Isolation and Identification of Arsenic Hyper-Tolerant Bacterium with Potential Plant Growth Promoting Properties from Soil
Abstract
1. Introduction
2. Materials and Methods
2.1. Selection of the Study Area
2.2. Physicochemical Analysis of the Soil Sample
2.3. Isolation and Characterization of Arsenic Hyper-Tolerant Bacteria
2.4. Scanning Electron Microscope Imaging
2.5. 16S rDNA Sequencing and Phylogenetic Analysis for Identification of the Bacterial Isolate
2.6. Determination of Heavy Metal Resistance
2.7. Determination of Minimum Inhibitory Concentration of Arsenic (III and V) and Chromium (III and VI)
2.8. Study of the Growth Curve
2.9. Bioremediation Test
2.10. Nitrogen Fixation and Phosphate/Potassium Solubilzation by the Bacterial Isolate for Plant Growth Promotion
2.11. Indole-3-Acetic Acid Production
2.12. Siderophore Production
3. Results and Discussion
3.1. Biochemical Characterization of the Garden Soil
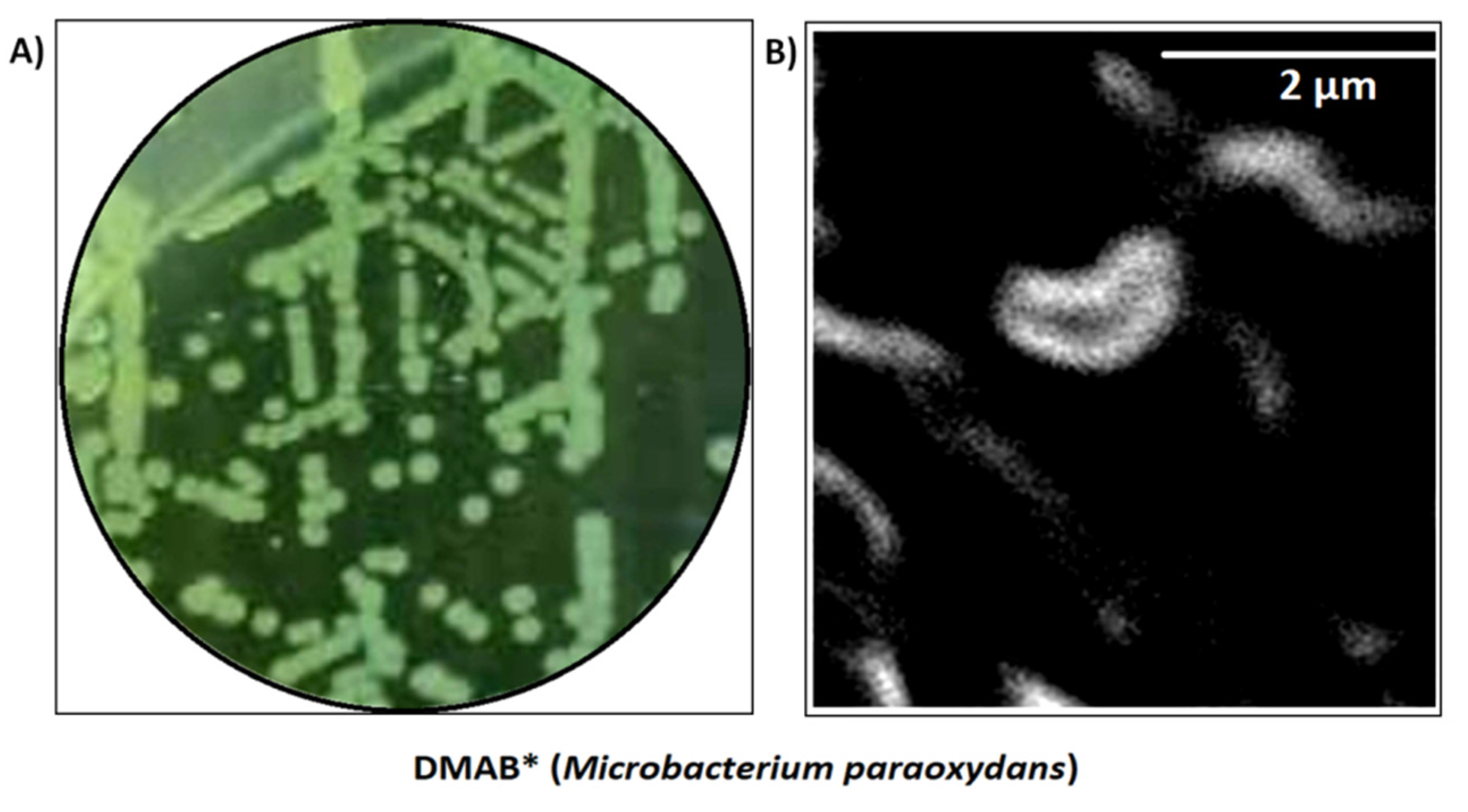
3.2. Isolation and Characterization of Arsenic Hyper-Tolerant Bacterium from the Garden Soil
3.3. 16S rDNA Sequencing and Phylogenetic Analysis Identified the Bacterium as Microbacterium Paraoxydans
3.4. Heavy Metal Tolerance and Susceptibility of the Isolated Bacterium
3.5. Growth Patterns of DMAB* in High Concentration of Arsenic (III) and Chromium (III)
3.6. Bioremediation of Arsenic Toxicity by DMAB*
3.7. Plant Growth-Promoting Properties Shown by DMAB*
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dangleben, N.L.; Skibola, C.F.; Smith, M.T. Arsenic immunotoxicity: A review. Environ. Health 2013, 12, 73. [Google Scholar] [CrossRef] [PubMed]
- Samal, A.C.; Kar, S.; Maity, J.P.; Santra, S.C. Arsenicosis and its relationship with nutritional status in two arsenic affected areas of West Bengal, India. J. Asian Earth Sci. 2013, 77, 303–310. [Google Scholar] [CrossRef]
- Guha Mazumder, D.N.; Haque, R.; Ghosh, N.; De, B.K.; Santra, A.; Chakraborty, D.; Smith, A.H. Arsenic levels in drinking water and the prevalence of skin lesions in West Bengal, India. Int. J. Epidemiol. 1998, 27, 871–877. [Google Scholar] [CrossRef]
- Milton, A.H.; Hasan, Z.; Shahidullah, S.M.; Sharmin, S.; Jakariya, M.D.; Rahman, M.; Dear, K.; Smith, W. Association between nutritional status and arsenicosis due to chronic arsenic exposure in Bangladesh. Int. J. Environ. Health Res. 2004, 4, 99–108. [Google Scholar] [CrossRef] [PubMed]
- Mondal, N.K.; Roy, P.; Das, B.; Data, J.K. Chronic arsenic toxicity and its relation with nutritional status: A case study in Purbasthali-II, Burdwan, west Bengal. Int. J. Environ. Sci. 2011, 2, 1103–1118. [Google Scholar] [CrossRef]
- Maharjan, M.; Watanabe, C.; Ahmad, S.A.; Umezaki, M.; Ohtsuka, R. Mutual interaction between nutritional status and chronic arsenic toxicity due to groundwater contamination in an area of Terai, lowland Nepal. J. Epidemiol. Community Health 2007, 61, 389–394. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.; Vahter, M.; Sohel, N.; Yunus, M.; Wahed, M.A.; Streatfield, P.K.; Ekstrom, E.C.; Persson, L.A. Arsenic exposure and age and sex-specific risk for skin lesions: A population-based case-referent study in Bangladesh. Environ. Health Perspect. 2006, 114, 1847–1852. [Google Scholar] [CrossRef]
