A Novel Robust Topological Denoising Method Based on Homotopy Theory for Virtual Colonoscopy
Abstract
:1. Introduction
- (1)
- Mathematical rigor. Our proposed method is based on rigorous mathematical theory in homotopy, which guarantees the computation of the non-trivial loops.
- (2)
- Novel framework. The proposed algorithm is novel and has been first applied to colon surface denoising in virtual colonoscopy.
- (3)
- Robust computation. Compared to the State-of-the-Art topological denoising method, our method is more robust, based on the experimental results.
2. Materials and Methods
2.1. Theoretic Background
2.2. Overview
2.3. Algorithms
- 1.
- Every face of a simplex from is also in .
- 2.
- The non-empty intersection of any two simplices is a face of both and .
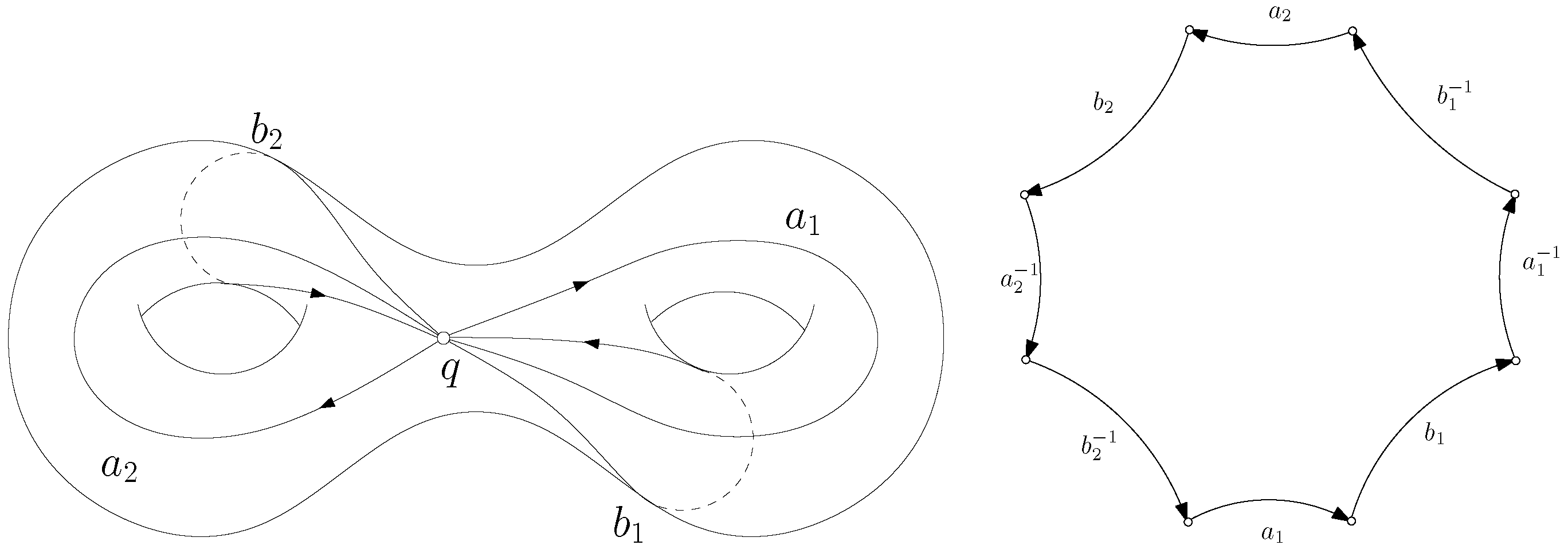
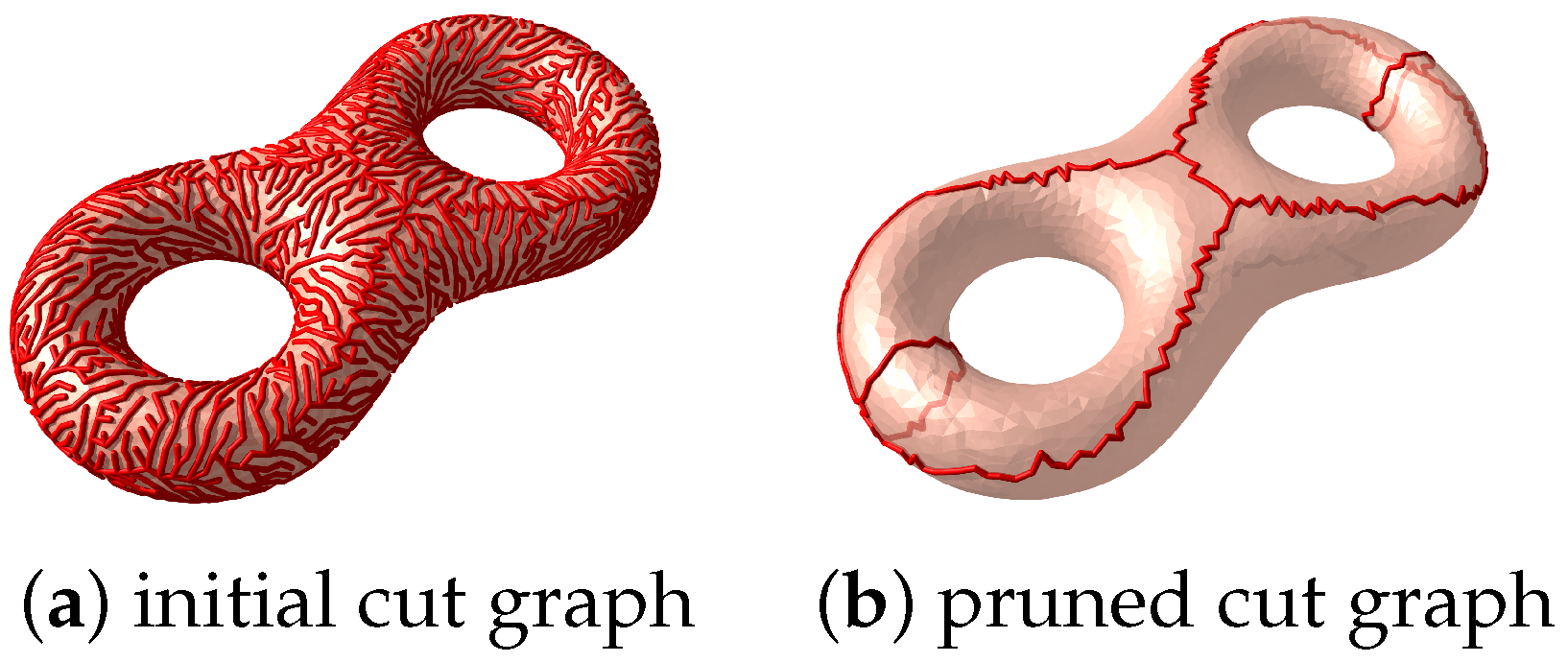
2.3.1. Cut Graph
| Algorithm 1 Algorithm for Cut Graph |
| Require: A closed triangle mesh M Ensure: is a cut graph of M 1: Compute the dual mesh of the input mesh M; 2: Compute a spanning tree of ; 3: The cut graph is given by ; 4: Prune all the leaves of recursively. |
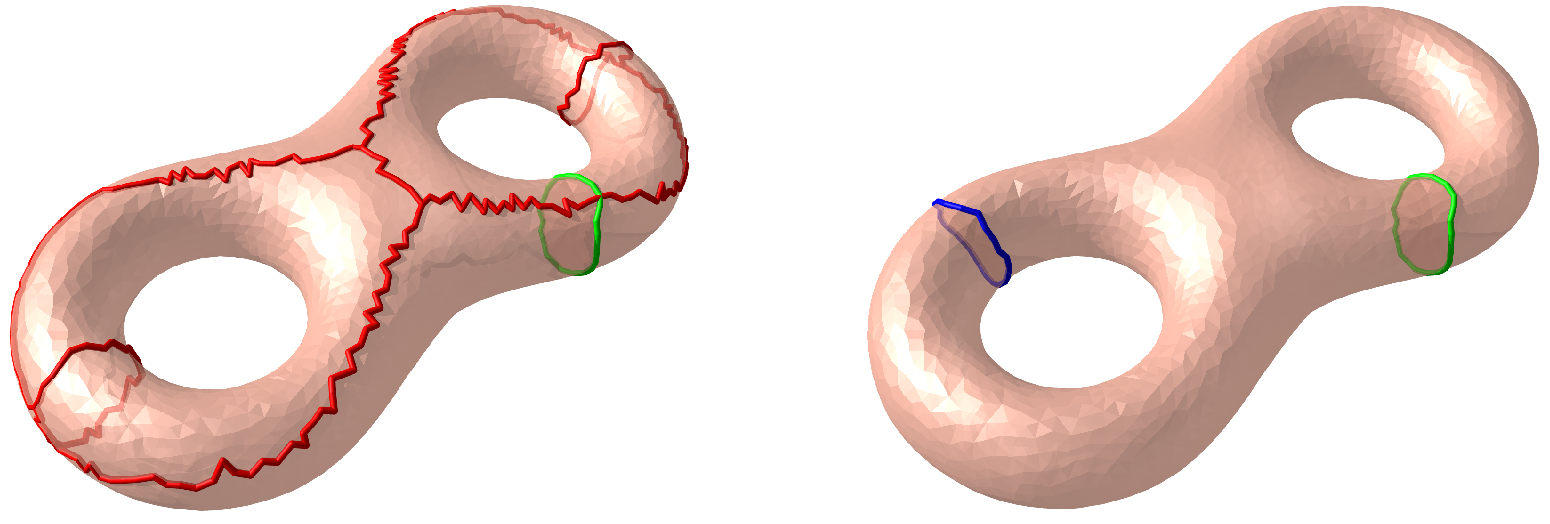
2.3.2. Shortest Loop
| Algorithm 2 Algorithm for Shortest Loop |
| Require: A closed triangle mesh M Ensure: The shortest non-trivial loop of M 1: Compute the cut graph of M, using Algorithm 1; 2: Slice M along , to obtain a simply connected mesh ; 3: for all vertex with do 4: Find all ; 5: for all pair on the boundary do 6: Compute the shortest path , using Dijkstra’s algorithm; 7: Find the loop corresponding to ; 8: end for 9: end for 10: Sort all the shortest loops in ascending order by their total lengths; 11: Return the shortest loop . |
2.3.3. Topological Surgery
| Algorithm 3 Algorithm for Topological Surgery |
| Require: A closed triangle mesh M, a non-trivial loop Ensure: A mesh N with one handle removed from M 1: Slice M along to obtain a mesh , ; 2: for all boundary component , do 3: ; 4: for all vertex do 5: ; 6: end for 7: ; 8: for all edge do 9: and form a triangle face ; 10: ; 11: end for 12: end for 13: . |
| Algorithm 4 Topological Denoising Algorithm |
