Abstract
Autotrophic acidophilic bacteria Acidithiobacillus ferrooxidans is a model species for studying metal bioleaching from low-grade sulfide ores and concentrates. Arsenopyrite gold-bearing concentrates are refractory and often processed using biohydrometallurgical approaches; therefore, it is important to develop methods to improve arsenopyrite bioleaching. In the present work, we have studied the possibility of improving arsenopyrite concentrate bioleaching by the strain of A. ferrooxidans. For this purpose, we have analyzed the genome of the strain A. ferrooxidans TFBk to reveal the genes potentially important in the bioleaching process. Genes determining resistance to arsenic, as well genes involved in the utilization of C1-compounds and resistance to oxidative stress, were revealed. Therefore, the possibility of increasing the rate of arsenopyrite concentrate bioleaching using C1-compounds (methanol and formate) was studied. Formate was able to increase both the biomass yield of the strain A. ferrooxidans TFBk as well as the bioleaching rate. In addition, the effect of redox potential increase by means of the addition of sodium persulfate in the medium on arsenopyrite concentrate bioleaching was studied. It was shown that the addition of 0.1% sodium persulfate stimulated strain growth, while a higher concentration inhibited it. Despite this, the rate of concentrate bioleaching increased in the presence of 0.5–1.0% of persulfate, which may be explained by the interactions of added oxidizer with concentrate components.
1. Introduction
Metal extraction from low-grade ores, refractory sulfide concentrates, and different mining wastes using pyrometallurgical approaches is often not profitable, while hydrometallurgical methods in this case are more suitable both from economic and technological points of view. In the last decades, bioleaching methods have been widely used. Bioleaching processes are based on microbially mediated destruction/oxidation of mineral components (mainly sulfide minerals) performed by acidophilic bacteria and archaea. Successful application of acidophilic microorganisms for mineral raw materials processing is possible since (1) their growth optima is in the low pH level range that also corresponds to the optima of active metal leaching; (2) most of the acidophilic microorganisms used for bioleaching are chemolitoautotrophs, i.e., they do not require organic nutrients for growth and, on the contrary, use components of sulfide minerals to gain energy for metabolism that in turn simplify its participation in technological processes [1,2].
Since the 1950s, iron- and sulfur-oxidizing microorganisms have been used on an industrial scale for bioleaching processes [1]. Bacteria Acidithiobacillus ferrooxidans (formerly Thiobacillus ferrooxidans) are first described as acidophilic bacteria oxidizing both ferrous iron and reduced sulfur compounds. These bacteria are model organisms, which are successfully used for bioleaching studies up to now due to their properties (pH optima < 2, capability of ferrous iron and sulfur oxidation, resistance to various metal and metalloid ions) [2].
In general, optimization of metal extraction is a complex task, which may be solved by different methods depending on the features of mineral raw materials and extracted metals. One of the most complex problems is the processing of arsenic-containing ores and concentrates. To date, a lot of different arsenic minerals have been described; arsenopyrite (FeAsS), enargite (Cu3AsS4), tennantite (Cu12As4S13), realgar (As4S4), and aurpigment (As2S3) are the most widespread ones [3]. Arsenopyrite is the most widespread arsenic mineral, which contains up to 45% arsenic [4,5,6] and gold in the form of fine particles in a crystal lattice [7]. Acid mine drainage (AMD) formed due to arsenopyrite biooxidation is a serious environmental issue as it is highly toxic due to the arsenic presence [6,8]. In this regard, the study of the resistance of leaching strains to arsenic and their application is one of the important areas of hydrometallurgy and microbial ecology.
In general, the main approaches for the intensification of metal bioleaching involve: (1) the selection of strains and study of their metabolic and genetic potential [9,10]; (2) the study of the use of stimulating additives for the activation of microorganisms and microbially mediated processes [11,12,13,14,15,16,17,18]; (3) the optimization of physicochemical conditions of the process [19].
In modern hydrometallurgy, the search for new industrial strains as well as attempts to intensify bioleaching by stimulating bacteria with introduced chemical compounds has continued for decades [15,16,17]. A. ferrooxidans is one of the major microorganisms in consortia used for the industrial leaching of various valuable metals, which is an obligate autotroph. Three decades ago, J. Pronk et al. [20] already showed that the growth of A. ferrooxidans (formerly Thiobacillus ferrooxidans) can be stimulated with formic acid. According to the literature data [20], formic acid as an organic compound was not used as a substrate for growth but provided some energy requirements. Moreover, the mentioned authors also proposed pre-cultivation of A. ferrooxidans with formate for subsequent leaching of metals from ores and applied for a patent regarding this approach [21]. Developing these works, we obtained similar results for neutrophilic leaching bacteria [15]. Moreover, it was shown that genomes of some representatives of the genus Acidithiobacillus contain the genes involved in the utilization of C1-compounds [10]. Thus, C1-compounds, including formate and methanol, may be considered stimulating additives that increase the activity of the bacteria of the genus Acidithiobacillus, involved in the bioleaching process.
Another approach, which may be used to improve the bioleaching of different ores and concentrates, is the application of additional oxidizing agents, which are able to increase redox potential and enhance the bioleaching rate [18,22]. Persulfate as a peroxide compound is a more powerful oxidizing agent than dissolved atmospheric oxygen. It was shown that persulfate may be used as a disinfectant, inhibiting bacterial growth and activity [23,24,25,26,27] and for the destruction of a wide range of pollutants (including processes of bioremediation in which persulfate is applied in combination with microorganisms) [28,29,30,31]. It has also been shown that persulfate interacts with different sulfide minerals as a strong oxidant [25,32,33,34]. Therefore, persulfate may be used for oxidative leaching sulfide ores and concentrates in different hydrometallurgical processes [32,33,34,35,36,37,38], including bioleaching of metals from metallurgical industry sludge [22]. Thus, the application of persulfate may be considered as an approach to increase the rate of bioleaching of sulfide ore and concentrates. At the same time, it can act as an agent with an antimicrobial effect. Therefore, its effect on the growth of different microorganisms possessing various systems involved in oxidative stress resistance should be studied.
The goal of the present work was to study acidophilic autotrophic strain Acidithiobacillus ferrooxidans TFBk and the possibility to increase its bioleaching activity. For this purpose, the genes determining arsenic and oxidative stress resistance, as well as utilization of C1-compounds, were analyzed, while the effect of C1-organic compounds of formate and methanol, as well as the effect of the additional oxidizing agent persulfate on the growth of A. ferrooxidans TFBk and bioleaching of metals from arsenopyrite concentrate, were studied.