- Paul, S.; Majumdar, S.; Giri, A.K. Genetic susceptibility to arsenic-induced skin lesions and health effects: A review. Genes Environ. 2015, 37, 23. [Google Scholar] [CrossRef] [PubMed]
- Shen, S.; Li, X.F.; Cullen, W.R.; Weinfeld, M.; Le, X.C. Arsenic Binding to Proteins. Chem. Rev. 2013, 113, 7769–7792. [Google Scholar] [CrossRef]
- Islam, K.; Qian, W.; Yu, H.J.; Chao, W.; Hua, N. Metabolism, toxicity and anticancer activities of Arsenic compounds. Oncotarget 2017, 8, 23905–23926. [Google Scholar] [CrossRef]
- Rahman, M.M.; Sengupta, M.K.; Ahamed, S.; Lodh, D.; Das, B.; Hossain, M.A.; Nayak, B.; Mukherjee, A.; Chakraborti, D.; Mukherjee, S.C.; et al. Murshidabad-One of the Nine Groundwater Arsenic-Affected Districts of West Bengal, India. Part I: Magnitude of Contamination and Population at Risk. Clin. Toxicol. 2005, 43, 823–834. [Google Scholar] [CrossRef] [PubMed]
- SOES. Groundwater Arsenic Contamination in West Bengal-India (20 Years Study); SOES: Kolkata, India, 2006. [Google Scholar]
- Shamim, S. Biosorption of Heavy Metals, Biosorption, Jan Derco and Branislav Vrana, IntechOpen. 2018. Available online: https://www.intechopen.com/chapters/58112 (accessed on 10 October 2022). [CrossRef]
- Katz, S.A.; Salem, H. The biological and environmental chemistry of chromium. In The Toxicology of Chromium and Its Compound; VCH Publishers Inc.: New York, NY, USA, 1994. [Google Scholar] [CrossRef]
- Brooks, R.R. Plants That Hyperaccumulate Heavy Metals: Their Role in Phytoremediation, Microbiology, Archaeology, Mineral Exploration and Phytomining; University Press: Cambridge, MA, USA; Publisher CAB International: Wallingford, UK, 1998; 80p, ISBN 9780851991566. Available online: https://www.cabi.org/bookshop/book/9780851992365/ (accessed on 10 October 2022).
- Xu, S.; Xu, R.; Nan, Z.; Chen, P. Bioadsorption of Arsenic from aqueous solution by the extremophilic bacterium Acidithiobacillus ferrooxidans DLC-5. Biocatal. Biotransform. 2018, 37, 35–43. [Google Scholar] [CrossRef]
- Mandal, D.; Biswas, S.; Seal, S.; Mandal, R.; Das, S.; Basu, A. Arsenic toxicity and its clinical manifestations in Murshidabad district with some potential remedial measures. In Microbes and Microbial Biotechnology for Green Remediation; Elsevier: Amsterdam, The Netherlands, 2022. [Google Scholar]
- Sharma, I. Bioremediation Techniques for Polluted Environment: Concept, Advantages, Limitations, and Prospects. In Trace Metals in the Environment—New Approaches and Recent Advances; Murillo-Tovar, M.A., Saldarriaga-Noreña, H., Saeid, A., Eds.; IntechOpen: London, UK, 2019. [Google Scholar] [CrossRef]
