1. Introduction
Networks are used to describe various types of complex systems, such as social networks, biological networks, and the Internet. In recent years, community detection has emerged as a crucial tool for revealing hidden functional and structural properties in complex networks. It involves the partitioning of networks into distinct communities, wherein nodes within the same community are more densely and closely connected, while connections between nodes in different communities are significantly sparser.
Community detection methods have found wide application across multiple scenarios. For example, in the e-Commerce domain, community-based recommendation systems can effectively identify users’ preferences for products and reviews [
1]. In the realm of social networks, relationships among individuals sharing common interests can be discovered through community detection methods [
2]. Consequently, many researchers have developed diverse community detection approaches. These include algorithms based on modularity optimization [
3], information theory-based algorithms [
4], dynamics-based algorithms [
5], and spectral clustering-based algorithms [
6]. Fortunato et al. [
7] provide a comprehensive review of these methods for community detection.
In recent years, scientists have proposed numerous metrics and methods for evaluating community detection. In 2004, Newman [
8] introduced the concept of modularity to model the community detection problem as an NP-hard optimization problem. In 2009, Andrea Lancichinetti et al. [
9] proposed community fitness to assess community detection quality, while Pizzuti [
10] introduced the community score for similar assessment purposes. Community detection methods can be categorized into three main groups according to their approach. The first category comprises traditional methods, including graph partitioning, hierarchical clustering, partitional clustering, and spectral clustering [
11]. The second category encompasses split-based approaches, which remove edges connecting nodes from different communities. The third category consists of optimization algorithms. Pizzuti et al. [
12] proposed the first multi-objective community detection algorithm, called MOGA-Net. Pizzuti et al. [
10] proposed a genetic algorithm for community detection through optimization of the community score objective function. Li et al. [
13] attempted to apply extended compact genetic algorithms to explore community structures in complex networks. Gong et al. [
14] proposed a modal tone algorithm called Meme-Net to optimize the density of the modules. Li et al. [
15] designed a multi-objective adaptive fast evolutionary algorithm for extracting communities from networks. Chen [
16] proposed a MODTLBO/D community detection algorithm based on multi-objective teaching–learning optimization combined with a decomposition mechanism. DYN-MODPSO [
17] modified and enhanced the traditional evolutionary clustering framework and particle swarm algorithm, rendering dynamic detection more effective and efficient. Gong et al. [
18] proposed a multi-objective discrete particle swarm optimization algorithm (MODPSO) to address network clustering problems. Yang et al. [
19] proposed a multi-objective optimization algorithm based on node classification, NCMOEA, which employs a hybrid representation of different node types and performs effectively.
Single-objective optimization methods have achieved considerable success in community detection. However, as described in [
20], several limitations exist when addressing this type of community detection problem. Single-objective optimization typically seeks larger communities within networks while overlooking smaller communities that actually exist. Moreover, the definition of community commonly exhibits characteristics of high cohesion and low coupling; thus, community detection inherently requires multiple objectives to compete simultaneously, which necessitates multi-objective optimization methods to achieve superior performance. Furthermore, multi-objective evolutionary algorithms (MOEAs) [
21,
22] can provide a set of Pareto-optimal solutions rather than merely a single solution obtained through traditional methods. Consequently, multi-objective optimization has been adopted to overcome the resolution limitation of modularity. Hence, a community detection algorithm based on multi-objective optimization is investigated in this study.
MOEA-based community detection algorithms require encoding and decoding processes. Two of the most extensively utilized encoding methods are locus-based encoding [
23,
24] and label-based encoding [
25]. However, when utilizing discrete variables, minor changes in genotype can lead to substantial changes in phenotype [
26]. To address these challenges, Sun et al. [
27] proposed a novel continuous coding method employing a neural network approach, wherein each individual is represented as a continuous random vector and the population space is transformed from discrete to continuous. Nevertheless, this approach overlooks the structural relationships between nodes, and the randomly generated initial populations are characterized by poor quality. Since initial population quality typically impacts optimization performance, this paper proposes a novel encoding scheme that integrates similarity between nodes and continuous coding to enhance community detection performance. Moreover, an adaptive multi-objective optimization framework based on surrogate model adaptive switching strategy has been adopted, which establishes the surrogate model between continuous coding and objective functions through the core node learning method.