2. Materials and Methods
2.1. Concentrate
The floatation arsenopyrite concentrate as well as information on the content of various metals and arsenic (Table 1) were provided by Bioprom Technologies Ltd. (Stepnogorsk, Republic of Kazakhstan). The amounts of metals, which are important from an industrial point of view, as well as arsenic, which is important as the main toxicant that interferes with bioleaching, were determined. The weight ratio of iron and arsenic in the concentrate showed a higher content of arsenic than its calculated presence according to the arsenopyrite formula FeAsS. Thus, the concentrate could also contain other arsenic minerals, for example: lollingite (FeAs2), orpiment (As2S3), realgar (AsS), etc.

Table 1.
Element content in arsenopyrite concentrate.
2.2. Microorganisms
We used the acidophilic chemolithoautotrophic strain A. ferrooxidans TFBk, isolated from the gold-bearing pyrite-arsenopyrite concentrate obtained from the ore of the Bakyrchik deposit (Republic of Kazakhstan), and deposited in the Laboratory of Chemolithotrophic Microorganisms of Winogradsky, Institute of Microbiology, Research Centre of Biotechnology RAS (Moscow, Russia) [39,40]. The strain was cultivated in the Silverman-Lundgren 9K medium [41] containing the following components (g/L): (NH4)2SO4-−3, KCl-0.1, K2HPO4−0.5, MgSO4·7H2O−0.5, Ca(NO3)2−0.01, FeSO4·7H2O−44.22, 5M H2SO4−4.5 mL/L. We used reagents produced by Sigma-Aldrich/Merck (Darmstadt, Germany) and Reakhim (Moscow, Russia).
Recently, the strain has also been deposited in the All-Russian Collection of Microorganisms (VKM) as A. ferrooxidans VKM B-3655 (IBPM RAS, Pushchino, Moscow region, Russia).
2.3. Molecular Genetic Analysis of Arsenic Resistance Genes
Genomic DNA was isolated from cells using the Fungal/Bacterial DNA Kit (ZymoResearch, Irvine, CA, USA) according to the manufacturer’s recommendation. Arsenic resistance genes were amplified by PCR using specific primers developed in the present work.
Based on the genomes of different strains of A. ferrooxidans available in the database NCBI, degenerate primers for this species were compiled:
- (1)
- arsC-F ATGAAAACCCYGRAMATCCTarsC-R GTGCCGATGCGCKCCAGTTC(PCR fragment length of arsC gene was about 500 bp)
- (2)
- arsB-F ATGCTGGCSGTMGYCATATTarsB-R TCAAGCCAGCGGCARCCACCA(PCR fragment length of arsB gene was about 1300 bp)
- (3)
- arsM-F ATGAGCMAACAGAAYGCCTGCTarsM-R AGCCACCAGGYTTGAKCGCCTC(PCR fragment length of arsM gene was about 860 bp).
2.4. Genome Analysis
Genomic DNA for the sequencing was extracted using the QIAamp DNA minikit (Qiagen, Hilden, Germany). The DNA libraries were constructed with the MGIEasy universal DNA library prep set, according to the protocol for the kit. Sequencing of genomic DNA was carried out using the DNBSEQ-G400 platform (MGI Tech, Shenzhen, China), with 150-bp paired-end reads. A total of 4,013,413 paired-end reads were obtained from strain TFBk. These raw sequence reads quality was checked with FastQC v.0.11.9 (Illumina, San Diego, CA, USA), and low-quality reads were trimmed with Trimmomatic v.0.39 [42], using the default settings for paired-end reads. These quality-filtered reads were then de novo assembled with SPAdes v.3.15.0 (CAB SPbU, St. Petersburg, Russia) using the default settings [43]. The resulting assembly was quality assessed with QUAST v5.0 (CAB SPbU, St. Petersburg, Russia) [44]. The genome coverages were as estimated by QualiMap 2 v.2.2.2 [45] and Bowtie 2 v.2.3.5.1 [46]. The estimated completeness was evaluated with CheckM v.1.1.3 [47]. The taxonomic position of the assembled genome was determined using GTDB-Tk v.1.5.0 [48], and the assembly’s average nucleotide identity (ANI) with closely related genomes was calculated with FastANI 2.0 [49]. Genome annotation was performed using the NCBI Prokaryotic Genome Annotation Pipeline (PGAP; v.4.7) (NCBI, Rockville, MD, USA) [50]. The presence of Clustered Regularly Interspaced Short Palindromic Repeats (CRISPRs) was evaluated with CRISPRCasFinder [51]. The reCOGnizer was used to identify the Clusters of Ortholog Groups (COGs) [52] and Pfam domains [53] in the predicted protein sequences. Transmembrane helices and signal peptides were predicted by the online bioinformatic tools TMHMM v.2.0 (Department of Health Technology, Copenhagen, Denmark) [54] and SignalP v.5.0 (Department of Health Technology, Copenhagen, Denmark) [55], respectively.
2.5. Growth of Bacteria and Bioleaching Experiments
The effect of sodium persulfate, as well as C1 compounds (formate and methanol), was studied using 9K medium supplemented with different concentrations of studied compounds. Experiments with these compounds included a comparison of growth in the 9K medium or leaching with and without (control) formate and methanol. Formate and methanol, as well as persulfate, were added at various concentrations. The inoculum was 10 mL of the starting culture, 106–107 cells/mL. Growth of the culture was determined by optical density (OD) measured using a Spekol 221 spectrophotometer (Carl Zeiss Industrielle Messtechnik GmbH, Jena, Germany) at a wavelength of 600 nm. Data show the average of three replicated experiments; variations were within 5%.
Bioleaching experiments were performed using 9K medium without ferrous sulfate supplemented with studied concentrate. A sample of a fine fraction (<1 mm) of a solid mineral raw material (solid phase, S) and a leaching solution (liquid phase, L) were placed in a 750 mL shake flask. Thus, the flask contained 200 mL of the 9K medium as the leach solution and 40 g of the concentrate, i.e., the solid and liquid phases were in ratio S:L = 1:5. All experiments were carried out in triplicate under aerobic conditions at 28 °C using the IBPM shaker (IBPM RAS, Pushchino, Russia) at 180 rpm. Thus, the experiments modeled stirred tank leaching at a constant temperature.
2.6. Chemical Analyzes
The content of leached iron in the solution was determined by spectrophotometry according to GOST 13195-73. The method is based on the formation of a complex compound during the interaction of iron ions with potassium ferricyanide in an acidic medium. The optical density (OD) was measured at a wavelength of 540 nm using Schimadzu UV-1800 spectrophotometer (Schimadzu, Kyoto, Japan). Iron (Fe3+) concentration in the solution was determined by the constructed calibration curve.