- Santana, M.M.; Gonzalez, J.M. High temperature microbial activity in upper soil layers. FEMS Microbiol. Lett. 2015, 362, fnv182. [Google Scholar] [CrossRef] [PubMed]
- Portillo, M.C.; Santana, M.; Gonzalez, J.M. Presence and potential role of thermophilic bacteria in temperate terrestrial environments. Naturwissenschaften 2012, 99, 43–53. [Google Scholar] [CrossRef] [PubMed]
- Manzoni, S.; Schimel, J.P.; Porporato, A. Responses of soil microbial communities to water stress:results from meta-analysis. Ecology 2012, 93, 930–938. [Google Scholar] [CrossRef]
- Biederman, J.A.; Scott, R.L.; Goulden, M.L.; Vargas, R.; Litvak, M.E.; Kolb, T.E.; Yepez, E.A.; Oechel, W.C.; Blanken, P.D.; Bell, T.W.; et al. Terrestrial carbon balance in a drier world: The effects of water availability in southwestern North America. Glob. Chang. Biol. 2016, 22, 1867–1879. [Google Scholar] [CrossRef] [PubMed]
- Grady, E.N.; MacDonald, J.; Liu, L.; Richman, A.; Yuan, Z.-C. Current knowledge and perspectives of Paenibacillus: A review. Microb. Cell Fact. 2016, 15, 203. [Google Scholar] [CrossRef]
- Das, H.K. Azotobacters as biofertilizer. Adv. Appl. Microbiol. 2019, 108, 1–43. [Google Scholar] [CrossRef]
- do Amaral, F.P.; Tuleski, T.R.; Pankievicz, V.C.S.; Melnyk, R.A.; Arkin, A.P.; Griffitts, J.; Tadra-Sfeir, M.Z.; de Souza, E.M.; Deutschbauer, A.; Monteiro, R.A.; et al. Diverse Bacterial Genes Modulate Plant Root Association by Beneficial Bacteria. mBio 2020, 11, 6. [Google Scholar] [CrossRef]
- Bhattacharya, P.; Chatterjee, D.; Nath, B.; Jana, J.; Jacks, G.; Vahter, M. High arsenic groundwater: Mobilization, metabolism and mitigation—An overview in the Bengal Delta Plain. Mol. Cell Biochem. 2003, 253, 347–355. [Google Scholar] [CrossRef]
- Chatterjee, D.; Halder, D.; Majumder, S.; Biswas, A.; Nath, B.; Bhattacharya, P.; Bhowmick, S.; Mukherjee-Goswami, A.; Saha, D.; Hazra, R.; et al. Assessment of arsenic exposure from groundwater and rice in Bengal Delta Region, West Bengal, India. Water Res. 2010, 44, 5803–5812. [Google Scholar] [CrossRef] [PubMed]
- Halder, D.; Bhowmick, S.; Biswas, A.; Chatterjee, D.; Nriagu, J.; Mazumder, D.N.G.; Šlejkovec, Z.; Jacks, G.; Bhattacharya, P. Risk of arsenic in exposure from drinking water and dietary components: Implications for risk management in rural Bengal. Environ. Sci. Technol. 2013, 47, 1120–1127. [Google Scholar] [CrossRef] [PubMed]
- Santra, S.C.; Samal, A.C.; Bhattacharya, P.; Banerjee, S.; Biswas, A.; Majumdar, J. Arsenic in Foodchain and Community Health Risk: A Study in Gangetic West Bengal. Procedia Environ. Sci. 2013, 18, 2–13. [Google Scholar] [CrossRef]
- Pelczar, M.J.; Bard, R.C.; Burnett, G.W.; Conn, H.J.; Demoss, R.D.; Euans, E.E.; Weiss, F.A.; Jennison, M.W.; Meckee, A.P.; Riker, A.J.; et al. Manual of Microbiological Methods. Society of American Bacteriology; McGraw Hill Book Company: New York, NY, USA, 1957; p. 315. [Google Scholar] [CrossRef]
