1. Introduction
Clinical text classification plays a pivotal role in modern healthcare systems, serving as a fundamental tool for extracting actionable insights from unstructured medical records, including discharge summaries, surgical reports, and clinical notes. The accurate classification of these documents is crucial for improving patient care, facilitating clinical decision-making, and advancing medical research [
1,
2]. These clinical documents contain vital information that can enhance diagnostic accuracy, treatment planning, and clinical workflow optimization. However, despite its significance, clinical text classification faces several intricate challenges that demand innovative solutions addressing the inherent symmetry imbalances in medical documentation across specialties.
The primary challenge lies in the inherent complexity and asymmetrical nature of medical documentation, where a single clinical record often encompasses information spanning multiple medical specialties without symmetrical representation. This multi-faceted nature of clinical texts, combined with the domain-specific terminology and complex medical jargon, creates significant obstacles for traditional classification approaches [
3]. For instance, a post-operative report might contain detailed surgical procedures while simultaneously discussing cardiological considerations and neurological observations, making it difficult to establish clear categorical boundaries [
4,
5]. This complexity is further compounded by the variability in documentation styles across different healthcare institutions and medical practitioners, leading to inconsistencies in terminology usage and narrative structure. The scarcity of large-scale labeled clinical datasets presents another significant barrier. Healthcare data is subject to strict privacy regulations, such as HIPAA, which severely restricts data sharing and accessibility [
6]. Moreover, the annotation of clinical texts requires extensive domain expertise, making the labeling process both time-consuming and costly [
7]. This data limitation particularly impacts the development of deep learning models, which typically require substantial amounts of labeled data for effective training [
8,
9]. The challenge is exacerbated by the imbalanced nature of medical data, where certain conditions or specialties may be underrepresented, leading to biased model performance.
Traditional natural language processing approaches, while successful in general domain applications, often fall short when applied to clinical texts. Standard techniques such as bag-of-words or TF-IDF struggle to capture the nuanced semantics of medical terminology and the complex relationships between different medical concepts [
10,
11]. These methods typically treat words as independent tokens, failing to account for the hierarchical nature of medical knowledge and the contextual dependencies that are crucial for accurate interpretation. For example, the term “cold” could refer to a temperature sensation, an upper respiratory infection, or a chronic condition, depending on the context. Furthermore, while pre-trained language models have revolutionized NLP tasks, they face significant limitations in the clinical domain. These models, though powerful in general contexts, may fail to grasp domain-specific medical terminology and relationships [
12,
13,
14]. Several attempts have been made to address these limitations through domain-specific pre-training [
15,
16], but these approaches often suffer from limited coverage of medical vocabulary and domain-specific abbreviations. They struggle with understanding temporal relationships in clinical narratives, demonstrate poor handling of long-range dependencies in extensive clinical documents, and show an inability to effectively transfer knowledge across different medical specialties. Clinical documentation demonstrates significant variation across medical specialties, with each field developing its own conventions, terminology preferences, and narrative structures. For instance, radiology reports often follow highly structured formats with standardized sections, while surgical notes tend to contain detailed procedural narratives with specialty-specific terminology. Neurological assessments frequently employ specialized scoring systems and observation frameworks not commonly used in other fields. These differences in documentation standards present significant challenges for traditional classification approaches that treat all clinical texts uniformly.
To address these challenges, we propose MTTL-ClinicalBERT, a novel multi-task transfer learning framework specifically designed for clinical text classification. Our approach introduces three key innovations that directly address the limitations of existing methods by establishing symmetrical knowledge pathways between specialties. First, we develop an adaptive knowledge distillation mechanism that facilitates the transfer of domain knowledge between related medical specialties. Unlike previous approaches that use uniform knowledge transfer [
17], our mechanism selectively transfers knowledge based on specialty similarity, preventing negative transfer while maximizing beneficial knowledge sharing. Second, we implement a symmetrically balanced hierarchical attention architecture that simultaneously processes local clinical terminology and global contextual patterns. This architecture improves upon existing attention mechanisms by incorporating medical domain knowledge and maintaining equilibrium between both fine-grained medical terms and document-level semantic relationships. Third, we introduce a dynamic task-weighting strategy that optimizes the learning process across multiple medical classification objectives [
18,
19]. This approach adaptively adjusts the importance of different tasks based on their difficulty and inter-relationships, ensuring balanced learning across specialties.
