treatment. Biomed. Pharmacother. 2021, 133, 111016. [Google Scholar] [CrossRef] [PubMed]
- Graham, T.A.; Sottoriva, A. Measuring cancer evolution from the genome. J. Pathol. 2017, 241, 183–191. [Google Scholar] [CrossRef] [PubMed]
- Turajlic, S.; Sottoriva, A.; Graham, T.; Swanton, C. Resolving genetic heterogeneity in cancer. Nat. Rev. Genet. 2019, 20, 404–416. [Google Scholar] [CrossRef]
- Saito, T.; Niida, A.; Uchi, R.; Hirata, H.; Komatsu, H.; Sakimura, S.; Hayashi, S.; Nambara, S.; Kuroda, Y.; Ito, S.; et al. A temporal shift of the evolutionary principle shaping intratumor heterogeneity in colorectal cancer. Nat. Commun. 2018, 9, 1–11. [Google Scholar] [CrossRef]
- Davis, A.; Gao, R.; Navin, N. Tumor evolution: Linear, branching, neutral or punctuated? Biochim. Biophys. Acta-(BBA) Cancer 2017, 1867, 151–161. [Google Scholar] [CrossRef]
- Frank, S.A. The Number of Neutral Mutants in an Expanding Luria-Delbrück Population Is Approximately Fréchet. 2022. Available online: https://papers.ssrn.com/sol3/papers.cfm?abstract_id=4219807 (accessed on 15 January 2023).
- Rodriguez-Brenes, I.A.; Wodarz, D.; Komarova, N.L. Cellular replication limits in the luria–delbrück mutation model. Phys. Nonlinear Phenom. 2016, 328, 44–51. [Google Scholar] [CrossRef]
- Frank, S.A. Numbers of mutations within multicellular bodies: Why it matters. Axioms 2022, 12, 12. [Google Scholar] [CrossRef]
- Frank, S.A.; Nowak, M.A. Developmental predisposition to cancer. Nature 2003, 422, 494. [Google Scholar] [CrossRef]
- Lesho, E.P.; Laguio-Vila, M. The slow-motion catastrophe of antimicrobial resistance and practical interventions for all prescribers. In Mayo Clinic Proceedings; Elsevier: Amsterdam, The Netherlands, 2019; Volume 94, pp. 1040–1047. [Google Scholar]
- Oakberg, E.F.; Luria, S. Mutations to sulfonamide resistance in Staphylococcusaureus. Genetics 1947, 32, 249. [Google Scholar] [CrossRef]
- Demerec, M. Production of staphylococcus strains resistant to various concentrations of penicillin. Proc. Natl. Acad. Sci. USA 1945, 31, 16–24. [Google Scholar] [CrossRef] [PubMed]
- Demerec, M. Origin of bacterial resistance to antibiotics. J. Bacteriol. 1948, 56, 63–74. [Google Scholar] [CrossRef] [PubMed]
- Cavalli, L. Genetic analysis of drug-resistance. Bull. World Health Organ. 1952, 6, 185. [Google Scholar] [PubMed]
- Creager, A.N. Adaptation or selection? old issues and new stakes in the postwar debates over bacterial drug resistance. Stud. Hist. Philos. Sci. Part Stud. Hist. Philos. Biol. Biomed. Sci. 2007, 38, 159–190. [Google Scholar] [CrossRef] [PubMed]
- Ventola, C.L. The antibiotic resistance crisis: Part 1: Causes and threats. Pharm. Ther. 2015, 40, 277. [Google Scholar]
- Sievert, D.; Kirby, A.; McDonald, L.C. The CDC response to antibiotic and antifungal resistance in the environment. Med 2021, 2, 365–369. [Google Scholar] [CrossRef]
- Carson, R. Silent Spring: Cambridge; Hamish Hamilton: London, UK, 1962. [Google Scholar]
- Epstein, L. Fifty years since silent spring. Annu. Rev. Phytopathol. 2014, 52, 377–402. [Google Scholar] [CrossRef]
- Daemmrich, A. A tale of two experts: Thalidomide and political engagement in the united states and west germany. Soc. Hist. Med. 2002, 15, 137–158. [Google Scholar] [CrossRef]
- McBride, W.G. Thalidomide and congenital abnormalities. Lancet 1961, 2, 291–293. [Google Scholar]
- O’Rourke, M. Sticky situation for teflon maker. Risk Manag. 2005, 52, 8. [Google Scholar]