- Brown, A.E.; Benson, H.J. Benson’s Microbiological Applications. Laboratory Manual in General Microbiology, Short Version, 10th ed.; The McGraw Hill Companies: New York, NY, USA, 2007; p. 50. [Google Scholar]
- Jukes, T.H.; Cantor, C.R. Evolution of Protein Molecules. In Mammalian Protein Metabolism; Munro, H.N., Ed.; Academic Press: New York, NY, USA, 1969; pp. 21–132. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- EFSA European Food Safety Authority. Guidance on the assessment of bacterial susceptibility to antimicrobial of human and veterinary importance. EFSA J. 2012, 10, 2740. [Google Scholar]
- Sher, S.; Sultan, S.; Rehman, A. Characterization of multiple metal resistant Bacillus licheniformis and its potential use in arsenic contaminated industrial wastewater. Appl. Water Sci. 2021, 11, 69. [Google Scholar] [CrossRef]
- Roy, P.; Mondal, N.K.; Das, B.; Das, K. Arsenic contamination in groundwater: A statistical modelling. J. Urban Environ. Eng. 2013, 7, 24–29. [Google Scholar] [CrossRef]
- Jensen, H.L. Nitrogen fixation in leguminous plants I. General characters of root-nodule bacteria isolated from species of Medicago and Trifolium in Australia. Proc. Linn. Soc. New South Wales 1942, 67, 98–108. [Google Scholar]
- Pikovskaya, R.I. Mobilization of phosphorus in soil in connection with the vital activity of some microbial species. Microbiology 1948, 17, 362–370. [Google Scholar]
- Ouertani, R.; Ouertani, A.; Mahjoubi, M.; Bousselmi, Y.; Najjari, A.; Cherif, H.; Chamkhi, A.; Mosbah, A.; Khdhira, H.; Sghaier, H.; et al. New Plant Growth-Promoting, Chromium-Detoxifying Microbacterium Species Isolated from a Tannery Wastewater: Performance and Genomic Insights. Front. Bioeng. Biotechnol. 2020, 8, 521. [Google Scholar] [CrossRef] [PubMed]
- Sun, F.; Ou, Q.; Wang, N.; Guo, Z.; Ou, Y.; Li, N.; Peng, C. Isolation and identification of potassium-solubilizing bacteria from Mikania micrantha rhizospheric soil and their effect on M. micrantha plants. Glob. Ecol. Conserv. 2020, 23, e01141. [Google Scholar] [CrossRef]
- Rahman, A.; Sitepu, I.R.; Tang, S.Y.; Hashidoko, Y. Salkowski’s reagent as a primary screening index for functionalities of rhizobacteria isolated from wild dipterocarpsapling growing naturally on medium-strongly acedictropical peatsoil. Biosci. Biotechnol. Biochem. 2010, 74, 2202–2208. [Google Scholar] [CrossRef]
- Payne, S.M. Detection, isolation and characterization of siderophore. Methods Enzymol. 1994, 235, 329–344. [Google Scholar] [CrossRef] [PubMed]
- Mandal, D.; Sonar, R.; Saha, I.; Ahmed, S.; Basu, A. Isolation and identification of arsenicresistant bacteria: A tool for bioremediation of arsenic toxicity. Int. J. Environ. Sci. Technol. 2021, 19, 9883–9900. [Google Scholar] [CrossRef]
- Prieto, D.M.; González, J.C.; Barral, M. Arsenic Mobility in As-Containing Soils from Geogenic Origin: Fractionation and Leachability. J. Chem. 2018, 2018, 7328203. [Google Scholar]
- Singh, A. Chemistry of Arsenic in Groundwater of Ganges–Brahmaputra River Basin. Curr. Sci. 2006, 91, 599–606. Available online: http://www.jstor.org/stable/24094363 (accessed on 10 October 2022).