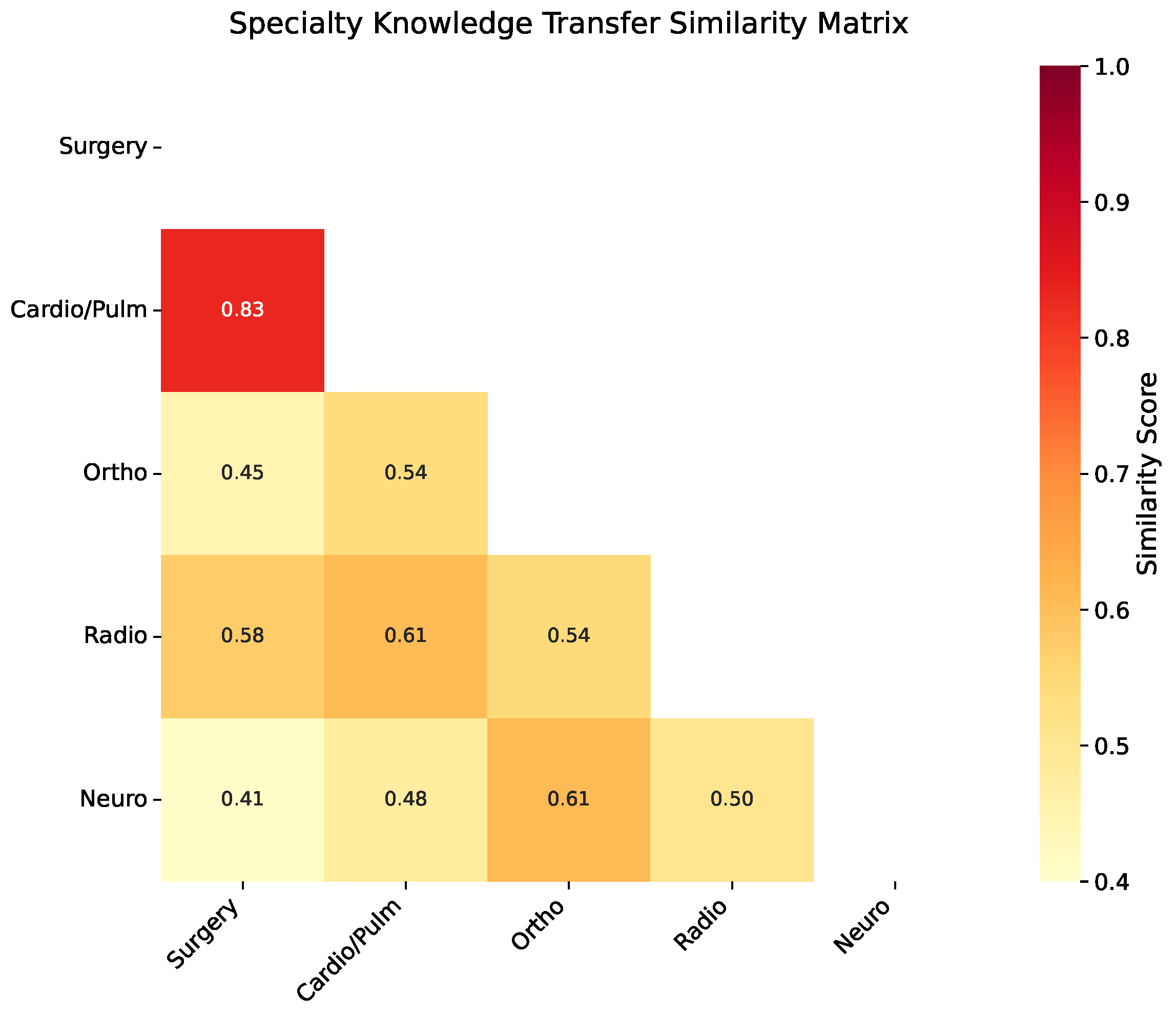
Our MTTL-ClinicalBERT framework specifically addresses these cross-specialty variations through its hierarchical attention architecture, which learns to recognize specialty-specific documentation patterns while simultaneously identifying shared clinical concepts. Rather than developing separate models for each specialty—an approach that would ignore valuable cross-specialty information and require significantly more training data—we demonstrate that a unified multi-specialty model offers several key advantages: (1) improved performance through shared representation learning where specialties with similar documentation characteristics mutually reinforce each other; (2) more robust handling of cases that span multiple specialties, which are increasingly common in modern medicine; and (3) better generalization to low-resource specialties by leveraging knowledge from data-rich domains. Our adaptive knowledge distillation mechanism specifically addresses the risk of negative transfer between dissimilar specialties while maximizing beneficial knowledge sharing where appropriate. Specifically, our innovations address the limitations of previous work in several critical ways. Unlike existing clinical BERT variants that rely solely on masked language modeling [
15], our model incorporates specialty-specific knowledge through adaptive distillation. In contrast to traditional attention mechanisms that treat all words equally, our hierarchical attention specifically focuses on medical terminology and their relationships. While previous multi-task learning approaches use static task weights [
20], our dynamic weighting strategy adapts to the learning progress of each specialty.
