1. Introduction
Statistical distributions are used to model real-world events. This has led to a lot of interest in generalizing classical distributions to obtain new distributions that accommodate various data forms better than the classical ones. Several methods have been developed to create classes of generalized distributions that are more flexible and better suited for modeling real-world data, such as the inverted method (Yassaee [
1]) and (Wang et al. [
2]), exponentiated class of distributions (Mudholkar and Srivastava [
3]), alpha-power transformation (Klakattawi [
4]), and transformed transformer (T-X distribution) (Alzaatreh [
5]).
One of the more common methods that has received the greatest interest in the statistical literature is the exponentiated technique, which is used to add new parameters to a baseline distribution. This leads to a flexible distribution for modeling a wide range of datasets. Mudholkar and Srivastava [
3] introduced the exponentiated Weibull family of probability distributions to extend the Weibull family by incorporating an additional shape parameter. Gupta et al. [
6] introduced the exponentiated gamma distribution, which produces non-monotonic and monotonic failure rates. Rao and Mbwambo [
7] introduced a new distribution called the exponentiated inverse Rayleigh (EIR) distribution, which is a generalization of the inverse Rayleigh distribution. They used the form suggested by Nadarajah and Kotz [
8] as follows:
where
is the reliability function of the baseline distribution and
is the power or additional shape parameter that aims to improve flexibility and overall performance to fit several datasets. The study of this distribution has been extended to incorporate more flexible models with a wider range of applications in real-life scenarios. Bashiru et al. [
9] developed a new lifetime distribution, termed the Topp–Leone exponentiated Gompertz inverse Rayleigh distribution, by compounding the Gompertz inverse Rayleigh model with the Topp–Leone exponentiated-G family. They investigated the statistical properties and demonstrated its applicability using two distinct datasets.
This paper presents a new extension of the EIR distribution, using the Marshall–Olkin (M-O) distribution as a generator, called the Marshall–Olkin exponentiated inverse Rayleigh (M-O-EIR) distribution. The main purpose of this paper is to obtain a flexible distribution that models asymmetric datasets using two methods of estimation, which are maximum likelihood and Bayesian methods. The following sections are arranged as follows.
Section 2 outlines an overview of the exponentiated inverse Rayleigh, its extended distribution, and the M-O-EIR distribution, and discusses relevant prior research.
Section 3 provides the definition of the Marshall–Olkin exponentiated inverse Rayleigh distribution and its associated derivation.
Section 4 studies the different statistical properties that are derived.
Section 5 illustrates the maximum likelihood method and its estimates for the unspecified parameters. Bayesian estimation assuming gamma independent prior distribution is presented in
Section 6.
Section 7 offers a Monte Carlo and Markov chain Monte Carlo (MCMC) simulation to assess the efficacy of the estimators for the M-O-EIR distribution.
Section 8 demonstrates the versatility of the new distribution in contrast to other distributions by utilizing two real datasets. Finally,
Section 9 provides some concluding remarks.
2. The Exponentiated Inverse Rayleigh Distribution
Let
X be a non-negative random variable having EIR distribution. Its cumulative distribution function (CDF) and probability density function (PDF) are, respectively, determined as
and
where
and
are shape and scale parameters, respectively.
The survival function of the EIR distribution is defined as
Marshall and Olkin [
10] proposed a general method for introducing a new positive shape parameter to an existing distribution. This resulted in the creation of a new family of distributions known as the Marshall–Olkin (M-O) family. It includes the baseline distribution as a special case, and it can also be used to model a wide range of data types. The CDF of the M-O family is defined as
and its PDF is given by
The survival function
is given by
where
and
is the shape parameter called a tilt parameter. For
, we obtain the baseline distribution, that is,
.
Many studies have used M-O to obtain new generalized distributions which show the flexibility of modeling data with non-monotone failure rates. MirMostafaee et al. [
11] introduced a novel expansion of the generalized Rayleigh distribution, termed the Marshall–Olkin extended generalized Rayleigh distribution. Afify et al. [
12] presented and analyzed the Marshall–Olkin additive Weibull distribution to accommodate various shapes of hazard rates, such as increasing, decreasing, bathtub, and unimodal patterns. ul Haq et al. [
13] proposed the M-O length-biased exponential distribution and estimated its parameters using the maximum likelihood estimation method.
Bantan et al. [
14] presented an extension of the inverse Lindley distribution, utilizing the Marshall–Olkin family of distributions called the generalized Marshall–Olkin inverse Lindley distribution. This new model provides enhanced flexibility for modeling lifetime data. Ahmadini et al. [
15] suggested a novel model known as the Marshall–Olkin Kumaraswamy moment exponential distribution. This distribution encompasses four specific sub-models. Aboraya et al. [
16] introduced and examined a novel four-parameter lifetime probability distribution known as the Marshall–Olkin Lehmann Lomax distribution. Mohamed et al. [
17] presented the M-O extended Gompertz Makeham lifetime distribution and derived the Bayesian and maximum likelihood estimators. Ozkan and Golbasi [
18] presented a sub-model of the family of generalized M-O distributions called the generalized M-O exponentiated exponential distribution, and studied its properties. Naz et al. [
19] investigated a new generalized K-family based on the Marshall–Olkin framework, using the Weibull distribution as the baseline. Their findings showed that the resulting Marshall–Olkin Weibull distribution displays a wider range of shapes for both its hazard rate and probability density functions. Lekhane et al. [
20] introduced a new generalized family of distributions known as the Exponentiated-Gompertz–Marshall–Olkin-G (EGMO-G) distribution. They explored various estimation techniques, including the maximum likelihood estimation, Cramér–von Mises method, weighted least squares, and least squares estimation. The performance of the proposed model was evaluated and compared with existing distributions using goodness-of-fit criteria. Alrweili and Alotaibi [
21] studied the Marshall–Olkin XLindley distribution using Bayesian and maximum likelihood methods, and applied them in three medical datasets.