In summary, the main contributions of this paper are summarized as follows:
(1) A continuous encoding scheme that effectively utilizes similarity between community network nodes has been developed. Through this encoding scheme, the discrete space of the original problem is transformed into a continuous space, which can better assume a certain level of symmetry where similar solutions have similar fitness values.
(2) A simple yet effective strategy capable of acquiring core nodes to compress the sample space of surrogate models has been designed. This approach effectively ensures the quality of initial populations and reduces the impact of randomness on the structure of nodes during the iteration process.
(3) A community detection algorithm framework based on adaptive selection of surrogate models is proposed. For different networks, the framework can adaptively select appropriate surrogate models to establish the relationship between continuous coding and objective functions. Meanwhile, during the iterative process, elite individuals from the population are selected to update the surrogate model, thereby ensuring precision throughout the optimization process.
Together, these contributions form an integrated framework for community detection that combines continuous encoding, surrogate modeling, and multi-objective evolutionary optimization. The proposed method addresses three core challenges: (1) enhancing the quality of the initial population through similarity-guided continuous encoding; (2) improving optimization efficiency via adaptive surrogate model selection; and (3) effectively balancing conflicting structural objectives using a Pareto-based multi-objective strategy. These components are seamlessly integrated into a unified algorithmic pipeline, as detailed in
Section 3.
3. The Proposed Algorithm
This section presents the proposed multi-objective community detection algorithm based on adaptive surrogate model selection. The method integrates similarity-guided continuous encoding, core node identification, surrogate-assisted optimization, and evolutionary operators into a unified framework. The key components include core node search, solution representation and initialization, objective function design, adaptive surrogate model selection, and the overall evolutionary process.
To address the multi-objective nature of community detection, the algorithm employs a similarity-aware encoding strategy. Specifically, node similarity is quantified using a diffusion kernel similarity matrix, from which similarity vectors are constructed to capture local structural proximity. These vectors are combined with randomly initialized continuous variables via Hadamard product operations, resulting in structure-informed individuals that populate the initial solution set.
The optimization is conducted within a multi-objective evolutionary framework, where surrogate models are adaptively selected to approximate the objective functions and guide the search process. Elite individuals are periodically used to update the surrogate model, ensuring prediction accuracy throughout iterations.
3.1. Core Node Search Strategy
The core node search strategy has been designed to efficiently identify nodes that exhibit high potential as community centers. Prior to determining the core nodes, an evaluation of each network node’s weight must be conducted. Node weights are assigned based on the underlying graph structure and are quantified by the node’s degree. The weight value attributed to each node corresponds to the cardinality of its respective neighborhood set.
The degree of node
v, denoted as
, is quantified by the cardinality of its neighborhood set.
The construction of the core node set is carried out through a two-step process, as described in Algorithm 1. First, candidate nodes are identified by selecting those whose weights exceed the average weight of all nodes in the network, forming the candidate core node set. Then, the core node set is iteratively built from these candidates. In each iteration, the node with the highest weight in the current candidate set is selected and added to the core node set. This node, together with all its incident edges, is removed from the network. After removal, only the weights of the remaining candidate nodes are updated, reflecting the structural changes caused by the deletion. The process repeats: the updated candidate set is examined, and the next node with the highest weight is selected as a new core node. This selection–removal–update cycle continues until no candidate nodes remain, at which point the construction of the core node set is complete.