| Require: A high-genus closed triangle mesh M Ensure: The mesh N with all handles removed from M 1: Compute the Euler number and the genus g of M; 2: Set ; 3: for do 4: Compute the shortest loop on , using Algorithm 2; 5: Perform topological surgery on along , to obtain , using Algorithm 3; 6: end for 7: Set . |
3. Results
3.1. Visual Evaluation on General Test Models
3.2. Topological Denoising for Two Colon Datasets
3.3. Colon Flattening in Virtual Colonoscopy
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Yao, J.; Li, J.; Summers, R. CT colonography computer-aided polyp detection using topographical height map. In Proceedings of the 2007 IEEE International Conference on Image Processing, San Antonio, TX, USA, 16–19 September 2007; Volume 5, pp. V-21–V-24. [Google Scholar]
- Lee, J.G.; Kim, J.H.; Kim, S.H.; Park, H.S.; Choi, B.I. A straightforward approach to computer-aided polyp detection using a polyp-specific volumetric feature in CT colonography. Comput. Biol. Med. 2011, 41, 790–801. [Google Scholar] [CrossRef]
- Umehara, K.; Näppi, J.J.; Hironaka, T.; Regge, D.; Ishida, T.; Yoshida, H. Deep ensemble learning of virtual endoluminal views for polyp detection in CT colonography. In Proceedings of the Medical Imaging 2017: Computer-Aided Diagnosis, SPIE, Orlando, FL, USA, 13–16 February 2017; Volume 10134, pp. 108–113. [Google Scholar]
- Ren, Y.; Ma, J.; Xiong, J.; Chen, Y.; Lu, L.; Zhao, J. Improved false positive reduction by novel morphological features for computer-aided polyp detection in CT colonography. IEEE J. Biomed. Health Inform. 2018, 23, 324–333. [Google Scholar] [CrossRef]
- Pomeroy, M.; Wang, Y.; Banerjee, A.; Abbasi, A.; Barish, M.; Sun, E.; Bucobo, J.C.; Pickhardt, P.J.; Liang, Z. Integration of optical and virtual colonoscopy images for enhanced classification of colorectal polyps. In Proceedings of the Medical Imaging 2020: Computer-Aided Diagnosis, SPIE, Houston, TX, USA, 16–19 February 2020; Volume 11314, pp. 139–144. [Google Scholar]
- Mirhosseini, S.; Gutenko, I.; Ojal, S.; Marino, J.; Kaufman, A. Immersive virtual colonoscopy. IEEE Trans. Vis. Comput. Graph. 2019, 25, 2011–2021. [Google Scholar] [CrossRef]