The content of sulfate, formed during the oxidation of sulfides of the arsenopyrite concentrate, was determined by ion chromatography. The analyzes were performed using a liquid ion chromatograph for anion detection (Metrohm, Herisau, Switzerland) controlled by 761 Compact IC PC equipped with injection valve for manual injection or an autosampler with a Metrohm 838 Advanced Sample Processor; 6.1010.300 Metrosep Anion (3 × 150 mm) chromatographic column, as well as with conductometric detector. The 844 UV/VIS Compact IC chromatograph was controlled using the IC Net software and PC. 15 mmol/L NaHCO3/2.0 mmol/L Na2CO3 in deionized water (Milli-Q) was used as an eluent (flow rate 1.0 mL/min).
The pH and redox potential (ORP) values were determined using an Ecotest-112 pH meter/potentiometer (Econix, Moscow, Russia).
2.7. Statistical Processing
Statistical data validation was performed using ANOVA method and MS 15.0.459.1506 Excel 2013 software (Microsoft, Redmond, WA, USA) software. The analyzes were carried out in triplicate. The results presented in the article showed a high level of reliability (p > 0.99).
3. Results
3.1. Molecular Genetic Analysis of the Strain
According to literature data and analyzes of known genomes of A. ferrooxidans strains, differences were found in terms of arsenic resistance and the ability to leach arsenopyrite, therefore, in recent years, research has shifted towards the selection of novel arsenic-resistant species and strains.
Microorganisms have several strategies for arsenic resistance and transformation: (1) the cytoplasmic (ars genes)/periplasmic (arr genes) reduction of As(V) and isolation of As(III); (2) the As(III) oxidation (aio genes) and As(V) release via the phosphate transport system; (3) the methylation of As(III) to the gaseous compound As(CH)3 (via ArsM), also called biomethylation. An analysis of complete bacterial genomes shows that a large number of phylogenetically diverse prokaryotes are able to transform As(V) and As(III) in various terrestrial and aquatic habitats, as well as in a wide range of environmental conditions [56,57,58]. Arsenotrophy, defined as the ability to grow by means of As(III) oxidation or As(V) reduction, requires membrane-associated proteins that transfer electrons from or to arsenic (AioBA and ArrAB, respectively). More common in many different bacteria is arsenic resistance based on the presence of Ars detoxification systems. In this process, As(V) is reduced intracellularly to As(III) by ArsC, a small 13–16 kDa protein. As(III) is then expelled from the cell by an efflux pump, ArsB or ACR3.
The “simplest” and most common system for protecting bacteria from arsenic is the Ars system. The resistance mechanisms encoded by the ars operon have been extensively studied. The operon configuration is different in different strains [59]. The simplest configuration (arsRBC) includes ArsR regulatory protein, which has an As(III)-specific binding site, an As(V) ArsC reductase, and an As(III) ArsB efflux pump. ArsC mediates As(V) reduction by glutaredoxin, glutathione, or thioredoxin. This detoxification system requires energy in the form of ATP [60]. ArsC is localized in the cytoplasm and can only reduce As(V) that enters cells; thus, it is unable to reduce As(V) outside the bacterial cells [61]. Two families of transmembrane efflux pumps, ArsB and ACR3, are known. The ACR3 type is more widely distributed in nature and occurs in bacteria, animals, and plants, while ArsB is present only in bacteria [62]. Another operon configuration (arsRDABC) contains the additional ArsA ATPase, which provides energy for ArsB, which is the chaperone for arsenic efflux through ArsAB. In the third operon configuration, the ars genes are located in two operons (arsRC and arsBH) transcribed in opposite directions. The function of ArsH is not completely clear: it is present in almost all Gram-negative bacteria carrying the operon and absent in Gram-positive bacteria.
By performing PCR with the designed primers, we obtained data that confirmed the presence of arsenic resistance genes in the strain A. ferrooxidans TFBk.
The results of the analysis for the presence of arsenic resistance genes are shown in Figure 1. The data obtained for arsenic resistance genes for A. ferrooxidans TFBk with degenerate primers were confirmed by the data obtained from the analysis of the complete genome of A. ferrooxidans TFBk obtained in the present study.
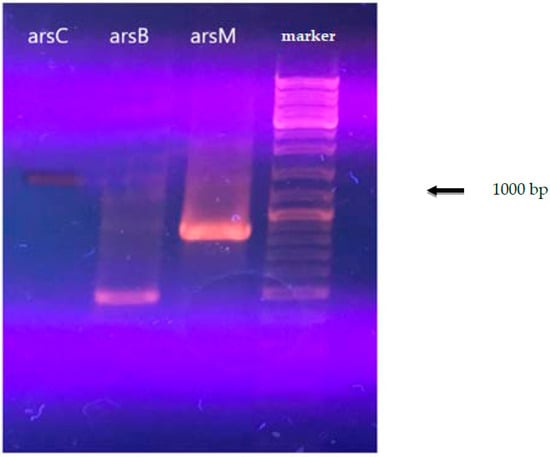
Figure 1.
Electrophoresis of the PCR product of A. ferrooxidans total DNA with primers for the arsC, arsM, and arsB genes.
The final assembled genome was 3,620,318 bp long and includes 159 scaffolds with an N50 value of 86,581 bp; the G+C content was 57.91%, and coverage was 280.61x. The completeness, estimated using CheckM v.1.1.3 software [47], was 99.34%, and the estimated contamination was 0.0%. The taxonomic position of the assembled genome determined using GTDB-Tk v.1.5.0 software [48] and average assembly nucleotide identity (ANI) with closely related genomes calculated using FastANI 2.0 software [49] confirmed that the TFBk strain belongs to the species A. ferrooxidans. Genome annotation performed using the NCBI Prokaryotic Genome Annotation Pipeline (PGAP; v.4.7) (NCBI, Rockville, MD, USA) [50] identified 3781 genes, 3558 protein coding sequences, 171 pseudogenes, and 52 RNA genes. The genome sequence parameters of the strain A. ferrooxidans TFBk are summarized in Table 2.

Table 2.
Results of the strain TFBk genome annotation.
