Isolation and Characterization of A Novel Iron–Sulfur Oxidizing Bacterium Acidithiobacillus Ferrooxidans YQ-N3 and its Applicability in Coal Biodesulfurization
Abstract
1. Introduction
2. Materials and Methods
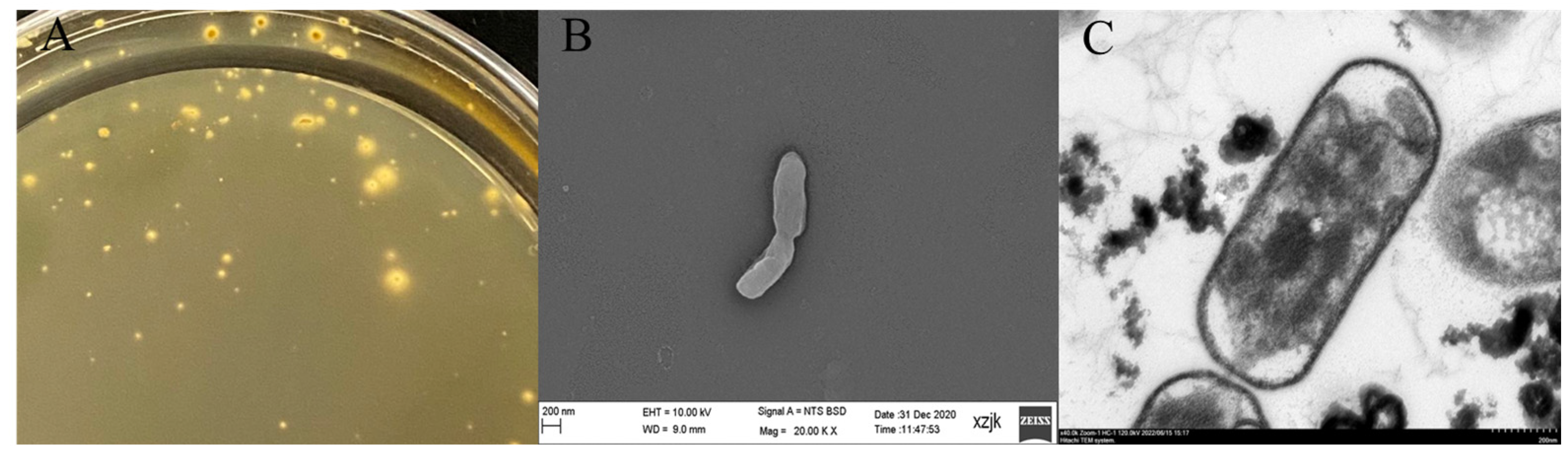
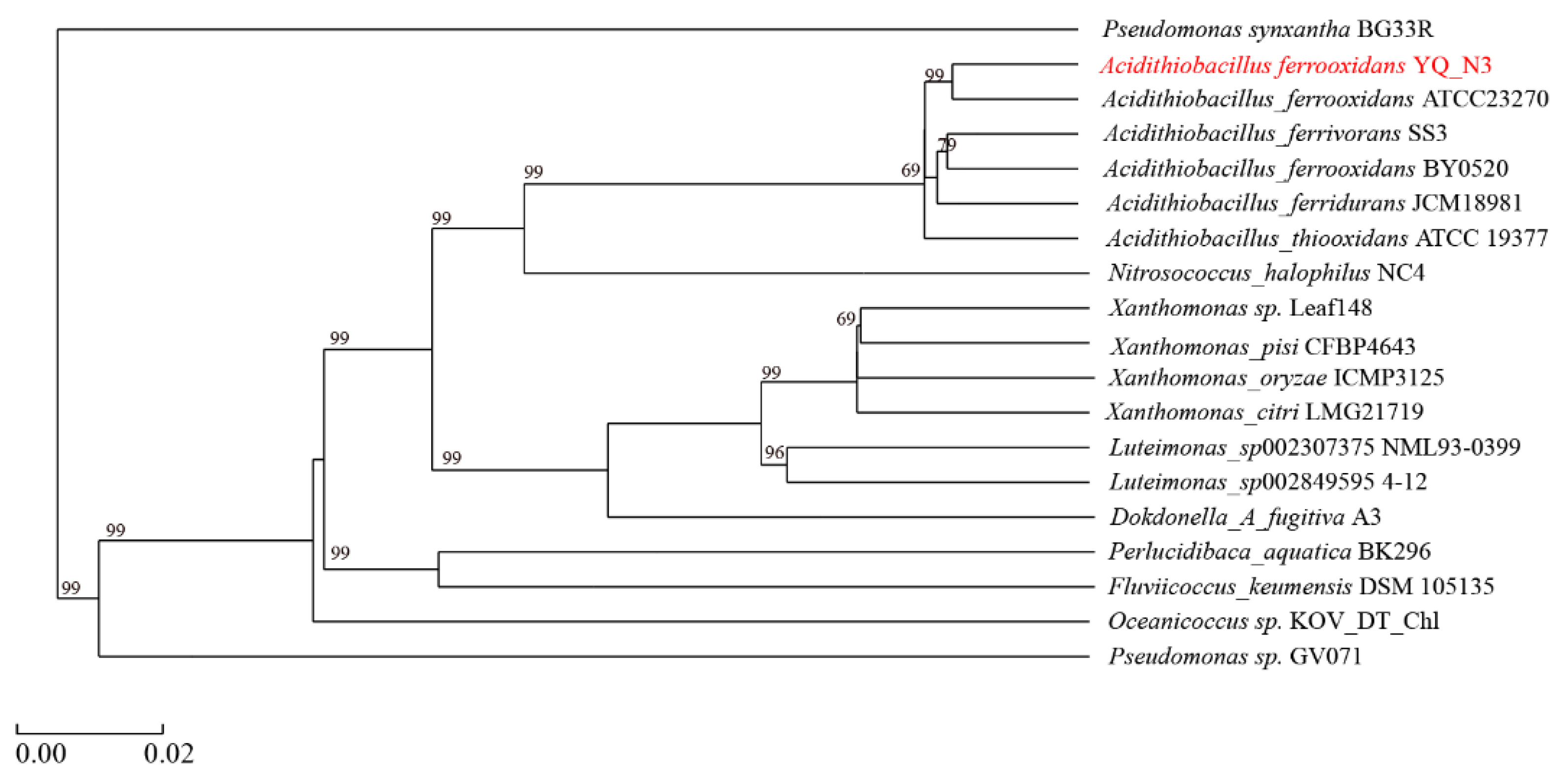
2.1. Isolation, Purification, and Identification
2.2. Whole genome Sequencing
2.3. Comparative Genomic Analysis
2.4. Oxidation Characteristics of Fe2+, S0, and FeS2
2.5. Coal Biodesulfurization Experiment
3. Results and Discussion
3.1. Growth Characteristics of A. ferrooxidans YQ-N3
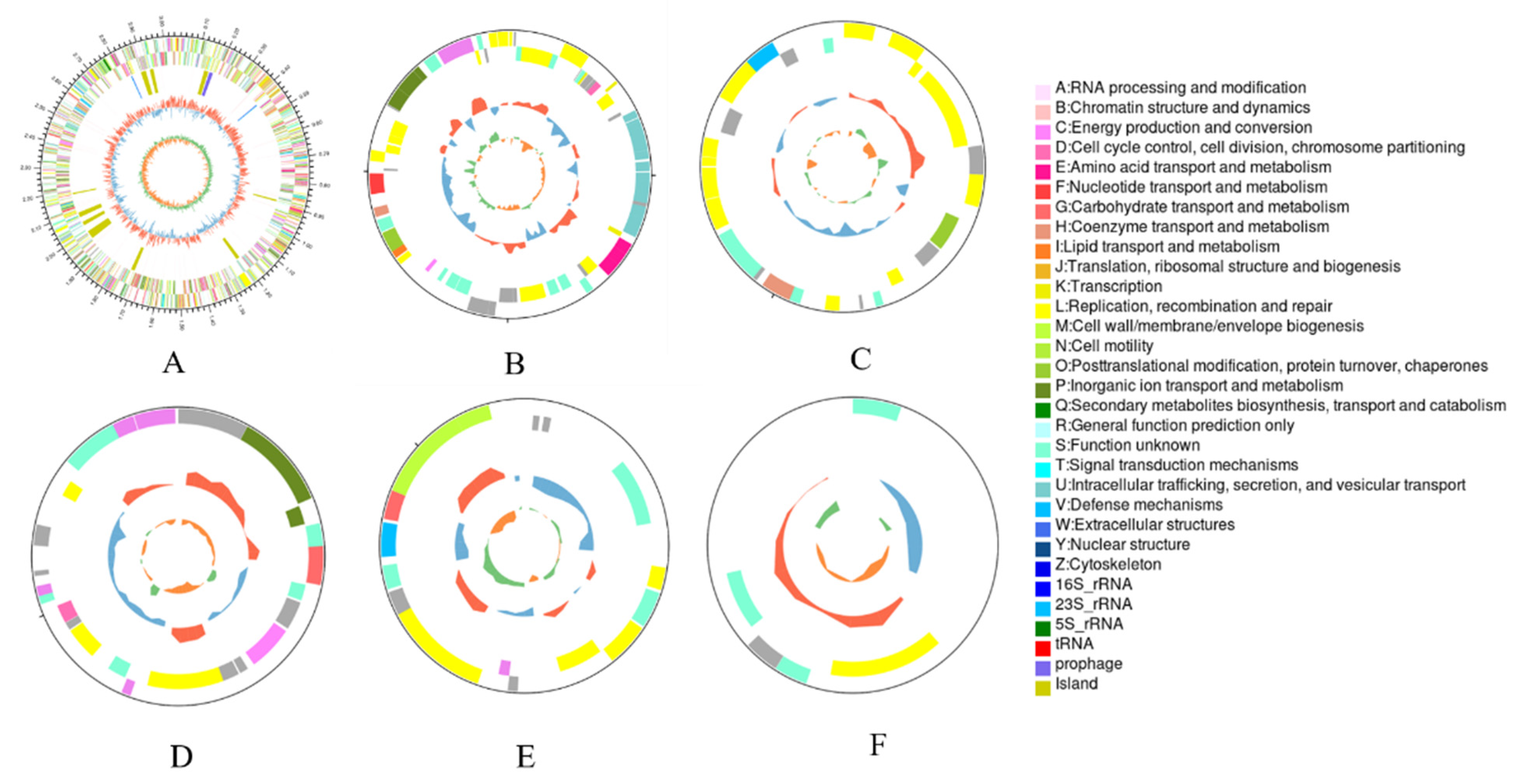
3.2. Genome Overview of A. ferrooxidans YQ-N3
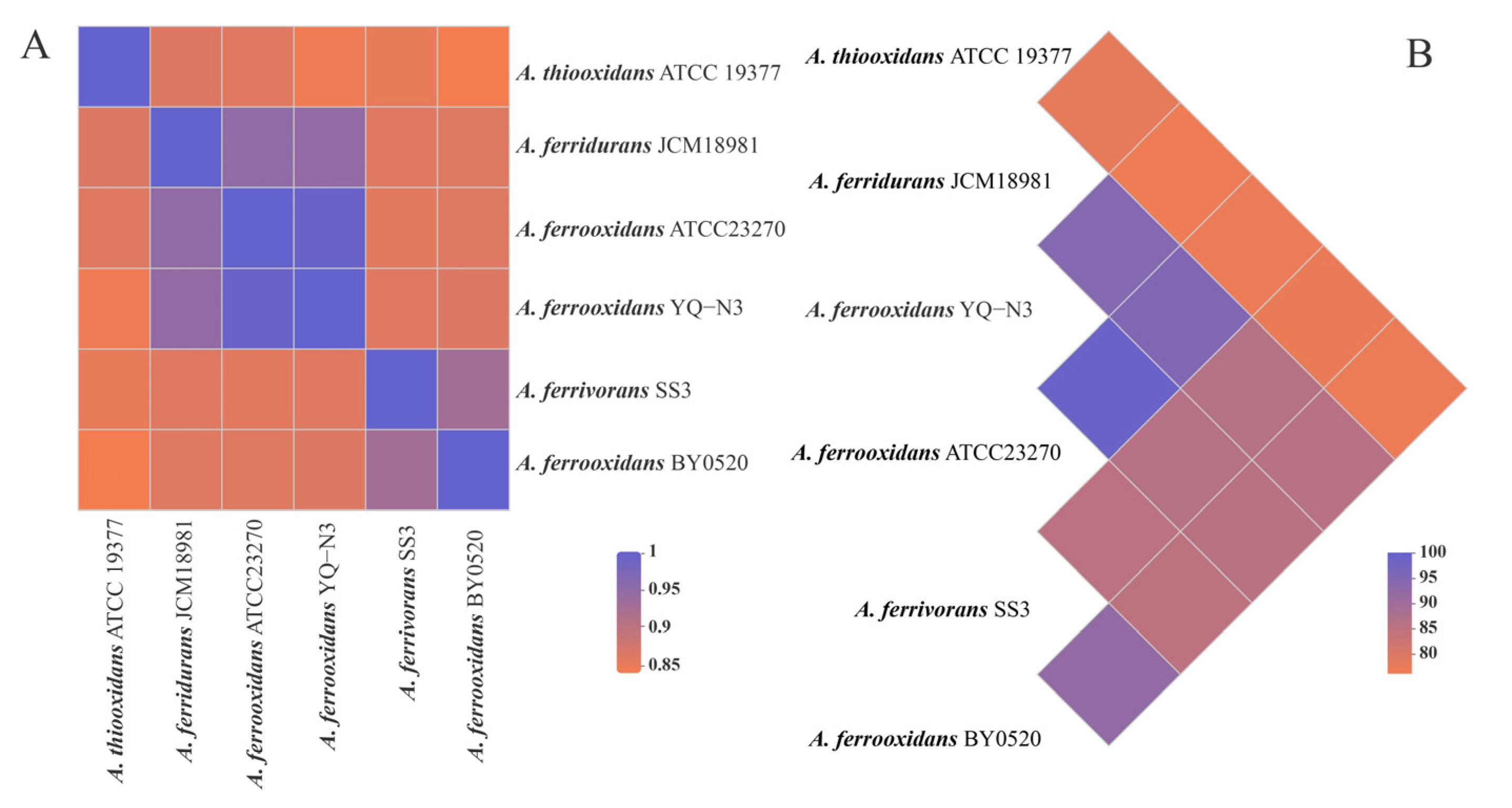
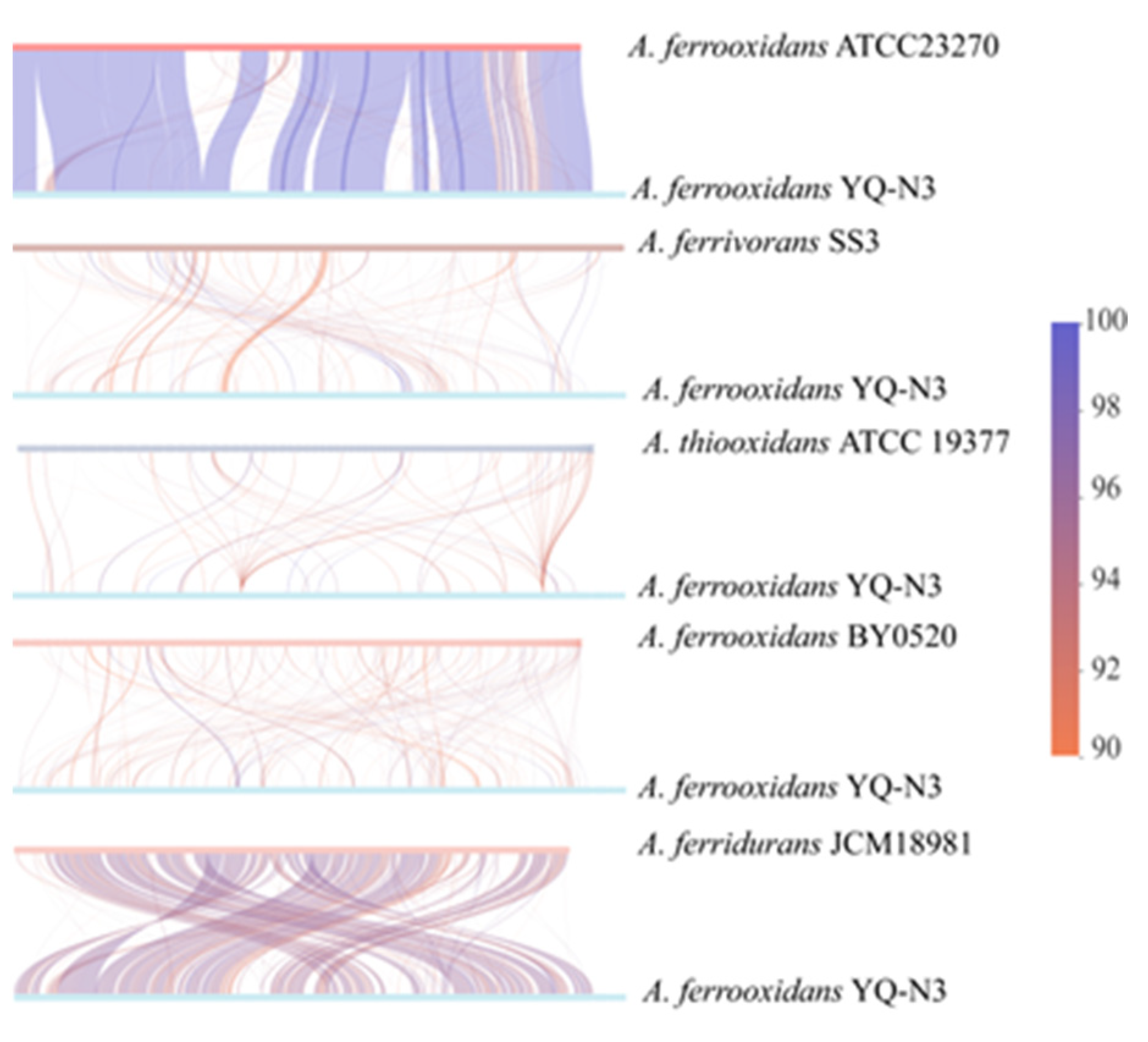
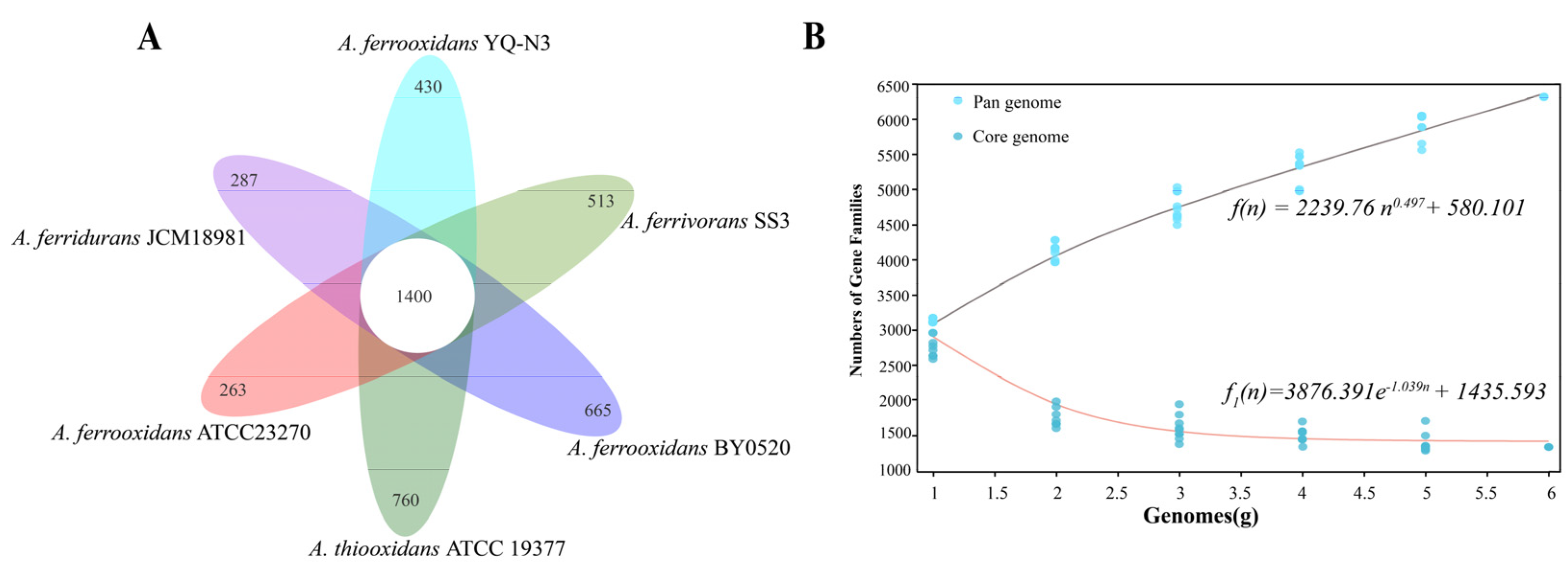
3.3. Comparative Genomic Analysis
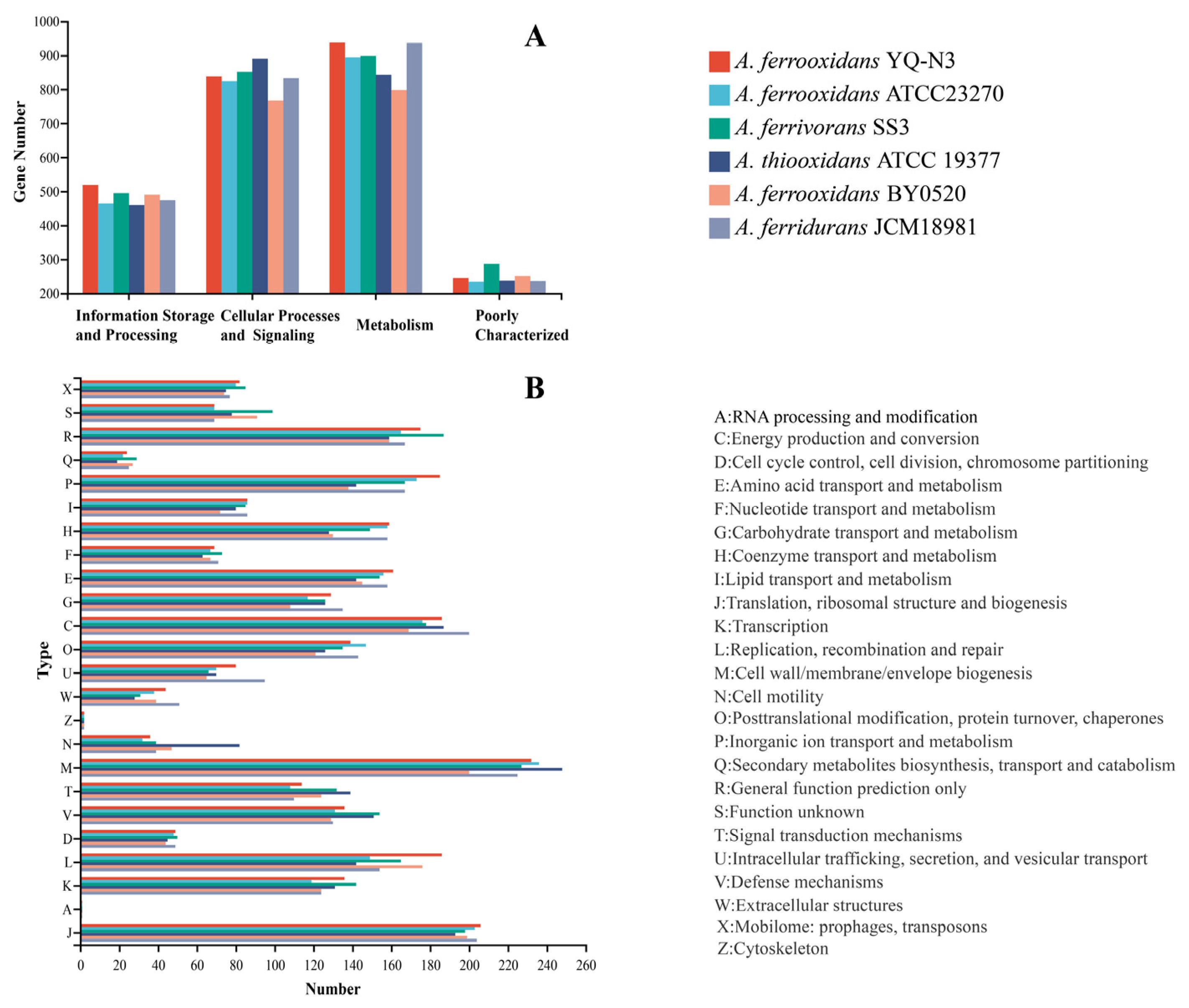
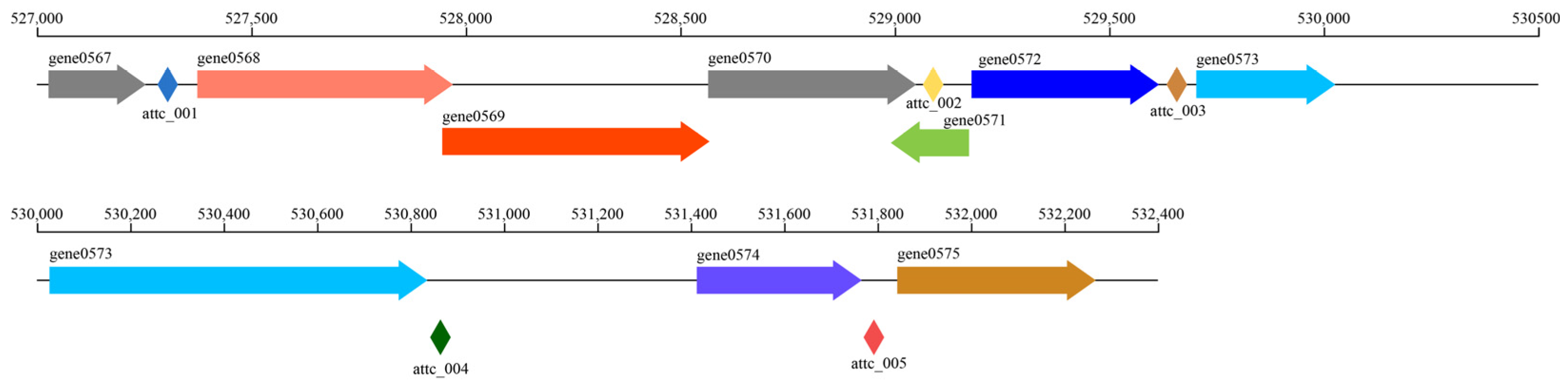
3.4. MEGs
3.5. Genes Associated with Iron and Sulfur Metabolism
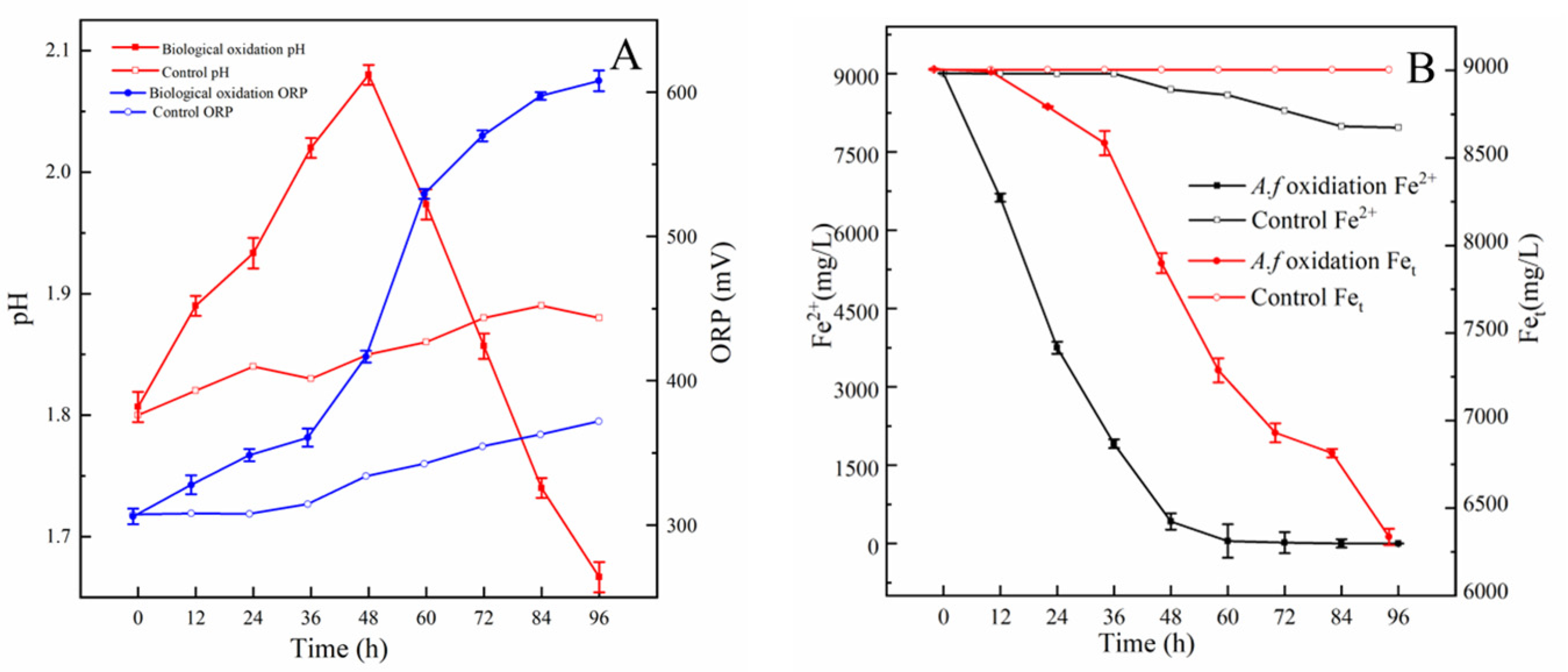
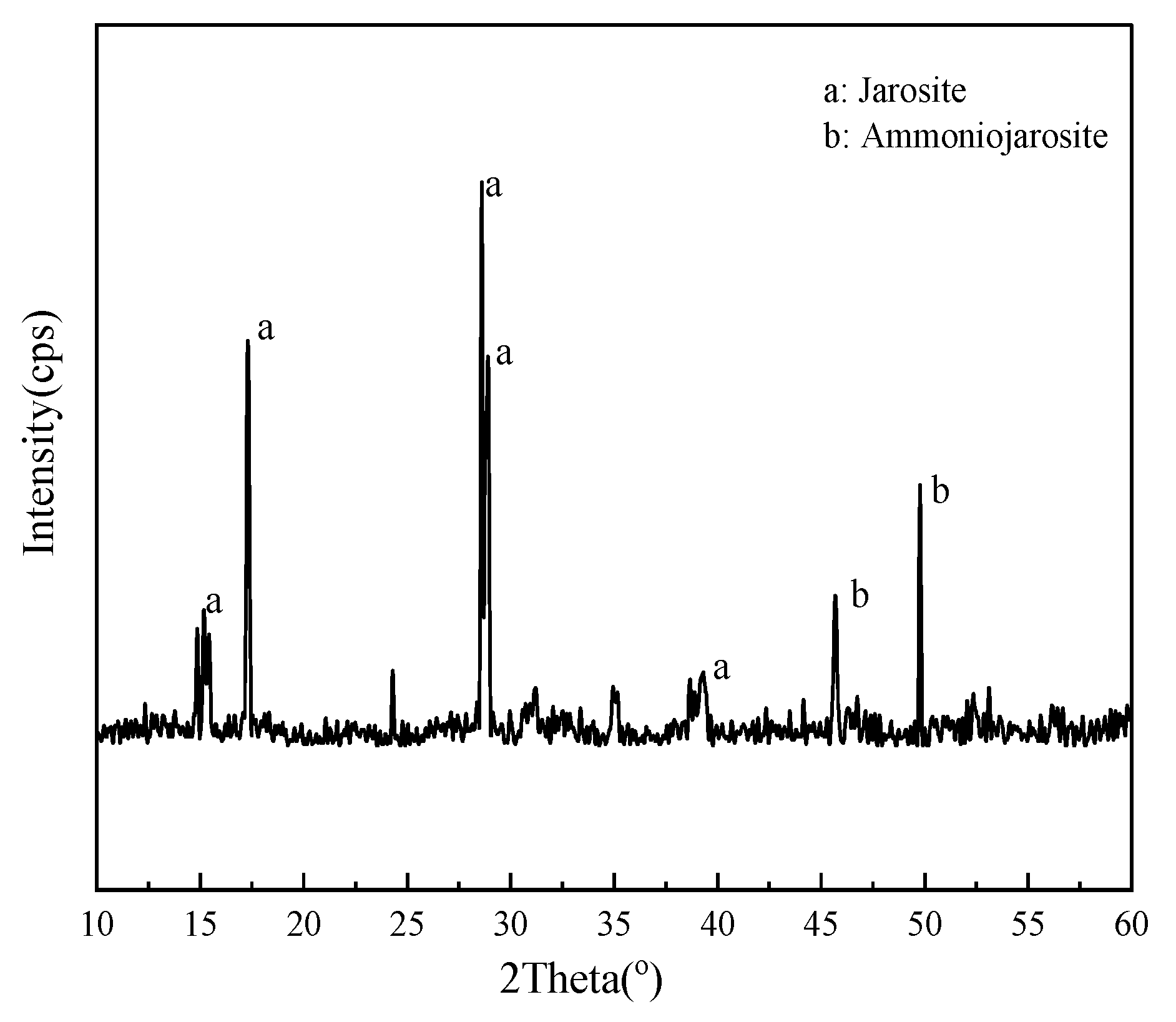
3.6. Oxidation of Fe2+, S0, and Pyrite
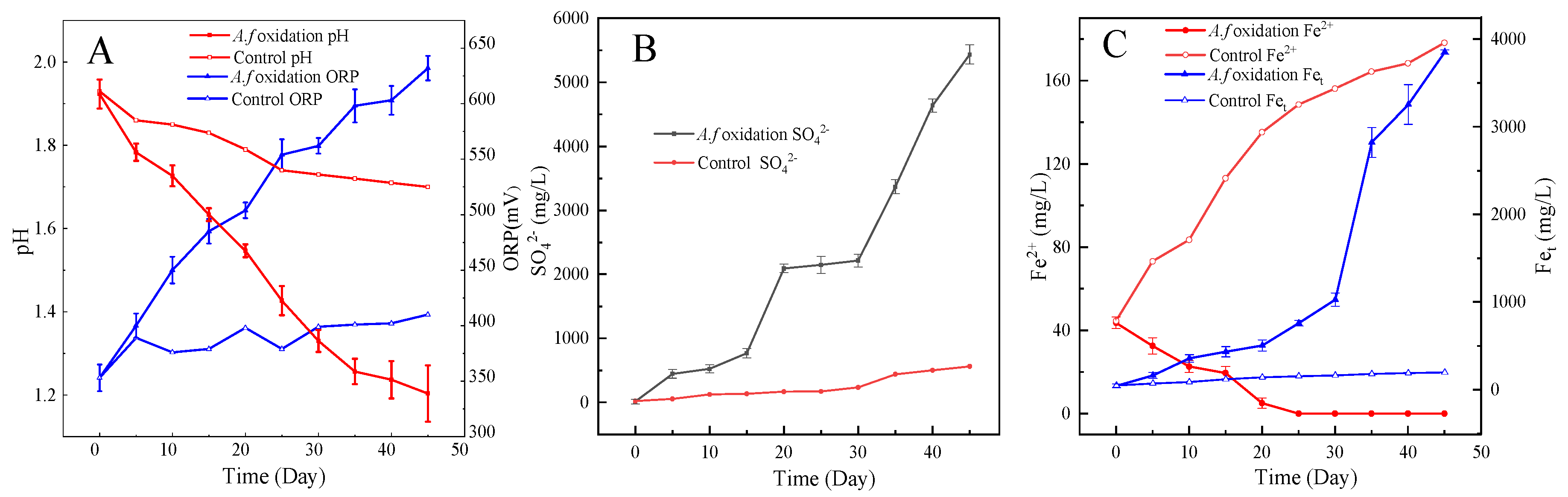
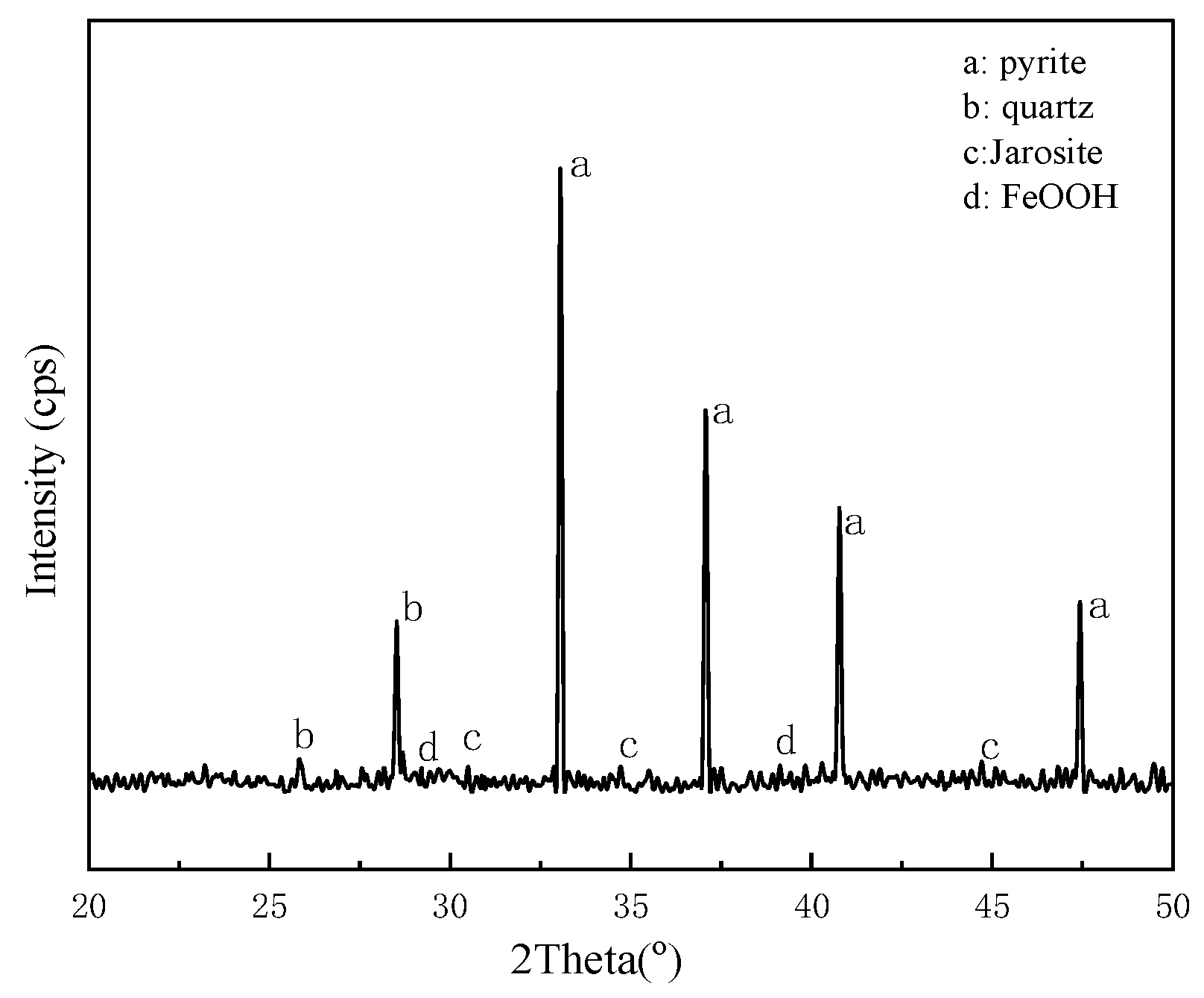
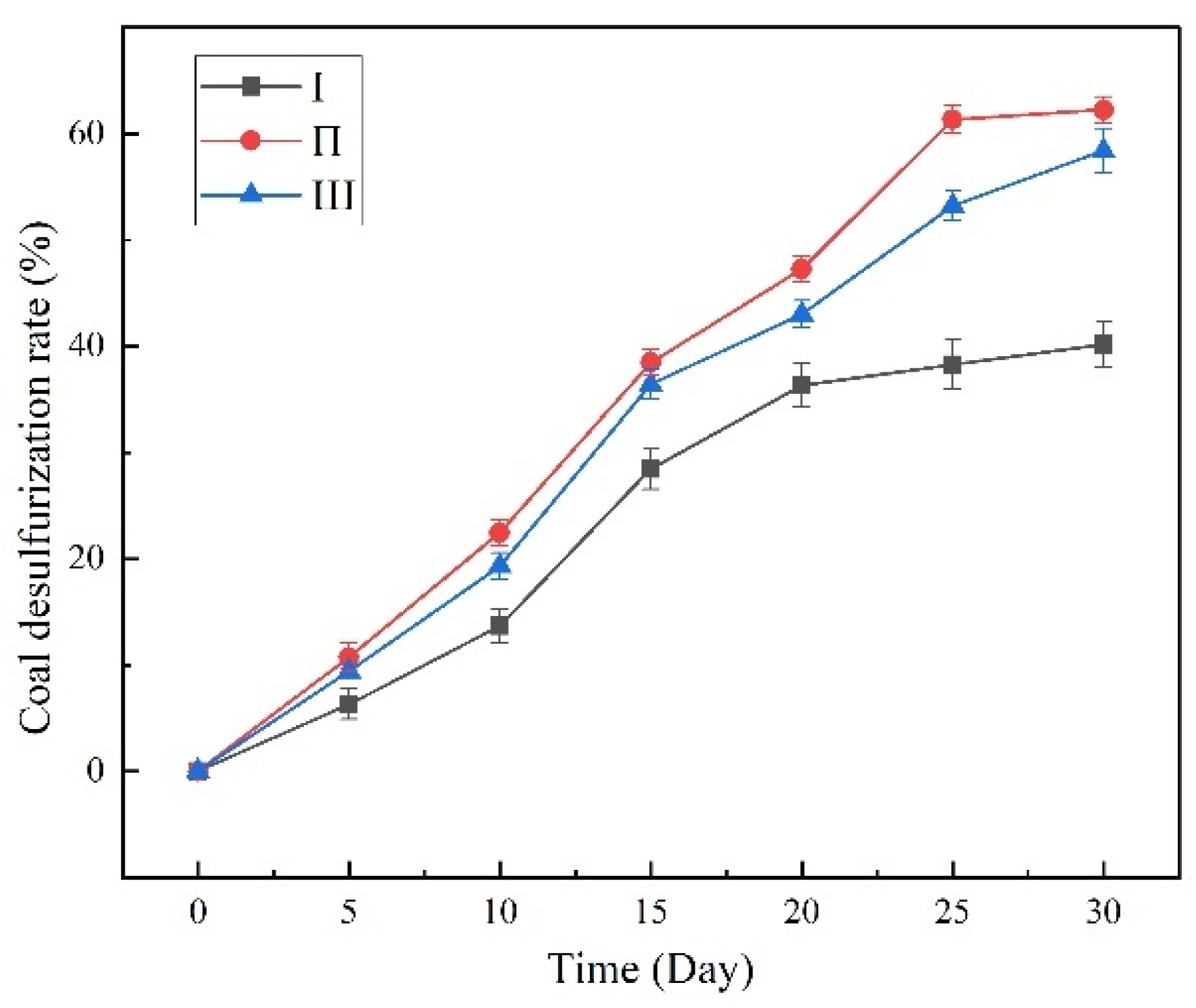
3.7. Desulfurization of Coal
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhan, Y.; Yang, M.R.; Zhang, S.; Zhao, D.; Duan, J.G.; Wang, W.D.; Yan, L. Iron and sulfur oxidation pathways of Acidithiobacillus ferrooxidans. World J. Microbiol. Biotechnol. 2019, 35, 12. [Google Scholar] [CrossRef] [PubMed]
- Moinier, D.; Byrne, D.; Amouric, A.; Bonnefoy, V. The Global Redox Responding RegB/RegA Signal Transduction System Regulates the Genes Involved in Ferrous Iron and Inorganic Sulfur Compound Oxidation of the Acidophilic Acidithiobacillus ferrooxidans. Front. Microbiol. 2017, 8, 16. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Yan, L.; Xing, W.J.; Chen, P.; Zhang, Y.; Wang, W.D. Acidithiobacillus ferrooxidans and its potential application. Extremophiles 2018, 22, 563–579. [Google Scholar] [CrossRef]
- Iakovleva, E.; Mäkilä, E.; Salonen, J.; Sitarz, M.; Wang, S.B.; Sillanpää, M. Acid mine drainage (AMD) treatment: Neutralization and toxic elements removal with unmodified and modified limestone. Ecol. Eng. 2014, 81, 30–40. [Google Scholar] [CrossRef]
- Valdes, J.; Pedroso, I.; Quatrini, R.; Dodson, R.J.; Tettelin, H.; Blake, R.; Eisen, J.A.; Holmes, D.S. Acidithiobacillus ferrooxidans metabolism: From genome sequence to industrial applications. BMC Genom. 2008, 9, 24. [Google Scholar] [CrossRef]