- Mathew, S.; Nadeem, S.; Kumari, S.; Kaufman, A. Augmenting colonoscopy using extended and directional cyclegan for lossy image translation. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 13–19 June 2020; pp. 4696–4705. [Google Scholar]
- Kotecha, S.; Vasudevan, A.; Holla, V.K.; Kumar, S.; Pruthviraja, D.; Latte, M.V. 3D visualization cloud based model to detect and classify the polyps according to their sizes for CT colonography. J. King Saud Univ.-Comput. Inf. Sci. 2022, 34, 4943–4955. [Google Scholar] [CrossRef]
- Nappi, J.J.; Hironaka, T.; Wu, D.; Yoshida, S.R.; Gupta, R.; Tachibana, R.; Taguchi, K.; Yoshida, H. Automated detection of colorectal polyps in photon-counting CT colonography. In Proceedings of the Medical Imaging 2023: Imaging Informatics for Healthcare, Research, and Applications, SPIE, San Diego, CA, USA, 19–24 February 2023; Volume 12469, pp. 187–191. [Google Scholar]
- Chen, S.; Li, Y.F.; Zhang, J. Vision processing for realtime 3-D data acquisition based on coded structured light. IEEE Trans. Image Process. 2008, 17, 167–176. [Google Scholar] [CrossRef]
- Bi, Z.; Wang, L. Advances in 3D data acquisition and processing for industrial applications. Robot. Comput. Integr. Manuf. 2010, 26, 403–413. [Google Scholar] [CrossRef]
- Henderson, P.; Tsiminaki, V.; Lampert, C.H. Leveraging 2d data to learn textured 3d mesh generation. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 13–19 June 2020; pp. 7498–7507. [Google Scholar]
- Glaßer, S.; Berg, P.; Neugebauer, M.; Preim, B. Reconstruction of 3D surface meshes for blood flow simulations of intracranial aneurysms. In Proceedings of the Conference of the German Society for Computer and Robotic Assisted Surgery, London, UK, 10–11 September 2015; pp. 163–168. [Google Scholar]
- Kong, F.; Wilson, N.; Shadden, S. A deep-learning approach for direct whole-heart mesh reconstruction. Med. Image Anal. 2021, 74, 102222. [Google Scholar] [CrossRef]
- Lee, S.; Chen, L.; Wang, J.; Liniger, A.; Kumar, S.; Yu, F. Uncertainty guided policy for active robotic 3d reconstruction using neural radiance fields. IEEE Robot. Autom. Lett. 2022, 7, 12070–12077. [Google Scholar] [CrossRef]