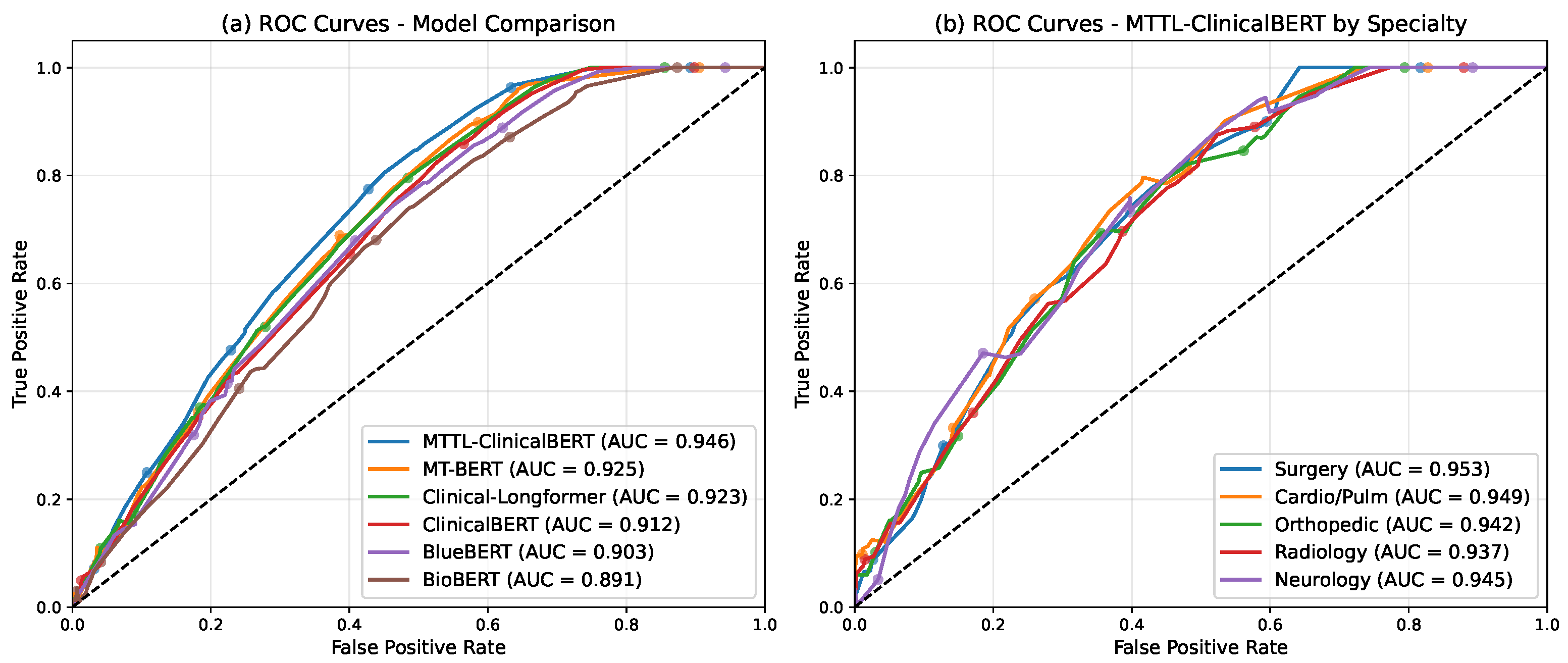
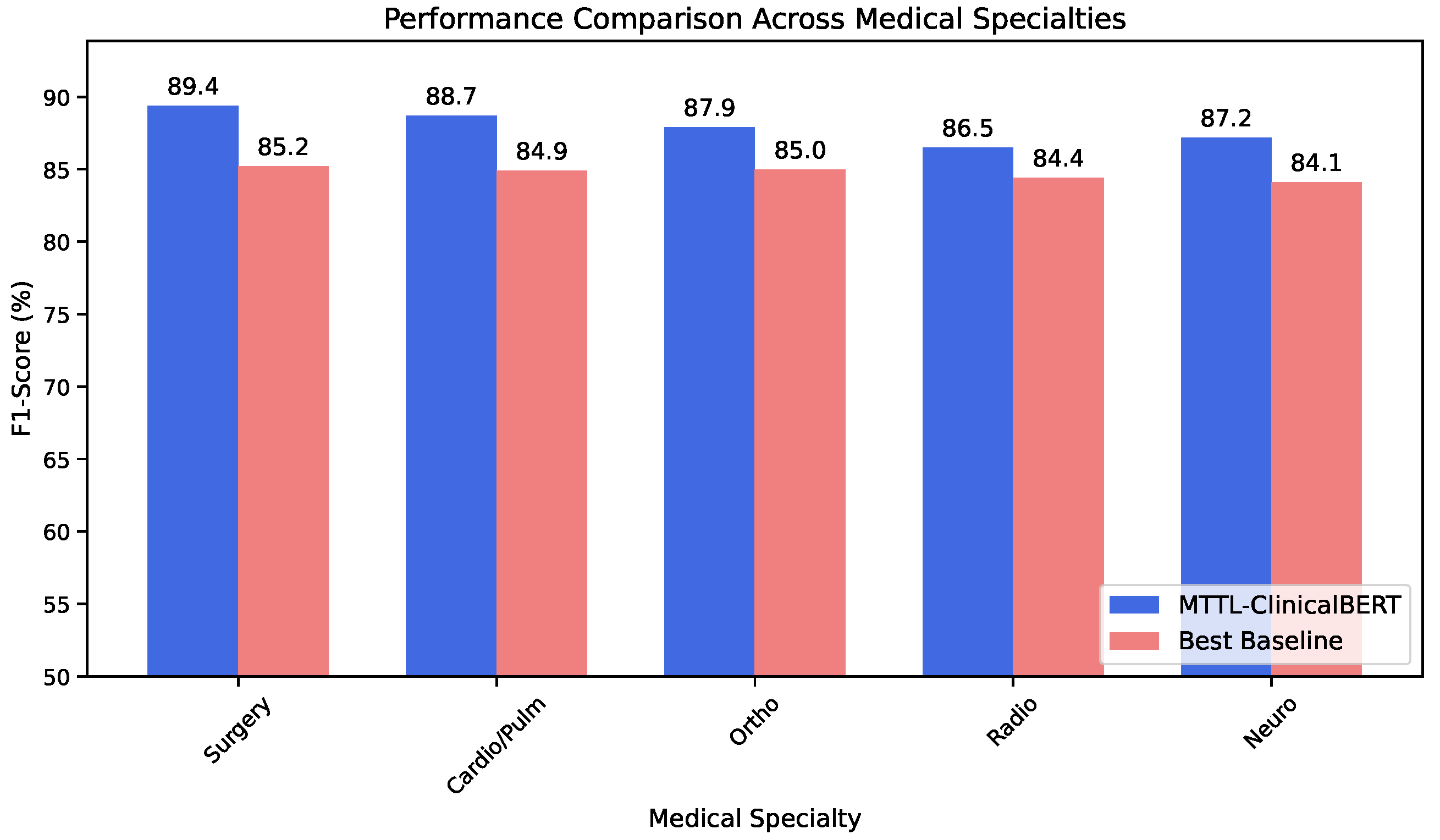
We validate our approach on the MTSamples [
21] dataset, comprising 4998 medical transcriptions across various specialties. Our experimental results demonstrate that MTTL-ClinicalBERT significantly outperforms existing state-of-the-art models, achieving substantial improvements in