- Lorenzo-Tallafigo, J.; Iglesias-González, N.; Mazuelos, A.; Romero, R.; Carranza, F. An alternative approach to recover lead, silver and gold from black gossan (polymetallic ore). Study of biological oxidation and lead recovery stages. J. Clean. Prod. 2019, 207, 510–521. [Google Scholar] [CrossRef]
- Nie, H.Y.; Yang, C.; Zhu, N.W.; Wu, P.X.; Zhang, T.; Zhang, Y.Q.; Xing, Y.J. Isolation of Acidithiobacillus ferrooxidans strain Z1 and its mechanism of bioleaching copper from waste printed circuit boards. J. Chem. Technol. Biotechnol. 2015, 90, 714–721. [Google Scholar] [CrossRef]
- Yang, M.R.; Zhan, Y.; Zhang, S.; Wang, W.D.; Yan, L. Biological materials formed by Acidithiobacillus ferrooxidans and their potential applications. 3 Biotech 2020, 10, 9. [Google Scholar] [CrossRef]
- Rout, P.G.; Mohanty, A.K.; Pradhan, N.; Biswal, S.K.; Behera, S.K. Study on the Reaction Mechanism of Oxidative Microbial Desulfurization of Organic Sulfur-Rich Coal. Geomicrobiol. J. 2022, 39, 210–218. [Google Scholar] [CrossRef]
- Lavalle, L.; Chiacchiarini, P.; Pogliani, C.; Donati, E. Isolation and characterization of acidophilic bacteria from Patagonia, Argentina. Process Biochem. 2005, 40, 1095–1099. [Google Scholar] [CrossRef]
- Kai, M.; Yano, T.; Fukumori, Y.; Yamanaka, T. Cytochrome oxidase of an acidophilic iron-oxidizing bacterium, Thiobacillus ferrooxidans, functions at pH 3.5. Biochem. Biophys. Res. Commun. 1989, 160, 839–843. [Google Scholar] [CrossRef] [PubMed]
- Rhoads, A.; Au, K.F. PacBio Sequencing and Its Applications. Genom. Proteom. Bioinform. 2015, 13, 278–289. [Google Scholar] [CrossRef] [PubMed]
- Delcher, A.L.; Bratke, K.A.; Powers, E.C.; Salzberg, S.L. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics 2007, 23, 673–679. [Google Scholar] [CrossRef]
- Besemer, J.; Borodovsky, M. GeneMark: Web software for gene finding in prokaryotes, eukaryotes and viruses. Nucleic Acids Res. 2005, 33, W451–W454. [Google Scholar] [CrossRef] [PubMed]
- Bertelli, C.; Laird, M.R.; Williams, K.P.; Lau, B.Y.; Hoad, G.; Winsor, G.L.; Brinkman, F.S.L.; Simon Fraser Univ Res Comp, G. IslandViewer 4: Expanded prediction of genomic islands for larger-scale datasets. Nucleic Acids Res. 2017, 45, W30–W35. [Google Scholar] [CrossRef] [PubMed]