- Chakraborty, M.; Mukherjee, A.; Ahmed, K.M. A Review of Groundwater Arsenic in the Bengal Basin, Bangladesh and India: From Source to Sink. Curr. Pollut. Rep. 2015, 1, 220–247. [Google Scholar] [CrossRef]
- Buss, S.N.; Starlin, R.; Iwen, P.C. Bacteremia caused by Microbacterium binotii in a patient with sickle cell anemia. J.. Clin. Microbiol. 2014, 52, 379–381. [Google Scholar] [CrossRef][Green Version]
- Qian, F.; An, L.; Wang, M.; Li, C.; Li, X. Isolation and characterization of a xanthan-degrading Microbacterium sp. strain XT11 from garden soil. J. Appl. Microbiol. 2007, 102, 1362–1371. [Google Scholar] [CrossRef]
- Jung, Y.; Lee, S.; Kim, N.; Yoon, H.; Jeong, G.; Lee, B.; Park, H.; Hur, M. Microbacterium caeni sp. nov., a novel species isolated from sludge in Yeosu, Korea. Res. Sq. 2022, PPR473869. [Google Scholar] [CrossRef]
- Hadjadj, L.; Rathored, J.; Keita, M.B.; Michelle, C.; Levasseur, A.; Raoult, D.; Fournier, P.; Rolain, J.; Bittar, F. Non contiguous-finished genome sequence and description of Microbacterium gorillae sp. nov. Stand Genom. Sci. 2016, 11, 32. [Google Scholar] [CrossRef]
- González Henao, S.; Ghneim-Herrera, T. Heavy Metals in Soils and the Remediation Potential of Bacteria Associated With the Plant Microbiome. Front. Environ. Sci. 2021, 9, 604216. [Google Scholar] [CrossRef]
- Kaushik, P.; Rawat, N.; Mathur, M.; Raghuvanshi, P.; Bhatnagar, P.; Swarnkar, H.; Flora, S. Arsenic Hyper-tolerance in Four Microbacterium Species Isolated from Soil Contaminated with Textile Effluent. Toxicol. Int. 2012, 19, 188–194. [Google Scholar] [CrossRef][Green Version]
- Mohd Bahari, Z.; Ibrahim, Z.; Jaafar, J.; Shahir, S. Draft Genome Sequence of Arsenic-Resistant Microbacterium sp. Strain SZ1 Isolated from Arsenic-Bearing Gold Ores. Genome Announc. 2017, 5, e01183-17. [Google Scholar] [CrossRef]
- Bermanec, V.; Paradžik, T.; Kazazić, S.P.; Venter, C.; Hrenović, J.; Vujaklija, D.; Duran, R.; Boev, I.; Boev, B. Novel arsenic hyper-resistant bacteria from an extreme environment, Crven Dol mine, Allchar, North Macedonia. J. Hazard Mater. 2021, 402, 123437. [Google Scholar] [CrossRef]
- Ben Fekih, I.; Zhang, C.; Li, Y.P.; Zhao, Y.; Alwathnani, H.A.; Saquib, Q.; Rensing, C.; Cervantes, C. Distribution of Arsenic Resistance Genes in Prokaryotes. Front. Microbiol. 2018, 9, 2473. [Google Scholar] [CrossRef]
- Koechler, S.; Cleiss-Arnold, J.; Proux, C.; Sismeiro, O.; Dillies, M.; Goulhen-Chollet, F.; Hommais, F.; Lièvremont, D.; Arsène-Ploetze, F.; Coppée, J.-Y.; et al. Multiple controls affect arsenite oxidase gene expression in Herminiimonas arsenicoxydans. BMC Microbiol. 2010, 10, 53. [Google Scholar] [CrossRef]
- Chang, J.S.; Yoon, I.H.; Lee, J.H.; Kim, K.-R.; An, J.; Kim, K.-W. Arsenic detoxification potential of aox genes in arsenite-oxidizing bacteria isolated from natural and constructed wetlands in the Republic of Korea. Environ. Geochem. Health 2010, 32, 95–105. [Google Scholar] [CrossRef]