- Di Angelo, L.; Di Stefano, P.; Guardiani, E. A review of computer-based methods for classification and reconstruction of 3D high-density scanned archaeological pottery. J. Cult. Herit. 2022, 56, 10–24. [Google Scholar] [CrossRef]
- Maboudi, M.; Homaei, M.; Song, S.; Malihi, S.; Saadatseresht, M.; Gerke, M. A Review on Viewpoints and Path Planning for UAV-Based 3D Reconstruction. IEEE J. Sel. Top. Appl. Earth Obs. Remote. Sens. 2023, 16, 5026–5048. [Google Scholar] [CrossRef]
- Erickson, J.; Whittlesey, K. Greedy optimal homotopy and homology generators. In Proceedings of the SODA, Vancouver, BC, Canada, 23–25 January 2005; Volume 5, pp. 1038–1046. [Google Scholar]
- Chen, C.; Freedman, D. Quantifying homology classes. arXiv 2008, arXiv:0802.2865. [Google Scholar]
- Dey, T.K.; Li, K.; Sun, J.; Cohen-Steiner, D. Computing geometry-aware handle and tunnel loops in 3d models. In ACM SIGGRAPH 2008 Papers; Association for Computing Machinery: New York, NY, USA, 2008; pp. 1–9. [Google Scholar]
- Zhou, Q.Y.; Ju, T.; Hu, S.M. Topology repair of solid models using skeletons. IEEE Trans. Vis. Comput. Graph. 2007, 13, 675–685. [Google Scholar] [CrossRef]
- Dey, T.K.; Li, K.; Sun, J. On computing handle and tunnel loops. In Proceedings of the 2007 International Conference on Cyberworlds (CW’07), Hannover, Germany, 24–26 October 2007; pp. 357–366. [Google Scholar]
- Dey, T.K.; Li, K.; Sun, J. Computing handle and tunnel loops with knot linking. Comput.-Aided Des. 2009, 41, 730–738. [Google Scholar] [CrossRef]
- Dey, T.K.; Fan, F.; Wang, Y. An efficient computation of handle and tunnel loops via Reeb graphs. ACM Trans. Graph. (TOG) 2013, 32, 1–10. [Google Scholar] [CrossRef]
- Weinrauch, A.; Seidel, H.P.; Mlakar, D.; Steinberger, M.; Zayer, R. A Variational Loop Shrinking Analogy for Handle and Tunnel Detection and Reeb Graph Construction on Surfaces. arXiv 2021, arXiv:2105.13168. [Google Scholar] [CrossRef]
- Gao, M.; Chen, C.; Zhang, S.; Qian, Z.; Metaxas, D.; Axel, L. Segmenting the papillary muscles and the trabeculae from high resolution cardiac CT through restoration of topological handles. In Proceedings of the 23rd International Conference on Information Processing in Medical Imaging, Asilomar, CA, USA, 28 June–3 July 2013; pp. 184–195. [Google Scholar]