- Fouts, D.E. Phage_Finder: Automated identification and classification of prophage regions in complete bacterial genome sequences. Nucleic Acids Res. 2006, 34, 5839–5851. [Google Scholar] [CrossRef] [PubMed]
- Bland, C.; Ramsey, T.L.; Sabree, F.; Lowe, M.; Brown, K.; Kyrpides, N.C.; Hugenholtz, P. CRISPR Recognition Tool (CRT): A tool for automatic detection of clustered regularly interspaced palindromic repeats. BMC Bioinform. 2007, 8, 8. [Google Scholar] [CrossRef]
- Cury, J.; Jove, T.; Touchon, M.; Neron, B.; Rocha, E.P.C. Identification and analysis of integrons and cassette arrays in bacterial genomes. Nucleic Acids Res. 2016, 44, 4539–4550. [Google Scholar] [CrossRef]
- Chen, J.J.; Liu, Y.L.; Diep, P.; Mahadevan, R. Genomic Analysis of a Newly Isolated Acidithiobacillus ferridurans JAGS Strain Reveals Its Adaptation to Acid Mine Drainage. Minerals 2021, 11, 14. [Google Scholar] [CrossRef]
- Akcil, A.; Ciftci, H.; Deveci, H. Role and contribution of pure and mixed cultures of mesophiles in bioleaching of a pyritic chalcopyrite concentrate. Miner. Eng. 2007, 20, 310–318. [Google Scholar] [CrossRef]
- Gonzalez-Toril, E.; Llobet-Brossa, E.; Casamayor, E.O.; Amann, R.; Amils, R. Microbial ecology of an extreme acidic environment, the Tinto river. Appl. Environ. Microbiol. 2003, 69, 6959. [Google Scholar] [CrossRef]
- Umanskii, A.B.; Klyushnikov, A.M. Bioleaching of low grade uranium ore containing pyrite using A. ferrooxidans and A. thiooxidans. J. Radioanal. Nucl. Chem. 2013, 295, 151–156. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, S.; Zhao, D.; Ni, Y.Q.; Wang, W.D.; Yan, L. Complete Genome Sequence of Acidithiobacillus ferrooxidans YNTRS-40, a Strain of the Ferrous Iron- and Sulfur-Oxidizing Acidophile. Microorganisms 2020, 8, 10. [Google Scholar] [CrossRef]
- Hinger, I.; Ansorge, R.; Mussmann, M.; Romano, S. Phylogenomic Analyses of Members of the Widespread Marine Heterotrophic Genus Pseudovibrio Suggest Distinct Evolutionary Trajectories and a Novel Genus, Polycladidibacter gen. nov. Appl. Environ. Microbiol. 2020, 86, 17. [Google Scholar] [CrossRef]
- Hemme, C.L.; Green, S.J.; Rishishwar, L.; Prakash, O.; Pettenato, A.; Chakraborty, R.; Deutschbauer, A.M.; Van Nostrand, J.D.; Wu, L.Y.; He, Z.L.; et al. Lateral Gene Transfer in a Heavy Metal-Contaminated-Groundwater Microbial Community. mBio 2016, 7, 14. [Google Scholar] [CrossRef] [PubMed]