Analysis of the A. ferrooxidans TFBk genome revealed the presence of the ars (arsenic resistance system) gene group (Figure 2). The arsC gene encodes arsenate reductase and is involved in the transformation of As(V) into As(III), which is then excreted by the ArsB arsenite pump. This mechanism provides arsenic resistance for bacteria, although it increases environmental toxicity. In prokaryotes, there are three families of ArsC arsenate reductases, distinguished on the basis of the protein structures, reduction mechanisms, and the location of catalytic cysteine residues: (i) glutathione-dependent (GSH)/glutaredoxin), (ii) thioredoxin-dependent (Trx)/thioredoxin-reductase), and (iii) mycothiol-dependent (MSH)/mycoredoxin, recently discovered in actinobacteria. A. ferrooxidans TFBk possesses dependent arsene reductases with 69% identity of the translated amino acid sequences. The arsC1 gene is surrounded by the genes ArsA, the arsenite/antimonite pump-driving ATPase, the arsenic metal chaperone ArsD, which carries trivalent metalloids to the ArsAB pump, the ArsR repressor, and the arsenite/antimonite: H+ antiporter ArsB. The second arsC2 gene is also in close proximity to the ars genes, namely the arsenic resistance protein arsH, the arsenite/antimonite: H+ antiporter arsB and repressor arsR.

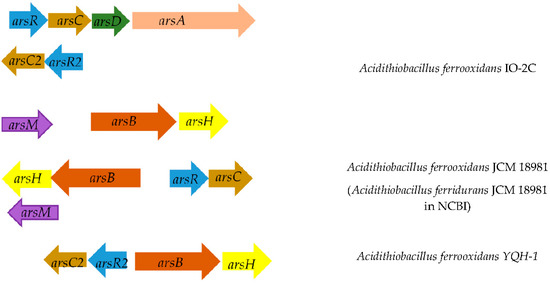
Figure 2.
Location of arsenic resistance genes on the chromosome in different strains of A. ferrooxidans. Below is the distance between genes. Genes encoding: ArsB, arsenite/antimonite: H+ antiporter; ArsR, transcriptional repressor of the arsenic resistance operon; ArsC, arsenate reductase; ArsD, transcriptional repressor and arsenic metal chaperone.
Thus, unlike other strains, A. ferrooxidans TFBk has all the genetic determinants for the operation of the arsenic resistance system in two variants. The genes responsible for other arsenic resistance strategies encoded by the arr and aio operons were not found in A. ferrooxidans TFBk.
In addition, A. ferrooxidans TFBk also has the arsM genes encoding putative arsenite methyltransferases, showing 95% identity. However, the arsI gene encoding the demethylase that breaks the As-C bond is absent. As shown by metagenomic studies, the arsM and arsI genes are often found in representatives of different species of the same community, forming an arsenate biogeochemical cycle in nature [63].
To better understand the possible mechanisms of formate and persulfate use, the presence of formate dehydrogenase genes and oxidative stress enzymes in the genome was studied. It can be assumed that formate dehydrogenase works according to the mechanism described for sulfurtransferase in E. coli [64]. Known strains of A. ferrooxidans have fdhF and fdhD genes in several variants (Table 3). They also have genes for the glutathione-dependent detoxification system, which detoxifies formaldehyde (the last one can be a by-product or produced during non-specific oxidation of methanol by alcohol dehydrogenase).

Table 3.
Distribution of formate dehydrogenase and oxidative stress enzymes in strains of A. ferrooxidans.
Bacteria can use superoxide dismutase and/or catalase to use or protect against peroxide compounds. As the study showed, all studied A. ferrooxidans strains have superoxide dismutase genes and no catalase genes. It can be assumed that superoxide dismutase plays the main role in the antioxidant defense of A. ferroxidans.
3.2. Bioleaching Stimulation with Organic C1 Compounds
Possible stimulation of acidophilic strain A. ferrooxidans TFBk was studied according to the scheme: growth was checked in a 9K medium with formate or methanol in comparison with the control. 5 mL of a 4-day culture of A. ferrooxidans TFBk was added as an inoculum.
The results on the effect of C1 compounds in the 9K medium without concentrate are shown in Figure 3 and Figure 4. The addition of methanol did not affect the growth of the bacteria (Figure 3). Formate concentration of 0.3% accelerated the growth of A. ferrooxidans TFBk only in the first three days compared to control, but did not provide an increase in final growth yield (Figure 4).
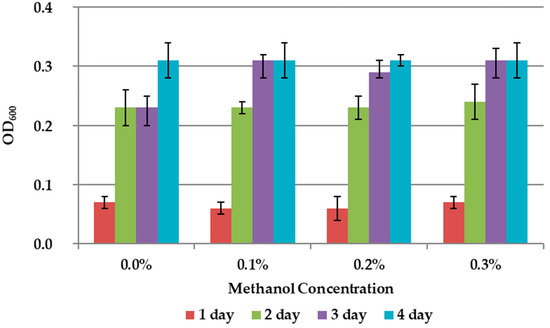
Figure 3.
Growth dynamic of A. ferrooxidans TFBk in 9K medium with different concentrations of methanol. The y-axis shows the optical density (OD600) of the medium with bacteria.
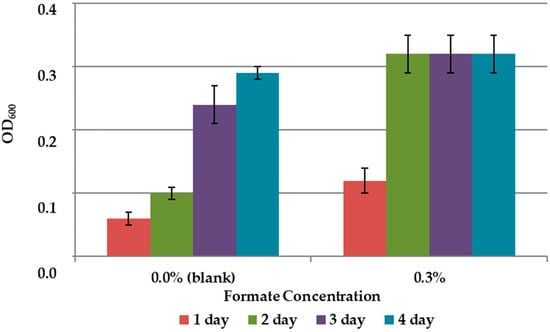
Figure 4.
Growth dynamics of A. ferrooxidans TFBk in 9K medium with various formate concentrations. The y-axis shows the optical density of the medium with bacteria.
Addition of formate and methanol during the arsenopyrite concentrate leaching by the bacteria A. ferrooxidans TFBk showed that the addition of methanol not only did not stimulate the process, but even inhibited it. The addition of formate accelerated the appearance of iron and sulfate in solution (Table 4). However, after two weeks of incubation, the content of iron and sulfates in the variant with formate and the control were almost equal. The decrease in the concentration of iron in the solution during the leaching of the arsenopyrite concentrate after 2 weeks suggests secondary precipitation reactions.

Table 4.
The effect of formate and methanol additions on the leaching of iron and sulfates from a sample of arsenopyrite concentrate by A. ferrooxidans TFBk bacteria. Control is the leaching in the medium without addition of formate and methanol.
3.3. Bioleaching Stimulation with Additional Oxidizing Agent, Persulfate
In patents devoted to biooxidation of gold-bearing mineral raw materials, it was noted that the intensification of the process can be achieved by increasing the redox potential (ORP) of the leaching medium [65,66]. At the same time, it is well known that the introduction of hydrogen peroxide into the medium can be accompanied by a sterilizing effect and inhibition of bacterial growth. We tested the possibility of stimulating bioleaching of the concentrate by the strain A. ferrooxidans TFBk by means of introducing a peroxide oxidant into the medium 9K that differs from hydrogen peroxide and is a more convenient form for transportation and storage: sodium persulfate.