| Algorithm 1 coreNode |
| Require: : weight of all nodes; |
| Ensure: : the core node set; |
| 1: ; |
| 2: Select the nodes with weights greater than the average weight as candidate core nodes; |
| % % Establishing the candidate core node set |
| 3: Select the node with the largest weight and add it to the core node set; |
| 4: Remove the selected node and its edges from the network; |
| 5: Update the weights of the remaining candidate core nodes; |
| 6: Repeat steps 3, 4, and 5 until the candidate core node set is empty; |
| 7: Return ; |
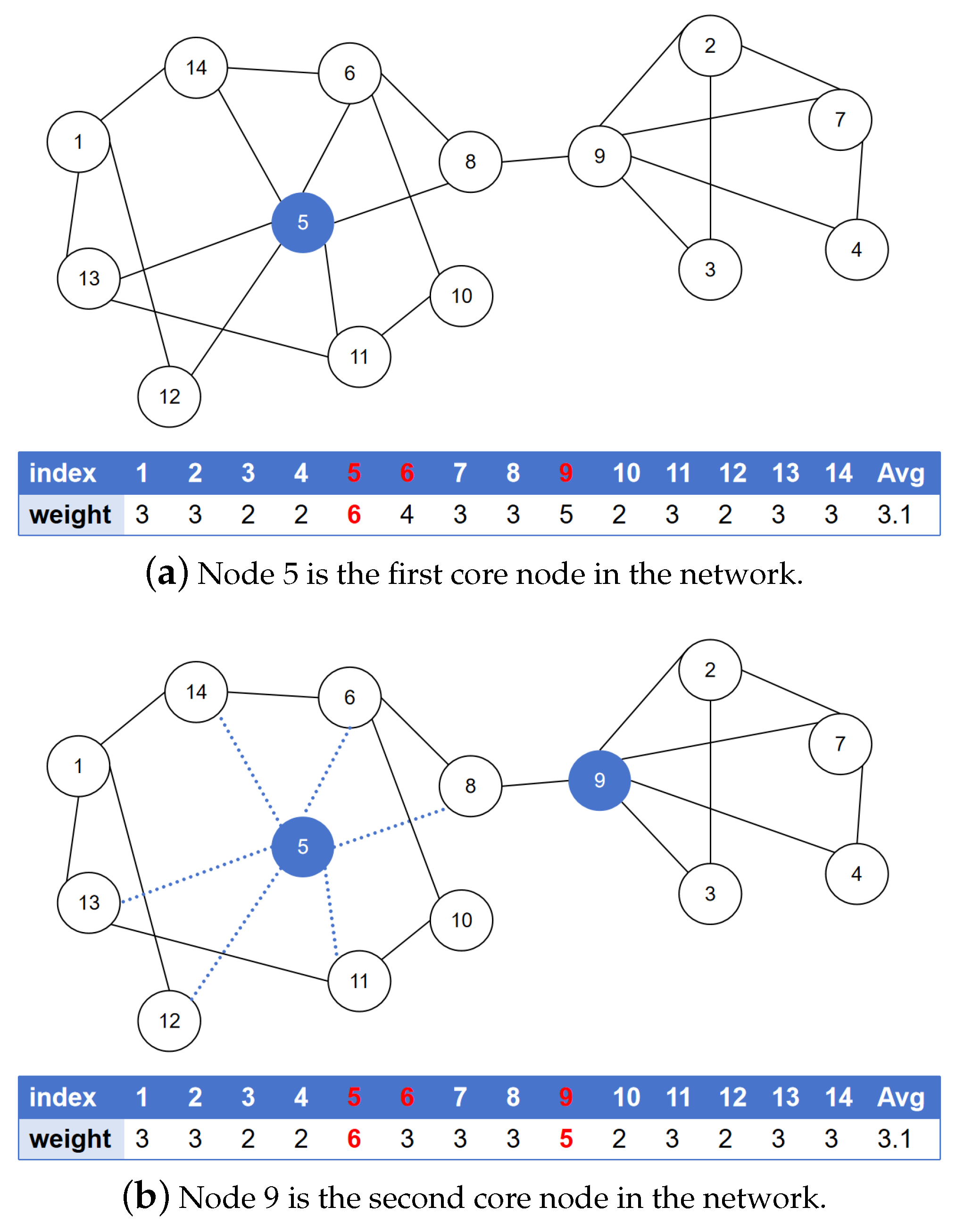
As illustrated in
Figure 1, the procedure commences with the calculation of each node’s weight based on its degree, resulting in
with a mean weight value of
. Based on these calculated weights, candidate core nodes are identified as those whose weights exceed the mean value of 3.1; consequently, the candidate core node set is established as
. In the subsequent step, node 5, which demonstrates the maximum weight among all candidates, is selected and incorporated into the core node set. Subsequently, this particular node and its associated edges are removed from the network, whereupon the weights of the remaining candidate core nodes undergo recalculation. Through systematic iteration of these procedural steps, the definitive core node set is determined as
.
3.2. Solution Representation and Initialization
Existing classical representation methods for network community detection are typically adapted from encodings utilized in evolutionary methodologies to address classical data clustering problems. These include label-based representation, locus-based representation, media-based representation, and permutation representation specifically designed for overlapping communities. Among these, label-based representation and locus-based representation [
35] are two commonly employed encoding methodologies.