- Acosta, M.; Beard, S.; Ponce, J.; Vera, M.; Mobarec, J.C.; Jerez, C.A. Identification of putative sulfurtransferase genes in the extremophilic Acidithiobacillus ferrooxidans ATCC 23270 genome: Structural and functional characterization of the proteins. Omics 2005, 9, 13–29. [Google Scholar] [CrossRef] [PubMed]
- Mealman, T.D.; Blackburn, N.J.; McEvoy, M.M. Metal export by CusCFBA, the periplasmic Cu(I)/Ag(I) transport system of Escherichia coli. Curr. Top. Membr. 2012, 69, 163–196. [Google Scholar]
- Chacon, K.N.; Perkins, J.; Mathe, Z.; Alwan, K.; Ho, E.N.; Ucisik, M.N.; Merz, K.M.; Blackburn, N.J. Trapping intermediates in metal transfer reactions of the CusCBAF export pump of Escherichia coli. Commun. Biol. 2018, 1, 11. [Google Scholar] [CrossRef]
- Almarcegui, R.J.; Navarro, C.A.; Paradela, A.; Albar, J.P.; von Bernath, D.; Jerez, C.A. Response to copper of Acidithiobacillus ferrooxidans ATCC 23270 grown in elemental sulfur. Res. Microbiol. 2014, 165, 761–772. [Google Scholar] [CrossRef]
- Wenbin, N.; Dejuan, Z.; Feifan, L.; Lei, Y.; Peng, C.; Xiaoxuan, Y.; Hongyu, L.; Nan, W.B.; Dejuan, L.Z.; Feifan, Y.; et al. Quorum-sensing system in Acidithiobacillus ferrooxidans involved in its resistance to Cu2+. Lett. Appl. Microbiol. 2011, 53, 84–91. [Google Scholar] [CrossRef]
- Popa, O.; Hazkani-Covo, E.; Landan, G.; Martin, W.; Dagan, T. Directed networks reveal genomic barriers and DNA repair bypasses to lateral gene transfer among prokaryotes. Genome Res. 2011, 21, 599–609. [Google Scholar] [CrossRef] [PubMed]
- Polz, M.F.; Alm, E.J.; Hanage, W.P. Horizontal gene transfer and the evolution of bacterial and archaeal population structure. Trends Genet. 2013, 29, 170–175. [Google Scholar] [CrossRef]
- Coutinho, T.J.D.; Franco, G.R.; Lobo, F.P. Homology-Independent Metrics for Comparative Genomics. Comp. Struct. Biotechnol. J. 2015, 13, 352–357. [Google Scholar] [CrossRef] [PubMed]
- Dominguez-Mirazo, M.; Jin, R.; Weitz, J.S. Functional and comparative genomic analysis of integrated prophage-like sequences in “candidatus liberibacter asiaticus”. mSphere 2019, 4, 409–419. [Google Scholar] [CrossRef] [PubMed]
- Cady, K.C.; Bondy-Denomy, J.; Heussler, G.E.; Davidson, A.R.; O’Toole, G.A. The CRISPR/Cas Adaptive Immune System of Pseudomonas aeruginosa Mediates Resistance to Naturally Occurring and Engineered Phages. J. Bacteriol. 2012, 194, 5728–5738. [Google Scholar] [CrossRef] [PubMed]
- Dopson, M.; Holmes, D.S. Metal resistance in acidophilic microorganisms and its significance for biotechnologies. Appl. Microbiol. Biotechnol. 2014, 98, 8133–8144. [Google Scholar] [CrossRef] [PubMed]