The results showed slight stimulation of biomass growth in 9K medium without mineral concentrate at 0.1% persulfate concentration and suppression of growth at higher concentrations (Figure 5).
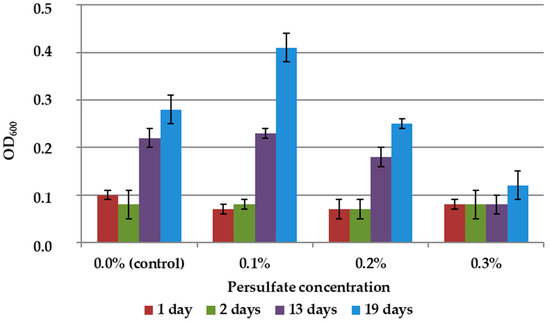
Figure 5.
Growth dynamics of A. ferrooxidans TFBk culture in 9K medium with various persulfate concentrations. The y-axis shows the optical density of the medium with bacteria.
At the same time, the effect of persulfate on bacteria in a liquid culture medium was not identical to its effect in the presence of leached concentrate, since the interaction of the oxidizing agent with the concentrate reduced its toxic effect and increased the availability of minerals for bacteria. We have previously shown that the addition of persulfate or persulfate in combination with organic acids can intensify the leaching of minerals [13]. The results of the experiments on the effect of persulfates on the bioleaching of arsenopyrite raw materials in a 9K medium when inoculating A. ferrooxidans TFBk are shown in Table 5.

Table 5.
Effect of sodium persulfate on the bioleaching of iron and sulfates from arsenopyrite flotation concentrate by A. ferrooxidans TFBk.
Thus, the rate of leaching of arsenopyrite raw materials was higher with the addition of persulfate than in the control, i.e., in the absence of an additional oxidizing agent (Table 4). At the same time, in full accordance with the inhibition data presented in Figure 5, microscopy revealed an altered cell morphology and a decrease in the number of unsorbed cells in suspension. The results obtained may mean that the chemical and biological leaching of mineral raw materials is intensified by persulfate due to the interaction of the oxidizing agent with minerals.
4. Discussion
There are different mechanisms of bacterial resistance to arsenic [67]. These strategies of defense are based primarily on the following genes/processes: (1) cytoplasmic (ars genes)/periplasmic (arr genes) As(V) reduction and As(III) excretion; (2) As(III) oxidation (aio genes) and As(V) release via the phosphate transport system; (3) methylation of As(III) to the gaseous compound As(CH)3 (via arsM), also called biomethylation. Our genetic analysis permitted us to determine which resistance mechanism is present in the strain A. ferrooxidans TFbk.
We carried out a genetic analysis of the A. ferrooxidans TFBk strain, originally isolated from the arsenopyrite deposit, and showed that it has a set of arsenic resistance genes. An analysis of the A. ferrooxidans TFBk genome revealed the presence of the ars gene group (arsenic resistance system). The arsC gene encodes arsene reductase and is involved in the transformation of As(V) into As(III), which is then excreted by the ArsB arsenite pump. A. ferrooxidans TFBk also possesses two thyredoxin-dependent arsene reductases. Thus, A. ferrooxidans TFBk has genetic determinants for resistance to arsenic in two variants. In addition, A. ferrooxidans TFBk also has the arsM genes encoding putative arsenite methyltransferases.
The applied genome-wide research methods provide more information than PCR amplification with degenerate primers, since they not only allow the detection of arsenic resistance genes, but also suggest possible bioleaching pathways. This approach is especially important when the leached mineral raw material is multicomponent (in our case, it contains not only arsenopyrite but also other mineral arsenic compounds). However, these primers can be useful when analyzing samples from different arsenic-contaminated environments to search for new strains with specific features. Thus, our work is one of the first attempts to use this approach for A. ferrooxidans for bioleaching intensification. At the same time, we expect that the development of this approach will make it possible to predict the phenotype and its applied use and, thus, will be included in the bioleaching methodology.
To better understand the possible mechanisms of formate and persulfate use, the presence of formate dehydrogenase genes and oxidative stress enzymes in the genome was studied. It can be assumed that formate dehydrogenase works according to the mechanism described for sulfurtransferase in E. coli [64]. All known strains of A. ferrooxidans have fdhF and fdhD genes in several variants (Table 5). Acidithiobacillus are acidophilic chemolithotrophs and assimilate carbon from carbon dioxide using the Calvin–Benson–Bassham cycle. Thus, an increase in the growth rate at the first stages with the addition of formate can be associated with formate dehydrogenase, oxidizing formate to CO2, which is assimilated by ribulose 1,5-bisphosphate carboxylase/oxygenase, i.e., formate can serve not only as a complementary source of energy, but also as a source of carbon. They also have genes for the glutathione-dependent detoxification system, which detoxifies formaldehyde (the last one can be a by-product or produced during non-specific oxidation of methanol by alcohol dehydrogenase). Bacteria can use superoxide dismutase and/or catalase to use or protect against peroxide compounds. As the study showed, all studied A. ferrooxidans strains have superoxide dismutase genes and no catalase genes. It can be assumed that superoxide dismutase plays the main role in the antioxidant defense of A. ferroxidans.
Despite the resistance of A. ferrooxidans TFBk to arsenic, the rate of the leaching of refractory arsenopyrite concentrates was comparatively low. In this regard, since the strain possessed by the genes involved in C1-compounds utilization and oxidative stress resistance, we investigated the possibility of intensifying bioleaching in two ways: (1) by means of stimulation with organic C1 compounds as energy substrates and (2) the introduction of an additional oxidizing agent, sodium persulfate.
The possibility of stimulating the growth of biomass and bioleaching of A. ferrooxidans TFBk metals by adding the organic C1 compound formate to the medium was confirmed. Previously, this ability was shown for another strain of this species [20]. It was shown that the organic C1 compound methanol did not stimulate A. ferrooxidans TFBk and could even inhibit growth.
The possibility of using additions of a peroxide-type oxidant, sodium persulfate, to intensify bioleaching by A. ferrooxidans TFBk bacteria was shown. The addition of persulfate to the 9K medium without arsenopyrite stimulated the growth of biomass only at low concentrations (0.1%). However, high concentrations (0.5–1.0%) markedly intensified chemical-biological leaching of arsenopyrite, possibly due to the oxidation/deliberation of the ore minerals by means of both direct oxidation of the minerals with persulfate and increase of Fe3+/Fe2+ ratio, which, in turn, lead to the increase in mineral oxidation rate.