- Achour-Rokbani, A.; Cordi, A.; Poupin, P.; Bauda, P.; Billard, P. Characterization of the ars gene cluster from extremely arsenic-resistant Microbacterium sp. strain A33. Appl. Environ. Microbiol. 2010, 76, 948–955. [Google Scholar] [CrossRef]
- Chowdhury, R.; Sen, A.K.; Karak, P.; Chatterjee, R.; Giri, A.K.; Chaudhuri, K. Isolation and characterization of an arsenic-resistant bacterium from a bore-well in West Bengal, India. Ann. Microbiol. 2009, 59, 253–258. [Google Scholar] [CrossRef]
- Dey, U.; Chatterjee, S.; Mondal, N.K. Isolation and characterization of arsenic-resistant bacteria and possible application in bioremediation. Biotechnol. Rep. 2016, 10, 1–7. [Google Scholar] [CrossRef]
- Sher, S.; Hussain, S.; Cheema, M.T.; Hussain, A.; Rehman, A. Efficient removal potential of Microbacterium sp. strain 1S1 against arsenite isolated from polluted environment. J. King Saud Univ.-Sci. 2022, 34, 102066. [Google Scholar] [CrossRef]
- Maizel, D.; Balverdi, P.; Rosen, B.; Sales, A.M.; Ferrero, M.A. Arsenic-hypertolerant and arsenic-reducing bacteria isolated from wells in Tucumán, Argentina. Can. J. Microbiol. 2018, 64, 876–886. [Google Scholar] [CrossRef]
- Waranusantigul, P.; Lee, H.; Kruatrachue, M.; Pokethitiyook, P.; Auesukaree, C. Isolation and characterization of lead-tolerant Ochrobactrum intermedium and its role in enhancing lead accumulation by Eucalyptus camaldulensis. Chemosphere 2011, 85, 584–590. [Google Scholar] [CrossRef] [PubMed]
- Singh, V.; Mishra, V. Microbial removal of Cr (VI) by a new bacterial strain isolated from the site contaminated with coal mine effluents. J. Environ. Chem. Eng. 2021, 9, 106279. [Google Scholar] [CrossRef]
- Stolz, J.F.; Oremland, R.S. Bacterial respiration of arsenic and selenium. FEMS Microbiol. Rev. 1999, 23, 615–627. [Google Scholar] [CrossRef]
- Oremland, R.S.; Stolz, J.F. The ecology of arsenic. Science 2003, 300, 939–944. [Google Scholar] [CrossRef]
- Stolz, J.F.; Basu, P.; Santini, J.M.; Oremland, R.S. Arsenic and selenium in microbial metabolism. Annu. Rev. Microbiol. 2006, 60, 107–130. [Google Scholar] [CrossRef]
- Mokashi, S.A.; Paknikar, K.M. Arsenic (III) oxidizing Microbacterium lacticum and its use in the treatment of arsenic contaminated groundwater. Lett. Appl. Microbiol. 2002, 34, 258–262. [Google Scholar] [CrossRef]
- Zhu, Y.G.; Rosen, B.P. Perspectives for genetic engineering for the photoremediation of arsenic contaminated environments: From imagination to reality? Curr. Opin. Biotechnol. 2009, 20, 220–224. [Google Scholar] [CrossRef] [PubMed]
- Chandraprabha, M.N.; Natarajan, K.A. Mechanism of arsenic tolerance and bioremoval of arsenic by Acidithiobacilus ferrooxidans. J. Biochem. Tech. 2011, 3, 257–265. Available online: https://jbiochemtech.com/en/article/mechanism-of-arsenic-tolerance-and-bioremoval-of-arsenic-by-acidithiobacilus-ferrooxidans (accessed on 10 October 2022).