- Nies, D.H. Efflux-mediated heavy metal resistance in prokaryotes. Fems Microbiol. Rev. 2003, 27, 313–339. [Google Scholar] [CrossRef]
- Hemme, C.L.; Deng, Y.; Gentry, T.J.; Fields, M.W.; Wu, L.Y.; Barua, S.; Barry, K.; Tringe, S.G.; Watson, D.B.; He, Z.L.; et al. Metagenomic insights into evolution of a heavy metal-contaminated groundwater microbial community. ISME J. 2010, 4, 660–672. [Google Scholar] [CrossRef]
- Hallberg, K.B.; Gonzalez-Toril, E.; Johnson, D.B. Acidithiobacillus ferrivorans, sp nov.; facultatively anaerobic, psychrotolerant iron-, and sulfur-oxidizing acidophiles isolated from metal mine-impacted environments. Extremophiles 2010, 14, 9–19. [Google Scholar] [CrossRef]
- Bonnefoy, V.; Holmes, D.S. Genomic insights into microbial iron oxidation and iron uptake strategies in extremely acidic environments. Environ. Microbiol. 2012, 14, 1597–1611. [Google Scholar] [CrossRef]
- Ponce, J.S.; Moinier, D.; Byrne, D.; Amouric, A.; Bonnefoy, V. Acidithiobacillus ferrooxidans oxidizes ferrous iron before sulfur likely through transcriptional regulation by the global redox responding RegBA signal transducing system. Hydrometallurgy 2012, 127, 187–194. [Google Scholar] [CrossRef]
- Johnson, D.B.; Hallberg, K.B. Carbon, Iron and Sulfur Metabolism in Acidophilic Micro-Organisms. In Advances in Microbial Physiology; Poole, R.K., Ed.; Academic Press Ltd-Elsevier Science Ltd: London, UK, 2009; Volume 54, pp. 201–255. [Google Scholar]
- Bird, L.J.; Bonnefoy, V.; Newman, D.K. Bioenergetic challenges of microbial iron metabolisms. Trends Microbiol. 2011, 19, 330–340. [Google Scholar] [CrossRef] [PubMed]
- Yarzabal, A.; Appia-Ayme, C.; Ratouchniak, J.; Bonnefoy, V. Regulation of the expression of the Acidithiobacillus ferrooxidans rus operon encoding two cytochromes c, a cytochrome oxidase and rusticyanin. Microbiology 2004, 150, 2113–2123. [Google Scholar] [CrossRef] [PubMed]
- Amouric, A.; Appia-Ayme, C.; Yarzabal, A.; Bonnefoy, V. Regulation of the iron and sulfur oxidation pathwayways in the acidophilic A. Ferrooxidans. Adv. Mater. Res. 2009, 71–73, 163–166. [Google Scholar] [CrossRef]
- Kucera, J.; Bouchal, P.; Lochman, J.; Potesil, D.; Janiczek, O.; Zdrahal, Z.; Mandl, M. Ferrous iron oxidation by sulfur-oxidizing Acidithiobacillus ferrooxidans and analysis of the process at the levels of transcription and protein synthesis. Antonie Van Leeuwenhoek 2013, 103, 905–919. [Google Scholar] [CrossRef]