5. Conclusions
The genome of the leaching strain A. ferrooxidans TFBk was studied. It has been shown that it has genes responsible for resistance to arsenic. It was also found that its set of genes was fundamentally different from known other representatives of the species A. ferrooxidans. In addition, an analysis of the strain was carried out to assess its potential resistance to adverse factors (oxidative stress in the presence of persulfate), as well as the ability to utilize C1 compounds.
In experiments with the strain A. ferrooxidans TFBk, the possibility of stimulating its growth and leaching activity with formate additives was shown. The possibility of stimulating autotrophic bacteria with organic C1-compounds is an interesting direction for general and applied microbiology.
In bioleaching experiments, regulation of the redox potential was carried out by the addition of sodium persulfate. It was shown that supplementation with this additional oxidizing agent at low concentrations leads to an intensification of bioleaching. The study showed that the strain A. ferrooxidans TFbk is a promising object for its use in the bioleaching of arsenic-containing ores and concentrates. In general, the study showed the possibility of intensifying the process of bioleaching, justified by the methods of genetic analysis.
Author Contributions
Conceptualization, T.A., M.V. and A.B.; validation, A.Y., T.A. and D.G.; investigation, A.Y. and O.R.; data curation, T.A.; writing—original draft preparation, review and editing, T.A., M.V. and A.B. All authors have read and agreed to the published version of the manuscript.
Funding
The study was partly supported by the Ministry of Science and Higher Education of the Russian Federation (Grant agreement no. 075-15-2021-1051).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Sequence data associated with the annotated genome have been deposited in the NCBI BioProject accession number PRJNA866167. This whole-genome shotgun project has been deposited at DDBJ/ENA/GenBank under the accession number JANJYU000000000. The version described in this paper is version JANJYU000000000.1.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Zimmerley, S.R.; Wilson, D.G.; Prater, J.D. Cyclic Leaching Process Employing Iron Oxidizing Bacteria. U.S. Patent 2,829,964, 8 April 1958. [Google Scholar]
- Natarajan, K.A. Bioleaching Mechanisms. In Biotechnology of Metals; Natarajan, K.A., Ed.; Elsevier: Amsterdam, The Netherlands, 2018; pp. 49–80. [Google Scholar]
- Diaz, A.J.; Serrano, J.; Leiva, E. Bioleaching of arsenic-bearing copper ores. Minerals 2018, 8, 215. [Google Scholar] [CrossRef]
- Mikhlin, Y.L.; Romanchenko, A.S.; Asanov, I.P. Oxidation of arsenopyrite and deposition of gold on the oxidized surfaces: A scanning probe microscopy, tunneling spectroscopy and XPS study. Geochim. Cosmochim. Acta. 2006, 70, 4874–4888. [Google Scholar] [CrossRef]
- Murciego, A.; Alvarez-Ayuso, E.; Pellitero, E.; Rodriguez, M.A.; Garcia-Sanchez, A.; Tamayo, A.; Rubio, J.; Rubio, F.; Rubin, J. Study of arsenopyrite weathering products in mine wastes from abandoned tungsten and tin exploitations. J. Hazard. Mater. 2011, 186, 590–601. [Google Scholar] [CrossRef] [PubMed]
- Paikaray, S. Arsenic geochemistry of acid mine drainage. Mine Water Environ. 2015, 34, 181–196. [Google Scholar] [CrossRef]
- Merkulova, M.; Mathon, O.; Glatzel, P.; Rovezzi, M.; Batanova, V.; Marion, P.; Boiron, M.-C.; Manceau, A. Revealing the chemical form of “invisible” gold in natural arsenian pyrite and arsenopyrite with high energy-resolution X-ray absorption spectroscopy. ACS Earth Space Chem. 2019, 3, 1905–1914. [Google Scholar] [CrossRef]
- Meng, X.; Zhao, H.; Sun, M.; Zhang, Y.; Zhang, Y.; Lv, X.; Kim, H.; Vainshtein, M.; Wang, S.; Qiu, G. The role of cupric ions in the oxidative dissolution process of marmatite: A dependence on Cu2+ concentration. Sci. Total Environ. 2019, 675, 213–223. [Google Scholar] [CrossRef]
- Li, S.-P.; Guo, N.; Wu, H.-Y.; Qiu, G.-Z.; Liu, X.-X. High efficient mixed culture screening and selected microbial community shift for bioleaching process. Trans. Nonferrous Met. Soc. China 2011, 21, 1383–1387. [Google Scholar] [CrossRef]
- Yin, Z.; Feng, S.; Tong, Y.; Yang, H. Adaptive mechanism of Acidithiobacillus thiooxidans CCTCC M 2012104 under stress during bioleaching of low-grade chalcopyrite based on physiological and comparative transcriptomic analysis. J. Ind. Microbiol. Biotechnol. 2019, 46, 1643–1656. [Google Scholar] [CrossRef]
- Yuehua, H.; Guanzhou, Q.; Jun, W.; Dianzuo, W. The effect of silver-bearing catalysts on bioleaching of chalcopyrite. Hydrometallurgy 2002, 64, 81–88. [Google Scholar] [CrossRef]
- Hernández, P.; Dorador, A.; Martínez, M.; Toro, N.; Castillo, J.; Ghorbani, Y. Use of seawater/brine and caliche’s salts as clean and environmentally friendly sources of chloride and nitrate ions for chalcopyrite concentrate leaching. Minerals 2020, 10, 477. [Google Scholar] [CrossRef]
- Ren, Z.; Krishnamoorthy, P.; Sanchez, P.Z.; Asselin, E.; Dixon, D.C.; Mora, N. Catalytic effect of ethylene thiourea on the leaching of chalcopyrite. Hydrometallurgy 2020, 196, 105410. [Google Scholar] [CrossRef]
- Abashina, T.N.; Yachkula, A.A.; Vainshtein, M.B. Prevention of sulfuric acid pollution: Intensification of metal leaching with organic acids. IOP Conf. Ser. Earth Environ. Sci. 2022, 981, 032029. [Google Scholar] [CrossRef]