- Tekdal, D.; Akça, I.; Küçükrecep, A.; Çetiner, S.; Hatipoğlu, R. Characterization of root-associated bacterial species of Leklek variety of common bean (Phaseolus sp.). Rom. Biotechnol. Lett. 2021, 26, 3008–3013. [Google Scholar] [CrossRef]
- Hernández-Pacheco, C.E.; Orozco-Mosqueda, M.D.C.; Flores, A.; Valencia-Cantero, E.; Santoyo, G. Tissue-specific diversity of bacterial endophytes in Mexican husk tomato plants (Physalis ixocarpa Brot. ex Horm.), and screening for their multiple plant growth-promoting activities. Curr. Res. Microb. Sci. 2021, 2, 100028. [Google Scholar] [CrossRef] [PubMed]
- Hirsch, A.M.; Monica, S.; Kaplan, D.; Sheva, B. Plant growth-promoting microorganisms and methods of use thereof. U.S. Patent 10212943B2, 26 February 2019. [Google Scholar]
- da Cunha, V.C.; Hanssen, I.; De Ceuster, T.J.J.; Carrion-Bravo, V.J.; Raaijmakers, J.M. Novel plant-growth promoting bacteria and the use thereof. Patent No. EP3311669A1, 25 April 2018. [Google Scholar]
- Khanna, A.; Raj, K.; Kumar, P.; Wati, L. Antagonistic and growth-promoting potential of multifarious bacterial endophytes against Fusarium wilt of chickpea. Egypt. J. Biol. Pest Control. 2022, 32, 1–9. [Google Scholar] [CrossRef]
- Song, Y.; Han, J.; Hu, Y. Diversity analysis of culturable bacteria in the root of tree peony (Paeonia ostii). In Plant Growth-Promoting Rhizobacteria (PGPR) for Sustainable Agriculture, Proceedings of the 2nd Asian PGPR Conference, Beijing, China, 21–24 August 2011. [Google Scholar]
- Anandaraj, M.; Bini, Y.K. Potential of PGPR application for seed spices with special reference to coriander and fenugreek in India. In Plant Growth-Promoting Rhizobacteria (PGPR) for Sustainable Agriculture, Proceedings of the 2nd Asian PGPR Conference, Beijing, China, 21–24 August 2011. [Google Scholar]
- Soto, J.; Ortiz, J.; Herrera, H.; Fuentes, A.; Almonacid, L.; Charles, T.C.; Arriagada, C. Enhanced Arsenic Tolerance in Triticum aestivum Inoculated with Arsenic-Resistant and Plant Growth Promoter Microorganisms from a Heavy Metal-Polluted Soil. Microorganisms 2019, 7, 348. [Google Scholar] [CrossRef] [PubMed]
- Alka, S.; Shahir, S.; Ibrahim, N.; Rahmad, N.; Haliba, N.; Abd Manan, F. Histological and proteome analyses of Microbacterium foliorum-mediated decrease in arsenic toxicity in Melastoma malabathricum. 3 Biotech 2021, 11, 336. [Google Scholar] [CrossRef]
- Alka, S.; Shahir, S.; Ibrahim, N.; Vo, D.N.; Abd Manan, F. Assessment of plant growth promotion properties and impact of Microbacterium foliorum for arsenic removal in Melastoma malabathricum. Bioremediation J. 2022, 26, 1–12. [Google Scholar] [CrossRef]
- Sun, Y.; Wang, X.; Liu, Y.; Duan, K.; Xia, Y.; Cai, Q.; Lou, L. Long term application of plant growth-promoting bacterium improved grain weight and reduced arsenic accumulation in rice grain: A comparison of 10 bacteria. Chemosphere 2022, 303 Pt 1, 135016. [Google Scholar] [CrossRef]
- Thongnok, S.; Siripornadulsil, W.; Siripornadulsil, S. AsIII-oxidizing and Cd-tolerant plant growth-promoting bacteria synergistically reduce arsenic translocation, toxicity and accumulation in KDML105 rice. Environ. Exp. Bot. 2021, 192, 104660. [Google Scholar] [CrossRef]