- Wakai, S.; Tsujita, M.; Kikumoto, M.; Manchur, M.A.; Kanao, T.; Kamimura, K. Purification and characterization of sulfide: Quinone oxidoreductase from an acidophilic iron-oxidizing bacterium, acidithiobacillus ferrooxidans. Biosci. Biotechnol. Biochem. 2007, 71, 2735–2742. [Google Scholar] [CrossRef]
- Liu, Y.D.; Ji, J.J.; Yu, R.L.; Qiu, G.Z. Expression, Purification and Molecular Modeling of Another HdrC from Acidithiobacillus ferrooxidans Which Binds Only One 4Fe-4S Cluster. Curr. Microbiol. 2012, 65, 416–423. [Google Scholar] [CrossRef]
- Offeddu, F.G.; Cama, J.; Soler, J.M.; Dávila, G.; McDowell, A.; Craciunescu, T. Processes affecting the efficiency of limestone in passive treatments for AMD: Column experiments. J. Environ. Chem. Eng. 2015, 3, 304–316. [Google Scholar] [CrossRef]
- Nazari, B.; Jorjani, E.; Hani, H.; Manfi, Z.; Riahi, A. Formation of jarosite and its effect on important ions for A. ferrooxidans bacteria. Trans. Nonferrous Met. Soc. China 2014, 24, 1152–1160. [Google Scholar] [CrossRef]
- Li, W.B.; Feng, Q.Y.; Liang, H.Q.; Chen, D.; Li, X.D. Passive treatment test of acid mine drainage from an abandoned coal mine in Kaili Guizhou, China. Water Sci. Technol. 2021, 84, 1981–1996. [Google Scholar]
- Konishi, Y.; Asai, S.; Yoshida, N. Growth Kinetics of Thiobacillus thiooxidans on the Surface of Elemental Sulfur. Appl. Environ. Microbiol. 1995, 61, 3617–3622. [Google Scholar] [CrossRef]
- Knickerbocker, C.; Nordstrom, D.K.; Southam, G. The role of “blebbing” in overcoming the hydrophobic barrier during biooxidation of elemental sulfur by Thiobacillus thiooxidans. Chem. Geol. 2000, 169, 425–433. [Google Scholar] [CrossRef]
- Kuang, J.L.; Huang, L.N.; Chen, L.X.; Hua, Z.S.; Li, S.J.; Hu, M.; Li, J.T.; Shu, W.S. Contemporary environmental variation determines microbial diversity patterns in acid mine drainage. ISME J. 2013, 7, 1038–1050. [Google Scholar] [CrossRef] [PubMed]
- Cardona, I.C.; Marquez, M.A. Biodesulfurization of two Colombian coals with native microorganisms. Fuel Process. Technol. 2009, 90, 1099–1106. [Google Scholar] [CrossRef]
- Yang, X.; Wang, S.; Liu, Y.; Zhang, Y. Identification and characterization of Acidithiobacillus ferrooxidans YY2 and its application in the biodesulfurization of coal. Can. J. Microbiol. 2015, 61, 65–71. [Google Scholar] [CrossRef]