- Abashina, T.; Yachkula, A.; Kaparullina, E.; Vainshtein, M. Intensification of nickel bioleaching with neutrophilic bacteria Guyparkeria halophila as an approach to limitation of sulfuric acid pollution. Microorganisms 2021, 9, 2461. [Google Scholar] [CrossRef] [PubMed]
- Aston, J.E.; Apel, W.A.; Lee, B.D.; Peyton, B.M. Growth effects and assimilation of organic acids in chemostat and batch cultures of Acidithiobacillus caldus. World J. Microbiol. Biotechnol. 2011, 27, 153–161. [Google Scholar] [CrossRef][Green Version]
- Bulaev, A.; Nechaeva, A.; Elkina, Y.; Melamud, V. Effect of carbon sources on pyrite-arsenopyrite concentrate bio-oxidation and growth of microbial population in stirred tank reactors. Microorganisms 2021, 9, 2350. [Google Scholar] [CrossRef]
- Zhang, X.; Feng, Y.-L.; Li, H.-R. Enhancement of bio-oxidation of refractory arsenopyritic gold ore by adding pyrolusite in bioleaching system. Trans. Nonferrous Met. Soc. China 2016, 26, 2479–2484. [Google Scholar] [CrossRef]
- Mahmoud, A.; Cezac, P.; Hoadley, A.F.A.; Contaminea, F.; D’Hugues, P. A review of sulfide minerals microbially assisted leaching in stirred tank reactors. Int. Biodeterior. Biodegrad. 2017, 119, 118–146. [Google Scholar] [CrossRef]
- Pronk, T.; Meijer, W.M.; Hazeu, W.; van Dijken, J.P.; Bos, P.; Kuenen, J.G. Growth of Thiobacillus ferrooxidans on formic acid. Appl. Environ. Microbiol. 1991, 57, 2057–2062. [Google Scholar] [CrossRef]
- Pronk, J.T.; van Dijken, J.P.; Bos, P.; Kuenen, J.G. High Yield Method of Growing Thiobacillus ferrooxidans on Formate. Patent ZA 923117B, 6 May 1991. [Google Scholar]
- Chen, C.; Li, H.; Cui, F.; Wang, Z.; Liu, X.; Jiang, G.; Cheng, T.; Bai, R.; Song, L. Novel combination of bioleaching and persulfate for the removal of heavy metals from metallurgical industry sludge. Environ. Sci. Pollut. Res. Int. 2022, 29, 33751–33763. [Google Scholar] [CrossRef]
- Tsitonaki, A.; Smets, B.F.; Bjerg, P.L. Effects of heat-activated persulfate oxidation on soil microorganisms. Water Res. 2008, 42, 1013–1022. [Google Scholar] [CrossRef]
- Afonyushkin, V.N.; Tabanyukhov, K.A.; Cherepushkina, V.S.; Khomenko, Y.S.; Tatarchuk, O.P. Effect of disinfectants based on potassium persulfate, hydrogen peroxide, glutaraldehyde and quaternary ammonium compounds on the genetic material of the pathogen bacteria specific to meat processing industry. Theory Pract. Meat Process. 2016, 1, 54–61. [Google Scholar] [CrossRef]
- Wu, B.; Gu, G.; Deng, S.; Liu, D.; Xiong, X. Efficient natural pyrrhotite activating persulfate for the degradation of O-isopropyl-N-ethyl thionocarbamate: Iron recycle mechanism and degradation pathway. Chemosphere 2019, 224, 120–127. [Google Scholar] [CrossRef]
- Qi, H.; Huang, Q.; Hung, Y.-C. Efficacy of activated persulfate in inactivating Escherichia coli O157:H7 and Listeria monocytogenes. Int. J. Food Microbiol. 2018, 284, 40–47. [Google Scholar] [CrossRef] [PubMed]
- Qi, H.; Huang, Q.; Hung, Y.-C. Efficacy of activated persulfate in pathogen inactivation: A further exploration. Food Res. Int. 2019, 120, 425–431. [Google Scholar] [CrossRef] [PubMed]
- Venâncio, J.P.F.; Rodrigues, C.S.D.; Nunes, O.C.; Madeira, L.M. Application of iron-activated persulfate for municipal wastewater disinfection. J. Hazard. Mater. 2022, 426, 127989. [Google Scholar] [PubMed]
- Lee, J.; von Gunten, U.; Kim, J.-H. Persulfate-based advanced oxidation: Critical assessment of opportunities and roadblocks. Environ. Sci. Technol. 2020, 54, 3064–3081. [Google Scholar] [CrossRef]
- Bajagain, R.; Lee, S.; Jeong, S.-W. Application of persulfate-oxidation foam spraying as a bioremediation pretreatment for diesel oil-contaminated soil. Chemosphere 2018, 207, 565–572. [Google Scholar] [CrossRef]
- Balaberda, A.; Ulrich, A.C. Persulfate oxidation coupled with biodegradation by Pseudomonas fluorescens enhances naphthenic acid remediation and toxicity reduction. Microorganisms 2021, 9, 1502. [Google Scholar] [CrossRef]
- Ma, J.; Tang, Y.; Yang, D.Q.; Pei, P. Kinetics of advanced oxidative leaching of pyrite in a potassium peroxydisulphate solution. J. S. Afr. Inst. Min. Metall. 2020, 120, 165–172. [Google Scholar] [CrossRef]
- Tang, Y.; Li, G.; Yang, Y.; Ma, J.; Zhi, Y.; Yao, Y.; Zheng, L.; Tuo, B. Oxidation of gold-bearing pyrite by ammonium persulfate. J. Sustain. Metall. 2021, 7, 1280–1292. [Google Scholar] [CrossRef]
- Dakubo, F.; Baygents, J.C.; Farrell, J. Peroxodisulfate assisted leaching of chalcopyrite. Hydrometallurgy 2012, 121, 68–73. [Google Scholar] [CrossRef]
- Turan, M.D.; Altundoğan, H.S. Leaching of a copper flotation concentrate with ammonium persulfate in an autoclave system. Int. J. Min. Met. Mater. 2014, 21, 862–870. [Google Scholar] [CrossRef]
- Liu, Z.-X.; Yin, Z.-L.; Hu, H.-P.; Chen, Q.-Y. Leaching kinetics of low-grade copper ore containing calcium-magnesium carbonate in ammonia-ammonium sulfate solution with persulfate. Trans. Nonferrous Met. Soc. China 2012, 22, 2822–2830. [Google Scholar] [CrossRef]
- Turan, M.D.; Arslanoğlu, H.; Altundoğan, H.S. Optimization of the leaching conditions of chalcopyrite concentrate using ammonium persulfate in an autoclave system. J. Taiwan Inst. Chem. Eng. 2015, 50, 49–55. [Google Scholar] [CrossRef]