In locus-based representation, the genotype of a node is considered as one of the nodes to which it is connected. For instance, in the sample network illustrated in
Figure 2, node 5 is connected to nodes 6, 7, and 8. Consequently, the genotype of node 5 could be represented as one of the following: 6, 7, or 8. In
Figure 2a, the individual genotype is given as
, which is derived by associating each node with one of its connected nodes. From the graphical representation, it can be observed that this genotype decodes the individual into two distinct communities:
and
.
In label-based representation, the individual genotype can be expressed as
, wherein each value represents the community to which a node belongs. As demonstrated in
Figure 2b, each node is assigned a community identifier as its genotype. The decoding process entails placing nodes with identical genotypes into the same community. It can be noted that both encoding methodologies ultimately result in discrete vectors.
Locus-based representation presents difficulties in designing evolutionary operators, whereas label-based representation encounters challenges in initializing high-quality individuals due to its inability to utilize similarity information between nodes. Furthermore, considering that continuous data is extensively employed in surrogate models to substitute the computation of the objective function in evolutionary algorithms, we have designed an improved continuous encoding strategy based on similarity matrices. This strategy aims to enhance the quality of both continuous encoding and the initial population. Algorithm 2 summarizes the pseudo code of the continuous encoding.
| Algorithm 2 Continuous encoding |
| Require: ,; |
| Ensure: A community partition ; |
| 1: Set ; |
| 2: for i = 1 to r do |
| 3: ; |
| 4: ; |
| 5: ; |
| 6: ; |
| 7: ; |
| 8: end for |
| 9: ; |
| 10: Return ; |
In Algorithm 2, a continuous-valued vector , where denotes the number of edges in the network, is taken as input. The vector is constructed by concatenation of r subvectors, where is a continuous vector that encodes the connections of node to its neighbors, with . Here, r denotes the number of nodes in the network. The length of each subvector is defined as , i.e., the degree of node . Each element in assigns a continuous value to the link connecting node to one of its neighbors. Let denote the set of neighboring nodes of . Meanwhile, the similarity between node and its neighbors is computed by extracting the corresponding entries from the diffusion kernel similarity matrix, and is represented as a vector .
During the initialization process, the influence of structurally related neighbors on node is amplified by computing the Hadamard product of and . The objective is to establish connections between node and its more influential neighbors at the initial stage. A detailed description of this improved continuous encoding approach is provided as follows.
For node
, the corresponding vector
, and the similarity matrix vector
. The Hadamard operator is initially applied to node
(line 1), resulting in a continuous vector
:
where ⊙ denotes the Hadamard product (element-wise multiplication).
Subsequently, a sigmoid function
is applied, which is defined as follows:
Each element of
is subjected to the sigmoid operation to obtain
(line 3). Thereafter, a softmax operation is performed on
to derive
(line 4), whereby softmax is expressed as
Based on
, the index
s is selected from the set of
neighbor nodes
:
The node constitutes the genotype of node . The node and its genotype are stored in . Decoding is then performed using a locus-based decoding method: by tracing links from each node to its selected neighbor , communities are formed by grouping nodes that are connected through mutual or transitive links. This process results in a discrete community partition by decoding .
Figure 3 illustrates the encoding process of a single node
. As observed in the figure, for each node
, a continuous value is associated with its neighbors. A node
is selected to be connected to node
through the Hadamard, sigmoid, softmax, argmax operations.
3.3. Objective Functions
In the multi-objective optimization process, two of the objectives are designated as negative ratio association (NRA) and ratio cut (RC) minimization. These particular objectives are capable of breaking the modularity constraints. Given an undirected network , where and , let the network be partitioned into k communities, denoted as , where each . For , define and , where . Consequently, the optimization problem can be formulated as
In the multi-objective optimization process, two of the objectives are designated as negative ratio association (NRA) and ratio cut (RC) minimization. These particular objectives are capable of breaking the modularity constraints. Given an undirected network , where and , let the network be partitioned into k communities, denoted as , where each and for .