- Wu, P.; Chen, C.; Wang, Y.; Zhang, S.; Yuan, C.; Qian, Z.; Metaxas, D.; Axel, L. Optimal topological cycles and their application in cardiac trabeculae restoration. In Proceedings of the 25th International Conference on Information Processing in Medical Imaging, Boone, NC, USA, 25–30 June 2017; pp. 80–92. [Google Scholar]
- Brezovsky, J.; Chovancova, E.; Gora, A.; Pavelka, A.; Biedermannova, L.; Damborsky, J. Software tools for identification, visualization and analysis of protein tunnels and channels. Biotechnol. Adv. 2013, 31, 38–49. [Google Scholar] [CrossRef]
- Paik, D.S.; Beaulieu, C.F.; Jeffrey Jr, R.B.; Karadi, C.A.; Napel, S. Visualization modes for CT colonography using cylindrical and planar map projections. J. Comput. Assist. Tomogr. 2000, 24, 179–188. [Google Scholar] [CrossRef]
- Vilanova Bartroli, A.; Wegenkittl, R.; König, A.; Gröller, E.; Sorantin, E. Virtual colon flattening. In Proceedings of the Data Visualization 2001: Proceedings of the Joint Eurographics—IEEE TCVG Symposium on Visualization in Ascona, Ascona, Switzerland, 28–30 May 2001; pp. 127–136. [Google Scholar]
- Balogh, E.; Sorantin, E.; Nyul, L.; Palagyi, K.; Kuba, A.; Werkgartner, G.; Spuller, E. Colon unraveling based on electronic field: Recent progress and future work. Proc. SPIE 2002, 4681, 713–721. [Google Scholar]
- Hong, W.; Gu, X.; Qiu, F.; Jin, M.; Kaufman, A. Conformal virtual colon flattening. In Proceedings of the 2006 ACM Symposium on Solid and Physical Modeling, Wales, UK, 6–8 June 2006; pp. 85–93. [Google Scholar]
- Hong, W.; Qiu, F. A pipeline for computer aided polyp detection. IEEE Trans. Vis. Comput. Graph. 2006, 12, 861–868. [Google Scholar] [CrossRef]
- Wang, Z.; Li, B.; Liang, Z. Feature-based texture display for detection of polyps on flattened colon volume. In Proceedings of the 2005 IEEE Engineering in Medicine and Biology 27th Annual Conference, Shanghai, China, 17–18 January 2006; pp. 3359–3362. [Google Scholar]