- Liu, Z.; Xiang, Y.; Yin, Z.; Wu, X.; Jiang, J.; Chen, Y.; Xiong, L. Oxidative leaching behavior of metalliferous black shale in acidic solution using persulfate as oxidant. Trans. Nonferrous Met. Soc. China 2016, 26, 565–574. [Google Scholar] [CrossRef]
- Kondratyeva, T.F.; Muntyan, L.N.; Karavaiko, G.I. Zinc- and arsenic-resistant strains of Thiobacillus ferrooxidans have increased copy numbers of chromosomal resistance genes. Microbiology 1995, 141, 1157–1162. [Google Scholar] [CrossRef] [PubMed]
- Kanaev, Z.K.; Bulaev, A.G.; Kanaev, A.T.; Kodrateva, T.F. Physiological properties of Acidithiobacillus ferrooxidans strains isolated from sulfide ore deposits in Kazakhstan. Microbiology 2015, 84, 370–376. [Google Scholar] [CrossRef]
- Silverman, M.P.; Lundgren, D.C. Study on the chemoautotrophic iron bacterium Ferrobacillus ferrooxidans. An improved medium and harvesting procedure for securing high cell yield. J. Bacteriol. 1959, 77, 642–647. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef] [PubMed]
- Okonechnikov, K.; Conesa, A.; García-Alcalde, F. Qualimap 2: Advanced multi-sample quality control for high-throughput sequencing data. Bioinformatics 2016, 32, 292–294. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Meth. 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef] [PubMed]
- Chaumeil, P.A.; Mussig, A.J.; Hugenholtz, P.; Parks, D.H. GTDB-Tk: A toolkit to classify genomes with the genome taxonomy database. Bioinformatics 2019, 36, 1925–1927. [Google Scholar] [CrossRef]
- Jain, C.; Rodriguez, R.L.M.; Phillippy, A.M.; Konstantinidis, K.T.; Aluru, S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 2018, 9, 5114. [Google Scholar] [CrossRef]
- Tatusova, T.; DiCuccio, M.; Badretdin, A.; Chetvernin, V.; Nawrocki, E.P.; Zaslavsky, L.; Lomsadze, A.; Pruitt, K.D.; Borodovsky, M.; Ostell, J. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016, 44, 6614–6624. [Google Scholar] [CrossRef]
- Couvin, D.; Bernheim, A.; Toffano-Nioche, C.; Touchon, M.; Michalik, J.; Néron, B.; Rocha, E.P.C.; Vergnaud, G.; Gautheret, D.; Pourcel, C. CRISPRCasFinder, an update of CRISRFinder, includes a portable version, enhanced performance and integrates search for Cas proteins. Nucleic Acids Res. 2018, 46, W246–W251. [Google Scholar] [CrossRef]
- Galperin, M.Y.; Makarova, K.S.; Wolf, Y.I.; Koonin, E.V. Expanded microbial genome coverage and improved protein family annotation in the COG database. Nucleic Acids Res. 2014, 43, D261–D269. [Google Scholar] [CrossRef]
- Finn, R.D.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Mistry, J.; Mitchell, A.L.; Potter, S.C.; Punta, M.; Qureshi, M.; Sangrador-Vegas, A.; et al. The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 2016, 44, D279–D285. [Google Scholar] [CrossRef]
- Krogh, A.; Larsson, B.; von Heijne, G.; Sonnhammer, E.L. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef] [PubMed]
- Almagro Armenteros, J.J.; Tsirigos, K.D.; Sønderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar] [CrossRef] [PubMed]
- Rhine, E.D.; Phelps, C.D.; Young, L.Y. Anaerobic arsenite oxidation by novel denitrifying isolates. Environ. Microbiol. 2006, 8, 899–908. [Google Scholar] [CrossRef] [PubMed]
- Handley, K.M.; Héry, M.; Lloyd, J.R. Redox cycling of arsenic by the hydrothermal marine bacterium Marinobacter santoriniensis. Environ. Microbiol. 2009, 11, 1601–1611. [Google Scholar] [CrossRef] [PubMed]
- Osborne, T.H.; Jamieson, H.E.; Hudson-Edwards, K.A.; Nordstrom, D.K.; Walker, S.R.; Ward, S.A.; Santini, J.M. Microbial oxidation of arsenite in a subartic environment: Diversity of arsenite oxidase genes and identification of a psychrotolerant arsenite oxidizer. BMC Microbiol. 2010, 10, 205. [Google Scholar] [CrossRef] [PubMed]
- Páez-Espino, D.; Tamames, J.; de Lorenzo, V.; Cánovas, D. Microbial responses to environmental arsenic. Biometals 2009, 22, 117–130. [Google Scholar] [CrossRef] [PubMed]
- Ordóñez, E.; Letek, M.; Valbuena, N.; Gil, J.A.; Mateos, L.M. Analysis of genes involved in arsenic resistance in Corynebacterium glutamicum ATCC 13032. Appl. Environ. Microbiol. 2005, 71, 6206–6215. [Google Scholar] [CrossRef]
- Langner, H.W.; Inskeep, W.P. Microbial reduction of arsenate in the presence of ferrihydrite. Environ. Sci. Technol. 2000, 34, 3131–3136. [Google Scholar] [CrossRef]
- Achour, A.R.; Bauda, P.; Billard, P. Diversity of arsenite transporter genes from arsenic resistant soil bacteria. Res. Microbiol. 2007, 158, 128–137. [Google Scholar] [CrossRef]
- Ben Fekih, I.; Zhang, C.; Li, Y.P.; Zhao, Y.; Alwathnani, H.A.; Saquib, Q.; Rensing, C.; Cervantes, C. Distribution of arsenic resistance genes in prokaryotes. Front Microbiol. 2018, 23, 2473. [Google Scholar] [CrossRef]
- Thomé, R.; Gust, A.; Toci, R.; Mendel, R.; Bittner, F.; Magalon, A.; Walburger, A. A sulfurtransferase is essential for activity of formate dehydrogenases in Escherichia coli. J. Biol. Chem. 2012, 287, 4671–4678. [Google Scholar] [CrossRef] [PubMed]
- Sekisov, A.G.; Piskunov, S.A.; Filatov, B.L. Method for leaching of gold-bearing complex ores. Patent RU 2,044,875, 27 September 1995. [Google Scholar]
- Bely, A.V.; Malashonok, A.P.; Leskiv, M.V.; Potylitsyn, N.V. Method for Bioleaching of Refractory Gold-Bearing Sulfide Flotation Concentrates. Patent RU 2,637,204, 30 November 2017. [Google Scholar]
- Andres, J.; Bertin, P.N. The microbial genomics of arsenic. FEMS Microbiol. Rev. 2016, 40, 299–322. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).