For
, define
, and let
denote the complement of
with respect to the node set
V. Consequently, the optimization problem can be formulated as
In this paper, NRA is modified to kernel K-means (KKM) as introduced in [
47]. Thus, the community detection problem is defined as
The rationale for defining the aforementioned objectives stems from the observation, as pointed out by [
47], that KKM represents a decreasing function of the number of communities, whereas RC exhibits the opposite behavior. In essence, these objectives constitute two conflicting criteria, considering that the right operand of KKM and RC can be interpreted as the sum of the density of connections within a community. Furthermore, RC can be conceptualized as the sum of the density of connections between distinct communities. The minimization of both KKM and RC ensures that intra-community connections are dense while inter-community connections remain sparse, which aligns with the fundamental characteristics of community structures in networks.
In the context of community detection, intra-community density and inter-community sparsity often exhibit conflicting tendencies. The proposed objective functions, KKM and RC, explicitly capture these two aspects. By optimizing both objectives simultaneously under the multi-objective paradigm, Pareto optimality provides a principled way to balance these structural trade-offs. Each Pareto-optimal solution corresponds to a network partition that represents a distinct trade-off between intra- and inter-community connections, enabling a comprehensive exploration of the solution space and uncovering community structures at different granularity levels.
3.4. Adaptive Selection of Surrogate Models
In the optimization process, continuous coding must first be decoded, followed by the calculation of the objective functions KKM and RC. To enhance computational efficiency, a surrogate model is constructed to establish the relationship between the continuous coding and the objective functions. In order to ensure the predictive accuracy of the constructed surrogate model, its performance is evaluated using Spearman and Kendall correlation coefficients.
Five distinct surrogate models have been collected from the literature, like Carts, SVR, Ridge, Knn, and Bayesian. Through ablation experiments, it has been observed that no single surrogate model consistently outperforms the others in terms of the aforementioned criteria across all datasets. Therefore, we propose a selection mechanism, termed adaptive selection (AC) of surrogate model, as described in Algorithm 3. It constructs all five types of surrogate models in each iteration and adaptively selects the optimal model through cross-validation.
| Algorithm 3 Adaptive selection of surrogate model |
| Require: : a sample space; |
| Ensure: : surrogate models; |
| 1: Constructing 5 surrogate models; |
| 2: Calculate kendall and spearman coefficients for each surrogate model; |
| 3: Cross-validation to select the optimal surrogate model; |
| 4: Return ; |
The AC-selected surrogate model is utilized in conjunction with NSGA-II to optimize both KKM and RC (prediction), resulting in a set of solutions as well as a Pareto frontier upon completion of an NSGA-II search round. Due to the variations in accuracy of the objective function values predicted by the surrogate model, the individuals positioned on the Pareto frontier are computed with precision; that is, subsequent to decoding the individuals, the original value is computed using the original function of the objective function to ensure evolutionary accuracy. These individuals are subsequently incorporated into the sample space to enhance the predictive accuracy of the surrogate model.
3.5. Overall Procedure of ACMOEA
For a given network G, the proposed algorithmic framework is partitioned into four distinct components. In the initial component, the primary objective is to determine the core node set and the sample space dimension for the surrogate model (lines 1–2). The core node set is identified based on the topological structure of network G, where c represents the cardinality of the core node set. The parameter denotes the number of samples required for constructing the surrogate model, is determined by c.
In the second component, population initialization is conducted using an improved continuous encoding strategy that integrates structural similarity information, as described in Algorithm 2. Specifically, a similarity matrix is first computed to quantify inter-nodal relationships within the network G. By combining with the continuous encoding process, each individual in the initial population is generated with enhanced guidance toward structurally meaningful neighbors. This results in higher-quality initial solutions for subsequent evolutionary optimization. Finally, the evaluation function values of all individuals in the population are calculated. The genotypes corresponding to the core nodes in the initial population are used as features, while their associated evaluation function values serve as labels. Together, these features and labels form the initial training dataset used for surrogate model construction.
The third component involves the training and updating of the surrogate model. The surrogate model training process necessitates a substantial number of samples to achieve superior predictive accuracy; therefore, the initial generations of evolution are utilized to accumulate the requisite samples for training. Once the sample count attains the target threshold, the first generation surrogate model is trained.
In the fourth component (lines 10–35), the optimization process is initiated. The algorithmic framework exhibits similarities with the NSGA-II framework [
21], wherein SELECT, NDS, and CD denote binary tournament selection, non-dominated sorting, and crowding distance, respectively.
During the evolutionary process, the crossover mutation operator of differential evolution is utilized to generate novel individuals. Three individuals,
,
, and
, are randomly selected from the mating pool for crossover operations.