- Gurijala, K.C.; Shi, R.; Zeng, W.; Gu, X.; Kaufman, A. Colon flattening using heat diffusion riemannian metric. IEEE Trans. Vis. Comput. Graph. 2013, 19, 2848–2857. [Google Scholar] [CrossRef]
- Wang, H.; Chen, Y.; Li, L.; Pan, H.; Gu, X.; Liang, Z. A novel colon wall flattening model for computed tomographic colonography: Method and validation. Comput. Methods Biomech. Biomed. Eng. Imaging Vis. 2015, 3, 213–221. [Google Scholar] [CrossRef]
- Kreiser, J.; Meuschke, M.; Mistelbauer, G.; Preim, B.; Ropinski, T. A survey of flattening-based medical visualization techniques. Comput. Graph. Forum 2018, 37, 597–624. [Google Scholar] [CrossRef]
- Eulzer, P.; Richter, K.; Meuschke, M.; Hundertmark, A.; Lawonn, K. Automatic Cutting and Flattening of Carotid Artery Geometries. In Proceedings of the VCBM, Paris, France, 22–24 September 2021; pp. 79–89. [Google Scholar]
- Zeng, W.; Shi, R.; Su, Z.; Gu, D.X. Efficient Topological Cleaning for Visual Colon Surface Flattening. In Abdomen Thoracic Imaging; Springer: Berlin/Heidelberg, Germany, 2014; pp. 421–441. [Google Scholar]
- Armstrong, M.A. Basic Topology; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2013. [Google Scholar]






| Mesh | #Faces | #Genus |
|---|---|---|
| Genus-6 model | 2 K | 6 |
| Double torus | 7 K | 2 |
| Knotty | 10 K | 2 |
| Genus-3 model | 12 K | 3 |
| Amphora | 20 K | 2 |
| LoveMe | 50 K | 3 |
| TwoKids | 80 K | 3 |
| Deco-cube | 120 K | 5 |
| Success? | All Loops Found? | #Vertices of All Loops | ||||
|---|---|---|---|---|---|---|
| Mesh (#Faces, #Genus) | PHM | Our | PHM | Our | PHM | Our |
| Colon1 (127 K, 67) | No | Yes | No | Yes | NA | 311 |
| Colon2 (44 K, 13) | Yes | Yes | Yes | Yes | 65 | 61 |
| Colon3 (158 K, 31) | No | Yes | No | Yes | NA | 141 |
| Colon4 (152 K, 8) | Yes | Yes | Yes | Yes | 56 | 28 |
| Colon5 (140 K, 38) | Yes | Yes | Yes | Yes | 660 | 170 |
| Colon6 (133 K, 30) | Yes | Yes | Yes | Yes | 177 | 141 |
| Colon7 (167 K, 14) | Yes | Yes | Yes | Yes | 161 | 66 |
| Colon8 (176 K, 13) | Yes | Yes | Yes | Yes | 131 | 67 |
| Colon9 (236 K, 7) | Yes | Yes | Yes | Yes | 24 | 23 |
| Colon10 (147 K, 19) | Yes | Yes | Yes | Yes | 118 | 74 |
| Success? | All Loops Found? | #Vertices of All Loops | ||||