- Yang, X.P.; Wang, S.M.; Liu, Y.J.; Liang, Y. A Comparative Study of the Biodesulfurization Efficiency of Acidithiobacillus ferrooxidans LY01 Cells Domesticated with Ferrous Iron and Pyrite. Geomicrobiol. J. 2016, 33, 488–493. [Google Scholar] [CrossRef]
- Newman, D.K. Feasting on Minerals. Science 2010, 327, 793–794. [Google Scholar] [CrossRef]
- Liu, F.W.; Lei, Y.S.; Shi, J.; Zhou, L.X.; Wu, Z.H.; Dong, Y.; Bi, W.L. Effect of microbial nutrients supply on coal bio-desulfurization. J. Hazard. Mater. 2020, 384, 10. [Google Scholar] [CrossRef]
- Arslan, V. The application of combined lignite cleaning processes, bacterial leaching and flotation, for reducing higher ash and sulfur contents. Int. J. Coal Prep. Util. 2022, 42, 2114–2126. [Google Scholar] [CrossRef]
- Silva, R.A.; Park, J.; Ilyas, S.; Borja, D.; Zhao, H.B.; Urik, M.; Rastegar, S.O.; Kim, H. Biodegradation mechanism of arsenopyrite mine tailing with Acidithiobacillus ferrooxidans and influence of ferric supplements. Int. Biodeterior. Biodegrad. 2020, 153, 6. [Google Scholar] [CrossRef]
- Schippers, A. Sand, Bacterial leaching of metal sulfides proceeds by two indirect mechanisms via thiosulfate or via polysulfides and sulfur. Appl. Environ. Microbiol. 1999, 65, 319–321. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.H.; Huang, X.Y.; He, H.; Tang, J.L.; Tao, X.X.; Huang, H.Z.; Haider, R.; Ali, M.I.; Jamal, A.; Huang, Z.X. Bioleaching Coal Gangue with a Mixed Culture of Acidithiobacillus ferrooxidans and Acidithiobacillus thiooxidans. Minerals 2021, 11, 11. [Google Scholar] [CrossRef]


| Group | Energy Source | Bacterial Inoculum |
|---|---|---|
| A.f Oxidation Fe2+ | FeSO4·7H2O (8.95 g) | 10% |
| Control 1 | FeSO4·7H2O (8.95 g) | 0% |
| A.f Oxidation S0 | S0 (0.5 g) | 10% |
| Control 2 | S0 (0.5 g) | 0% |
| A.f Oxidation FeS2 | FeS2 (5 g) | 10% |
| Control 3 | FeS2 (5 g) | 0% |
| Coal Samples | Total S | Sp, Ad | So, Ad | Ss, Ad |
|---|---|---|---|---|
| Ι | 3.46% | 1.02% | 0.81% | 1.63% |
| II | 3.02% | 1.27% | 0.83% | 0.92% |
| III | 2.77% | 0.83% | 1.42% | 0.52% |
| Location | Sequence Length (Bp) | GC Content (%) | Accession Number | Accession Strain Name | Accession Plasmid Name | Identity (%) |
|---|---|---|---|---|---|---|
| Plasmid E | 7910 | 52.4 | - | - | - | - |
| Plasmid D | 23,017 | 60.69 | NC_015188.1 | Acidiphilium multivorum AIU301 | pACMV4 | 88.265 |
| Plasmid C | 29,178 | 62.67 | NC_015178.1 | Acidiphilium multivorum AIU301 | pACMV1 | 88.293 |