- Wilson, J. ‘The Devil We Know:’How Dupont Poisoned the World with Teflon; Organic Consumer Association: Finland, MN, USA, 2019. [Google Scholar]
- Ames, B.N. The detection of chemical mutagens with enteric bacteria. In Chemical Mutagens; Springer: Berlin/Heidelberg, Germany, 1971; pp. 267–282. [Google Scholar]
- Ames, B.N. Identifying environmental chemicals causing mutations and cancer. Science 1979, 204, 587–593. [Google Scholar]
- Ames, B.N.; Lee, F.D.; Durston, W.E. An improved bacterial test system for the detection and classification of mutagens and carcinogens. Proc. Natl. Acad. Sci. USA 1973, 70, 782–786. [Google Scholar] [CrossRef] [PubMed]
- Ames, B.N.; McCann, J.; Yamasaki, E. Methods for detecting carcinogens and mutagens with the salmonella/mammalianmicrosome mutagenicity test. Mutat. Res. 1975, 31, 347–364. [Google Scholar] [CrossRef] [PubMed]
- McCann, J.; Choi, E.; Yamasaki, E.; Ames, B.N. Detection of carcinogens as mutagens in the salmonella/microsome test: Assay of 300 chemicals. Proc. Natl. Acad. Sci. USA 1975, 72, 5135–5139. [Google Scholar] [CrossRef]
- Claxton, L.D.; Umbuzeiro, G.d.; DeMarini, D.M. The Salmonella mutagenicity assay: The stethoscope of genetic toxicology for the 21st century. Environ. Health Perspect. 2010, 118, 1515–1522. [Google Scholar] [CrossRef] [PubMed]
- Bridges, B. The fluctuation test. Arch. Toxicol. 1980, 46, 41–44. [Google Scholar] [CrossRef] [PubMed]
- Parry, J.M. The use of yeast cultures for the detection of environmental mutagens using a fluctuation test. Mutat. Res. Mutagen. Relat. Subj. 1977, 46, 165–175. [Google Scholar] [CrossRef] [PubMed]
- Collings, B.J.; Margolin, B.H.; Oehlert, G.W. Analyses for binomial data, with application to the fluctuation test for mutagenicity. Biometrics 1981, 775–794. [Google Scholar] [CrossRef]
- Kim, B.S.; Margolin, B.H. Statistical methods for the Ames Salmonella assay: A review. Mutat. Res. Mutat. Res. 1999, 436, 113–122. [Google Scholar] [CrossRef]
- Piegorsch, W.W.; Simmons, S.J.; Margolin, B.H.; Zeiger, E.; Gidrol, X.M.; Gee, P. Statistical modeling and analyses of a base-specific salmonella mutagenicity assay. Mutat. Res. Toxicol. Environ. 2000, 467, 11–19. [Google Scholar] [CrossRef]
- Kauffmann, K.; Werner, F.; Deitert, A.; Finklenburg, J.; Brendt, J.; Schiwy, A.; Hollert, H.; Büchs, J. Optimization of the ames RAMOS test allows for a reproducible high-throughput mutagenicity test. Sci. Total Environ. 2020, 717, 137168. [Google Scholar] [CrossRef]
- Wlodkowic, D.; Jansen, M. High-Throughput Screening Paradigms in Ecotoxicity Testing: Emerging Prospects and Ongoing Challenges; Chemosphere: Los Angeles, CA, USA, 2022; p. 135929. [Google Scholar]
- Graham, M.J.; Frederick, J.; Byars-Winston, A.; Hunter, A.-B.; Handelsman, J. Increasing persistence of college students in STEM. Science 2013, 341, 1455–1456. [Google Scholar] [CrossRef] [PubMed]
- National Research Council. Transforming Undergraduate Education in Science, Mathematics, Engineering, and Technology; National Academies Press: Washington, DC, USA, 1999. [Google Scholar]
- Freeman, S.; Eddy, S.L.; McDonough, M.; Smith, M.K.; Okoroafor, N.; Jordt, H.; Wenderoth, M.P. Active learning increases student performance in science, engineering, and mathematics. Proc. Natl. Acad. Sci. USA 2014, 111, 8410–8415. [Google Scholar] [CrossRef] [PubMed]
- Olson, S.; Riordan, D.G. Engage to Excel: Producing One Million Additional College Graduates with Degrees in Science, Technology, Engineering, and Mathematics Report to the President; Executive Office of the President: Washington, DC, USA, 2012.