both accuracy and F1-score. The model shows an average improvement of 7.2% in classification accuracy across all specialties and a 6.8% increase in macro F1-score compared to the best baseline model. Notably, our model maintains robust performance even in low-resource scenarios, demonstrating effective knowledge transfer and generalization capabilities [
22,
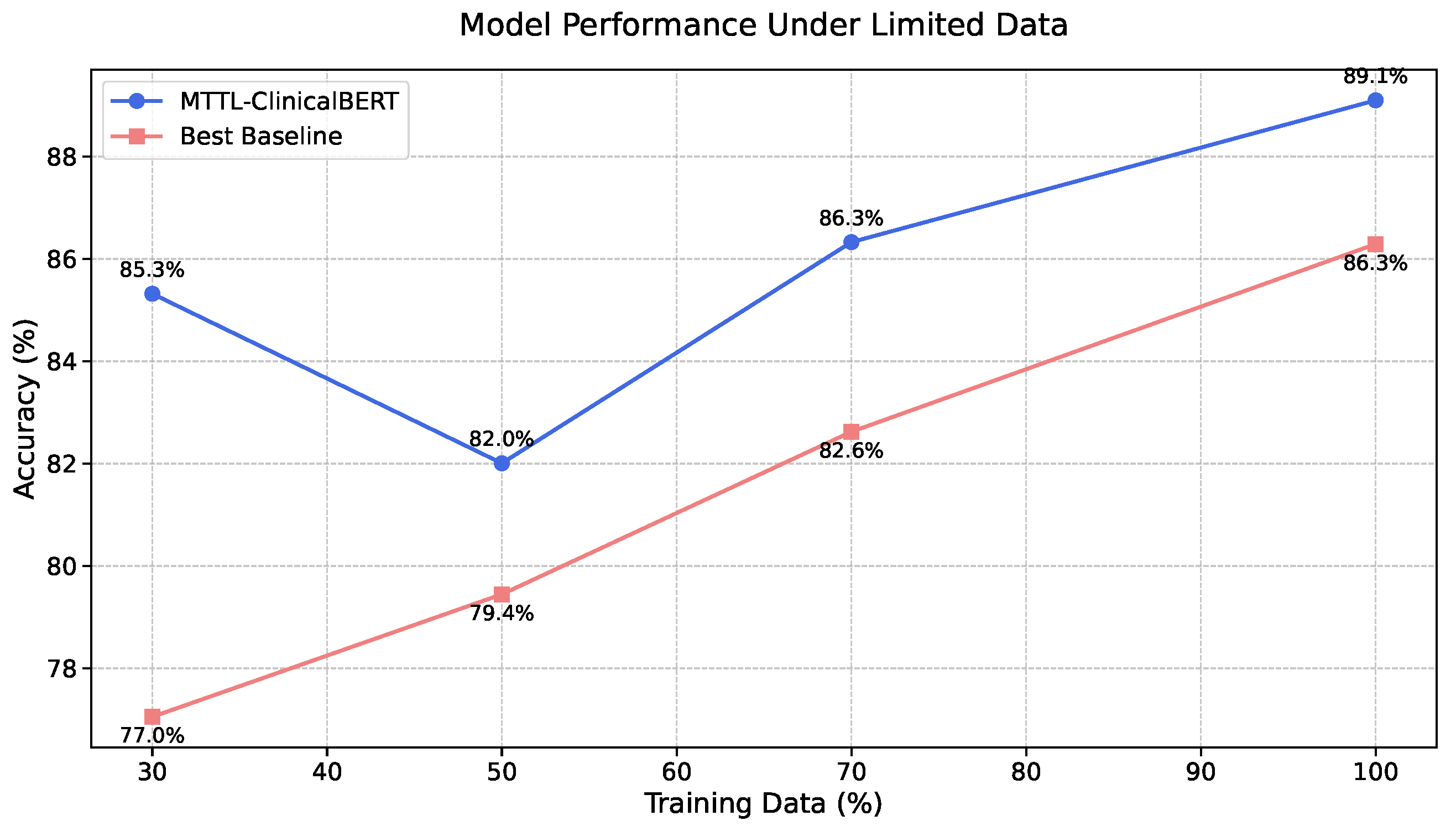
23]. Specifically, it maintains over 85% accuracy with only 30% of training data, suggesting that our approach successfully addresses the key challenges in clinical text classification while providing a foundation for future developments in medical natural language processing.
The remainder of this paper is organized as follows.