,
, and
are continuous encodings of individuals in the population, each corresponding to a network partition candidate. The formula for differential operation is expressed as follows:
where
represents a random number within the interval
, and
F and
denote the scaling factor and crossover probability, respectively. Differential evolution (DE) plays a central role in the optimization component of the algorithm. As shown in Equation (
13), DE generates new individuals by performing vector operations on continuous encodings of network partitions. This mechanism is particularly effective in exploring the high-dimensional Pareto front formed by KKM and RC. Through its mutation and crossover strategies, DE promotes diversity while guiding the population toward non-dominated solutions, making it well-suited for discovering multiple structurally distinct yet high-quality community partitions.
Each generational population undergoes sampling and integration into the training set. Upon accumulation of a sufficient number of samples, corresponding surrogate models are trained for the objective functions KKM and RC, respectively. Following the acquisition of the surrogate model, in subsequent optimization iterations, a subset of individuals employs the surrogate model for objective function prediction, while a smaller subset utilizes the original function for precise computation. Concurrently, these individuals are incorporated into the training set, and the surrogate model undergoes real-time updates to ensure predictive accuracy. Ultimately, the definitive community segmentation results are derived from the individuals situated on the Pareto frontier. Algorithm 4 presents the pseudo-code of ACMOEA, and the offspring generation operator is shown in Algorithm 5.
| Algorithm 4 Community detection algorithm based on adaptive selection of surrogate models |
| Require: : the adjacency matrix of the graph; w: the weights of all nodes; : |
| Population size; : maximum number of assessments; parameters of DE and PM: |
| F, , , ; |
| Ensure: : A set of Pareto front solutions; |
| 1: Get the core node and the size of the sample space: ; |
| 2: Initialize the population of size using the improved continuous encoding strategy described in Algorithm 2; |
| 3: Compute the node similarity matrix using the diffusion kernel method; |
| 4: Calculate the objective function of the population ; |
| 5: Undominated ordering of populations and calculation of crowding distances ; |
| 6: Get the features in the initial sample space as well as the labels ; |
| 7: while Number of real evaluations less than do |
| 8: Conduct crossover and mutation on by Algorithm 5; |
| 9: if the sample size is insufficient then |
| 10: Calculate the objective function; |
| 11: Merge samples into sample space ; |
| 12: else if then |
| 13: Perform Algorithm 3 to obtain the appropriate surrogate model; |
| 14: Calculate the value of the objective function of by means of the surrogate modeling; |
| 15: else |
| 16: Calculate the value of the objective function of by surrogate model; |
| 17: Select quality individuals from ; |
| 18: Compute the objective function for the population ; |
| 19: Update the sample space and surrogate model ; |
| 20: end if |
| 21: ; |
| 22: Undominated ordering of population and computation of crowding distance: ; |
| 23: Environmental selection yields new populations ; |
| 24: end while |
| 25: Return the Pareto frontier and its corresponding individual; |
| Algorithm 5 Crossover and mutation |
| Require: : population; |
| Ensure: : offspring; |
| 1: ; |
| 2: for do |
| 3: Creating mating pools through binary tournaments: ; |
| 4: Randomly select from ; |
| 5: DE and PM operations; |
| 6: ; |
| 7: end for |
| 8: Return offspring ; |
5. Conclusions
In this paper, we have proposed a community detection algorithm based on an adaptive switching surrogate model, termed ACMOEA-Net. The proposed approach employs a multi-objective optimization strategy to address the complex challenge of community detection in network structures. Our method simultaneously minimizes two conflicting objective functions, namely KKM and RC, thereby obtaining a partitioned structure characterized by dense intra-community connectivity and sparse inter-community connections.
To enhance computational efficiency, continuous coding has been implemented for community detection, wherein the KKM and RC objectives are predicted using a surrogate model, thus significantly reducing the computational resources required for the decoding process. Furthermore, the continuous coding mechanism has been improved through the initialization of populations based on a similarity matrix, which facilitates more effective exploration of the solution space.
The performance of the ACMOEA-Net algorithm has been extensively validated through comprehensive experiments conducted on both synthetic networks and five real-world network datasets. The experimental results have been systematically compared with seven state-of-the-art community detection algorithms, demonstrating the efficacy of our proposed approach.