- National Research Council; Singer, S.R.; Nielsen, N.R.; Schweingruber, H.A. Discipline-Based Education Research: Understanding and Improving Learning in Undergraduate Science and Engineering; National Academies Press: Washington, DC, USA, 2012. [Google Scholar]
- Barnett, J.H.; Lodder, J.; Pengelley, D. Teaching and learning mathematics from primary historical sources. PRIMUS 2016, 26, 1–18. [Google Scholar] [CrossRef]
- Kaiser, G.; Schwarz, B.; Buchholtz, N. Authentic modelling problems in mathematics education. In Trends in Teaching and Learning of Mathematical Modelling; Kaiser, G., Blum, W., Ferri, R.B., Stillman, G., Eds.; Springer: Dordrecht, The Netherlands, 2011; pp. 591–601. [Google Scholar]
- Kjeldsen, T.H.; Clark, K.M.; Jankvist, U.T. Developing Historical Awareness through the Use of Primary Sources in the Teaching and Learning of Mathematics; Springer International Publishing: Cham, Switzerland, 2022; pp. 45–68. [Google Scholar]
- Hoskins, S.G.; Lopatto, D.; Stevens, L.M. The CREATE approach to primary literature shifts undergraduates’ self-assessed ability to read and analyze journal articles, attitudes about science, and epistemological beliefs. CBE-Life Sci. Educ. 2011, 10, 368–378. [Google Scholar] [CrossRef] [PubMed]
- Kenyon, K.L.; Cosentino, B.J.; Gottesman, A.J.; Onorato, M.E.; Hoque, J.; Hoskins, S.G. From CREATE workshop to course implementation: Examining downstream impacts on teaching practices and student learning at 4-year institutions. BioScience 2019, 69, 47–58. [Google Scholar] [CrossRef] [PubMed]
- Dolan, E.; Weaver, G. A Guide to Course-Based Undergraduate Research; Macmillan Higher Education: London, UK, 2021. [Google Scholar]
- Vance-Chalcraft, H.D.; Hurlbert, A.H.; Styrsky, J.N.; Gates, T.A.; Bowser, G.; Hitchcock, C.B.; Reyes, M.A.; Cooper, C.B. Citizen science in postsecondary education: Current practices and knowledge gaps. BioScience 2022, 72, 276–288. [Google Scholar] [CrossRef]
- Waterman, M.A.; Stanley, E.D. Investigative case-based learning: Teaching scientifically while connecting science to society. In Invention and Impact: Building Excellence in Undergraduate Science, Technology, Engineering and Mathematics (STEM) Education, Successful Pedagogies; AAAS: Washington, DC, USA, 2004; pp. 55–60. [Google Scholar]
- Mustaffa, N.; Ismail, Z.; Tasir, Z.; Said, M.N. The impacts of implementing problem-based learning (PBL) in mathematics: A review of literature. Int. J. Acad. Res. Bus. Soc. Sci. 2016, 6, 490–503. [Google Scholar] [CrossRef]
- Rothstein, D.; Santana, L.; Minigan, A.P. Making questions flow. Educ. Leadersh. 2015, 73, 70–75. [Google Scholar]
- Brown, S.I.; Walter, M.I. Problem Posing: Reflections and Applications; Psychology Press: London, UK, 2014. [Google Scholar]
- Cai, J.; Hwang, S.; Jiang, C.; Silber, S. Problem-posing research in mathematics education: Some answered and unanswered questions. In Mathematical Problem Posing; Springer: Berlin/Heidelberg, Germany, 2015; pp. 3–34. [Google Scholar]