|---|---|---|---|---|---|---|
| Mesh (#Faces, #Genus) | PHM | Our | PHM | Our | PHM | Our |
| Colon1 (1091 K, 4) | Yes | Yes | Yes | Yes | 77 | 24 |
| Colon2 (1679 K, 6) | Yes | Yes | Yes | Yes | 115 | 65 |
| Colon3 (1679 K, 6) | Yes | Yes | Yes | Yes | 118 | 65 |
| Colon4 (1528 K, 26) | Yes | Yes | Yes | Yes | 1104 | 492 |
| Colon5 (1181 K, 17) | Yes | Yes | Yes | Yes | 231 | 119 |
| Colon6 (1350 K, 12) | Yes | Yes | Yes | Yes | 321 | 124 |
| Colon7 (1185 K, 3) | Yes | Yes | Yes | Yes | 60 | 36 |
| Colon8 (1144 K, 9) | Yes | Yes | Yes | Yes | 158 | 107 |
| Colon9 (1389 K, 17) | Yes | Yes | Yes | Yes | 675 | 436 |
| Colon10 (1259 K, 2) | Yes | Yes | Yes | Yes | 11 | 11 |
| Colon11 (1204 K, 4) | Yes | Yes | Yes | Yes | 116 | 58 |
| Colon12 (1692 K, 2) | Yes | Yes | Yes | Yes | 122 | 36 |
| Colon13 (1236 K, 11) | Yes | Yes | Yes | Yes | 206 | 129 |
| Colon14 (1300 K, 11) | Yes | Yes | Yes | Yes | 298 | 250 |
| Colon15 (1428 K, 17) | Yes | Yes | Yes | Yes | 326 | 213 |
| Colon16 (1631 K, 22) | No | Yes | No | Yes | NA | 410 |
| Colon17 (1531 K, 8) | Yes | Yes | Yes | Yes | 409 | 277 |
| Colon18 (1774 K, 11) | Yes | Yes | Yes | Yes | 210 | 83 |
| Colon19 (1482 K, 18) | Yes | Yes | Yes | Yes | 483 | 454 |
| Colon20 (1202 K, 19) | No | Yes | No | Yes | NA | 177 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, M.; Chen, W.; Lei, N.; Gu, X. A Novel Robust Topological Denoising Method Based on Homotopy Theory for Virtual Colonoscopy. Axioms 2023, 12, 942. https://doi.org/10.3390/axioms12100942
Ma M, Chen W, Lei N, Gu X. A Novel Robust Topological Denoising Method Based on Homotopy Theory for Virtual Colonoscopy. Axioms. 2023; 12(10):942. https://doi.org/10.3390/axioms12100942
Chicago/Turabian StyleMa, Ming, Wei Chen, Na Lei, and Xianfeng Gu. 2023. "A Novel Robust Topological Denoising Method Based on Homotopy Theory for Virtual Colonoscopy" Axioms 12, no. 10: 942. https://doi.org/10.3390/axioms12100942
APA StyleMa, M., Chen, W., Lei, N., & Gu, X. (2023). A Novel Robust Topological Denoising Method Based on Homotopy Theory for Virtual Colonoscopy. Axioms, 12(10), 942. https://doi.org/10.3390/axioms12100942