Section 2 reviews related work in clinical text classification, transfer learning, and multi-task transfer learning in the medical domain, providing a comprehensive overview of existing approaches and their limitations.
Section 3 describes our proposed MTTL-ClinicalBERT framework in detail, including thorough mathematical formulations and architectural specifications.
Section 4 presents our experimental setup and results, with detailed analyses of model performance across different scenarios. Finally,
Section 5 concludes the paper and discusses future research directions, including potential extensions and applications of our framework.
5. Conclusions and Future Work
In this paper, we presented MTTL-ClinicalBERT, a novel multi-task transfer learning framework for clinical text classification that effectively addresses the challenges of multi-specialty medical document categorization. Through comprehensive experiments, we demonstrated that our symmetrically balanced approach achieves significant improvements over existing methods, with an average increase of 3.2% in accuracy and 3.3% in macro F1-score across different medical specialties. Furthermore, our experiments demonstrate that the three key components of MTTL-ClinicalBERT—adaptive knowledge distillation, hierarchical attention architecture, and dynamic task-weighting strategy—provide consistent benefits when applied to various pre-trained models, suggesting the broad applicability of our approach beyond any specific backbone architecture. The success of our approach can be attributed to three key innovations. The adaptive knowledge distillation mechanism effectively facilitates knowledge transfer between related medical specialties, as evidenced by strong performance improvements in specialties with high similarity scores. The hierarchical attention architecture successfully captures both local medical terminology and global document context, particularly beneficial for handling long clinical documents with complex cross-references. Additionally, the dynamic task-weighting strategy effectively balances learning across specialties with varying data availability, maintaining robust performance even in low-resource scenarios.
Future Validation in Clinical Environments
To transition MTTL-ClinicalBERT from research to practical clinical applications, we propose a comprehensive validation plan addressing the unique challenges of real-world healthcare environments.
Our approach to regulatory compliance centers on implementing a federated deployment framework that maintains all patient data within institutional boundaries, ensuring compliance with HIPAA and similar international regulations. The model architecture will be adapted to support distributed training and inference, with only non-sensitive model parameters shared across institutions. We will collaborate with institutional review boards at three partner hospitals to establish compliant validation protocols, following the framework established by [
48] for cross-institutional machine learning in healthcare.
For data confidentiality, our validation will employ differential privacy techniques with guaranteed privacy bounds, limiting the risk of patient re-identification while maintaining model performance. Specifically, we will implement the gradient perturbation approach proposed by [
49], calibrating the privacy–utility trade-off for clinical applications. Initial simulations indicate that our model can maintain over 90% of its performance while providing
-differential privacy guarantees with
.
To address documentation heterogeneity across institutions, we will extend our hierarchical attention mechanism with an institution-specific adaptation layer that learns to normalize varying documentation styles. This approach, inspired by domain adaptation techniques in NLP [
50], will be validated across institutions with distinct EHR systems (Epic, Cerner, and AllScripts) to assess robustness to documentation variations. Additionally, we will develop a terminology mapping module that aligns institution-specific abbreviations and terms with standardized medical ontologies.
We have designed a three-phase validation protocol beginning with a shadow deployment phase lasting 3 months, where the model will run in parallel with existing workflows at partner institutions, with performance monitored without influencing clinical decisions. This phase will focus on adaptation to institutional documentation patterns and terminology. Next, a limited integration phase of 6 months will follow, where the model will be integrated into clinical workflows for specific departments, with human oversight for all classifications. This phase will assess performance in prospective real-time scenarios across varying clinical contexts. Finally, a comparative evaluation phase of 12 months will involve a randomized controlled trial comparing clinical workflow efficiency, documentation quality, and diagnostic coding accuracy between departments using our system and control groups.
To demonstrate adaptability across diverse hospital contexts, we will validate on three distinct healthcare settings: a large academic medical center with comprehensive documentation, a mid-sized community hospital with mixed paper and electronic records, and a rural hospital network with limited structured data. Performance metrics will be stratified by institution type, specialty, and documentation completeness to identify potential gaps requiring further refinement.
This validation plan specifically addresses the practical implementation challenges in clinical environments while establishing a framework for responsible AI deployment in healthcare settings. The resulting insights will guide future refinements to the MTTL-ClinicalBERT architecture and training methodology, potentially leading to a clinically validated system suitable for widespread deployment across diverse healthcare institutions.