- Jungck, J.R. A problem posing approach to biology education. Am. Biol. Teach. 1985, 47, 264–266. [Google Scholar] [CrossRef]
- Peterson, N.S.; Jungck, J.R. Problem-posing, problem-solving and persuasion in biology education. Acad. Comput. 1988, 2, 14–17. [Google Scholar]
- Robeva, R.S.; Jungck, J.R.; Gross, L.J. Changing the nature of quantitative biology education: Data science as a driver. Bull. Math. Biol. 2020, 82, 1–30. [Google Scholar] [CrossRef] [PubMed]
- Blaschke, L.M. Heutagogy and lifelong learning: A review of heutagogical practice and self-determined learning. Int. Rev. Res. Open Distrib. Learn. 2012, 13, 56–71. [Google Scholar] [CrossRef]
- Stains, M.; Harshman, J.; Barker, M.K.; Chasteen, S.V.; Cole, R.; DeChenne-Peters, S.E.; Jr, M.K.E.; Esson, J.M.; Knight, J.K.; Laski, F.A.; et al. Anatomy of STEM teaching in North American universities. Science 2018, 359, 1468–1470. [Google Scholar] [CrossRef] [PubMed]
- Cajori, F. The Teaching and History of Mathematics in the United States; Number 3; US Government Printing Office: Washington, DC, USA, 1890.
- Herbart, J.F. The Science of Education: Its General Principles Deduced from Its Aim and the Aesthetic Revelation of the World; DC Heath & Company: Lexington, MA, USA, 1895. [Google Scholar]
- Knowles, M.S. The Modern Practice of Adult Education; andragogy versus pedagogy; The Association Press: New York, NY, USA, 1970. [Google Scholar]
- Schwab, J.J. The Teaching of Science: The Teaching of Science as Enquiry; Harvard University Press: Cambridge, MA, USA, 1966; Volume 253. [Google Scholar]
- Bruner, J.S. “The process of education” revisited. Phi Delta Kappan 1971, 53, 18–21. [Google Scholar]
- Hase, S. Heutagogy and e-learning in the workplace: Some challenges and opportunities. Impact J. Appl. Res. Workplace-Learn. 2009, 1, 43–52. [Google Scholar]
- Robeva, R. Algebraic and Discrete Mathematical Methods for Modern Biology; Academic Press: Cambridge, MA, USA, 2015. [Google Scholar]
- Robeva, R.; Hodge, T. Mathematical Concepts and Methods in Modern Biology: Using Modern Discrete Models; Academic Press: Cambridge, MA, USA, 2013. [Google Scholar]
- Robeva, R.; Macauley, M. Algebraic and Combinatorial Computational Biology; Academic Press: Cambridge, MA, USA, 2018. [Google Scholar]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Robeva, R.S.; Jungck, J.R. Fascination with Fluctuation: Luria and Delbrück’s Legacy. Axioms 2023, 12, 280. https://doi.org/10.3390/axioms12030280
Robeva RS, Jungck JR. Fascination with Fluctuation: Luria and Delbrück’s Legacy. Axioms. 2023; 12(3):280. https://doi.org/10.3390/axioms12030280
Chicago/Turabian StyleRobeva, Raina S., and John R. Jungck. 2023. "Fascination with Fluctuation: Luria and Delbrück’s Legacy" Axioms 12, no. 3: 280. https://doi.org/10.3390/axioms12030280
APA StyleRobeva, R. S., & Jungck, J. R. (2023). Fascination with Fluctuation: Luria and Delbrück’s Legacy. Axioms, 12(3), 280. https://doi.org/10.3390/axioms12030280