| Plasmid B | 34,460 | 60.54 | NC_009470.1 | Acidiphilium cryptum JF-5 | pACRY04 | 96.491 |
| Plasmid A | 79,659 | 61.64 | NC_009469.1 | Acidiphilium cryptum JF-5 | pACRY03 | 99.949 |
| Strain | Geographic Origin | Genome Size (Mb) | GC% | Level | CDS | Genes |
|---|---|---|---|---|---|---|
| A. ferrooxidans YQ-N3 | Shanxi, China | 3.22 | 58.7 | Complete | 3195 | 3252 |
| A. ferrooxidans ATCC23270 | Bituminous coal mine effluent | 2.98 | 58.8 | Complete | 2927 | 3087 |
| A. ferrooxidans BY0502 | Gansu, China | 2.98 | 56.8 | Contig | 3026 | 3186 |
| A. ferridurans JCM18981 | Okayama, Japan | 2.98 | 58.4 | Complete | 2802 | 3043 |
| A. thiooxidans ATCC 19377 | - | 3.42 | 53 | Contig | 3498 | 3584 |
| A. ferrivorans SS3 | Norilsk, Russia | 3.20 | 56.5 | Complete | 3089 | 3200 |
| Strain | Geographic Origin | Highest Desulfurization Rate for Coal | Desulfurization Reaction Time | References |
|---|---|---|---|---|
| A. ferrooxidans YY2 | Guizhou, China | 75% | 30 days | [56] |
| A. ferrooxidans LY01 | Guizhou, China | 67.8% | 13 days | [57] |
| A. ferrooxidans | Johannesburg, South Africa | 79% | 14 days | [58] |
| A. ferrooxidans LX5 | China | 31.6% | 32 days | [59] |
| A. ferrooxidans DSM 583 | - | 30.84% | 12 h | [60] |
| A. ferrooxidans YQ-N3 | Shanxi, China | 62.25% | 30 days | This study |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, W.; Feng, Q.; Li, Z. Isolation and Characterization of A Novel Iron–Sulfur Oxidizing Bacterium Acidithiobacillus Ferrooxidans YQ-N3 and its Applicability in Coal Biodesulfurization. Minerals 2023, 13, 95. https://doi.org/10.3390/min13010095
Li W, Feng Q, Li Z. Isolation and Characterization of A Novel Iron–Sulfur Oxidizing Bacterium Acidithiobacillus Ferrooxidans YQ-N3 and its Applicability in Coal Biodesulfurization. Minerals. 2023; 13(1):95. https://doi.org/10.3390/min13010095
Chicago/Turabian StyleLi, Wenbo, Qiyan Feng, and Ze Li. 2023. "Isolation and Characterization of A Novel Iron–Sulfur Oxidizing Bacterium Acidithiobacillus Ferrooxidans YQ-N3 and its Applicability in Coal Biodesulfurization" Minerals 13, no. 1: 95. https://doi.org/10.3390/min13010095
APA StyleLi, W., Feng, Q., & Li, Z. (2023). Isolation and Characterization of A Novel Iron–Sulfur Oxidizing Bacterium Acidithiobacillus Ferrooxidans YQ-N3 and its Applicability in Coal Biodesulfurization. Minerals, 13(1), 95. https://doi.org/10.3390/min13010095