| Physicochemical Parameters | Concentration (mg/L) |
|---|---|
| pH | 7.5 |
| Iron | 0.03 |
| Phosphate | 0.02 |
| Arsenic | ~0.1 |
| Fluoride | 0 |
| Colony Morphological Profile | |
| Form | Round |
| Color | Yellow-pigmented |
| Texture | Mucoid |
| Size | Moderate (about 1 mm) |
| Microscopic Morphological Profile | |
| Gram’s nature | Gram-positive |
| Acid-fast | Negative |
| Shape | Rod |
| Size | 1.45 µM |
| Antimicrobial Agent | Concentration Used (µg/mL) | Susceptibility or Resistance |
|---|---|---|
| Ampicillin | 100 | Susceptible |
| Bleomycin | 40 | Resistant |
| Chloramphenicol | 25 | Susceptible |
| Ciprofloxacin | 5 | Susceptible |
| Gentamycin | 16 | Susceptible |
| Kanamycin | 50 | Susceptible |
| Ofloxacin | 2 | Susceptible |
| Rifampicin | 100 | Susceptible |
| Streptomycin | 50 | Susceptible |
| Tetracycline | 10 | Susceptible |
| Vancomycin | 30 | Susceptible |
| Carbohydrates | Fermentation Ability |
|---|---|
| Glucose | + |
| Fructose | + |
| Mannose | − |
| Sucrose | + |
| Lactose | − |
| Mannitol | + |
| Inositol | − |
| Heavy Metal | Maximum Tolerance Limit |
|---|---|
| Iron (Fe2+) | 10 mM |
| Cobalt (Co2+) | 3 mM |
| Zinc (Zn2+) | 5 mM |
| Copper (Cu2+) | 5 mM |
| Lead (Pb2+) | 2 mM |
| Manganese (Mn2+) | 5 mM |
| Nickel (Ni2+) | 1 mM |
| Mercury (Hg2+) | 0.5 mM |
| Cadmium (Cd2+) | 1 mM |
| Heavy Metal | Minimum Inhibitory Concentration (MIC) |
|---|---|
| Arsenite (As3+) | 36.95 mM |
| Arsenate (As5+) | 280.44 mM |
| Chromium (Cr3+) | 63 mM |
| Chromium (Cr6+) | 1 mM |
| Plant growth Promoting Properties | Observation |
|---|---|
| # IAA production | + (0.77 µg/mL) |
| * Siderophore production | + (21.16%) |
| Atmospheric nitrogen fixation | + |
| Insoluble phosphate solubilization | + |
| Insoluble potassium solublization | − |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mandal, D.; Aghababaei, M.; Das, S.K.; Majumder, S.; Chatterjee, D.; Basu, A. Isolation and Identification of Arsenic Hyper-Tolerant Bacterium with Potential Plant Growth Promoting Properties from Soil. Minerals 2022, 12, 1452. https://doi.org/10.3390/min12111452
Mandal D, Aghababaei M, Das SK, Majumder S, Chatterjee D, Basu A. Isolation and Identification of Arsenic Hyper-Tolerant Bacterium with Potential Plant Growth Promoting Properties from Soil. Minerals. 2022; 12(11):1452. https://doi.org/10.3390/min12111452
Chicago/Turabian StyleMandal, Debjani, Mina Aghababaei, Sadhan Kr Das, Santanu Majumder, Debashis Chatterjee, and Abhishek Basu. 2022. "Isolation and Identification of Arsenic Hyper-Tolerant Bacterium with Potential Plant Growth Promoting Properties from Soil" Minerals 12, no. 11: 1452. https://doi.org/10.3390/min12111452
APA StyleMandal, D., Aghababaei, M., Das, S. K., Majumder, S., Chatterjee, D., & Basu, A. (2022). Isolation and Identification of Arsenic Hyper-Tolerant Bacterium with Potential Plant Growth Promoting Properties from Soil. Minerals, 12(11), 1452. https://doi.org/10.3390/min12111452