3. The Marshall–Olkin Exponentiated Inverse Rayleigh Distribution
In this section, the three-parameter M-O-EIR distribution will be presented. Let
X be a random variable of the M-O-EIR distribution. Then, by substituting Equation (
1) in Equation (
3), we obtain the CDF of M-O-EIR distribution as follows:
The corresponding PDF and hazard rate function (HRF) of the M-O-EIR distribution are, respectively, given by
and
where
and
are shape parameters and
is the scale parameter. The M-O-EIR distribution becomes an EIR distribution when
= 1.
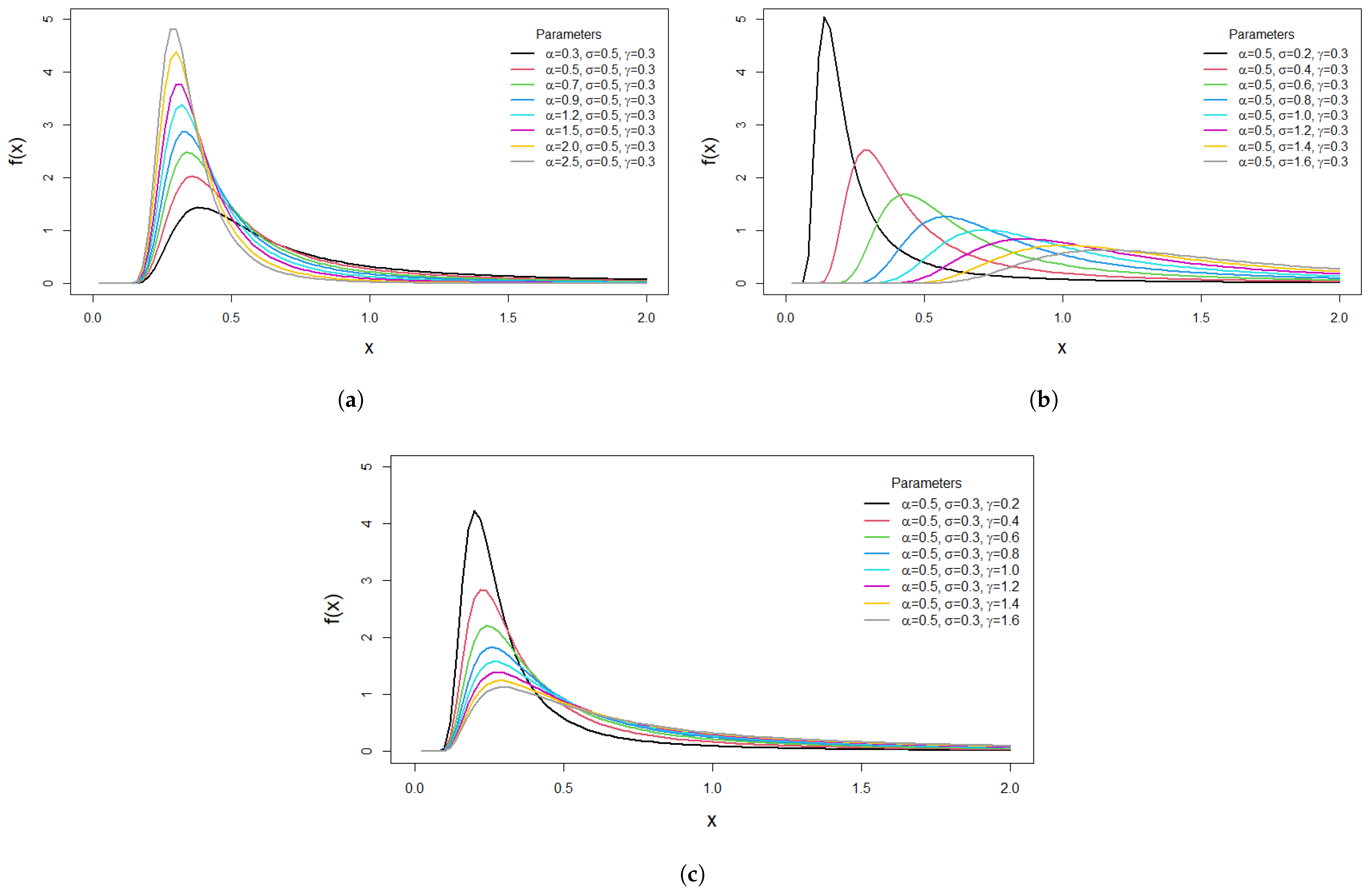
The plots in
Figure 1a–c illustrate the PDF plots when changing one of the parameters and fixing the others.
Figure 1a,c show different potential PDF shapes for the M-O-EIR distribution for various values of the shape parameters
and
. It is obvious that the distribution is unimodal and right skewed, which is suitable in modeling asymmetric data. In
Figure 1b, when the scale parameter
increases while the shape parameters
and
are fixed, the distribution expands to the right and its peak decreases. The plots in
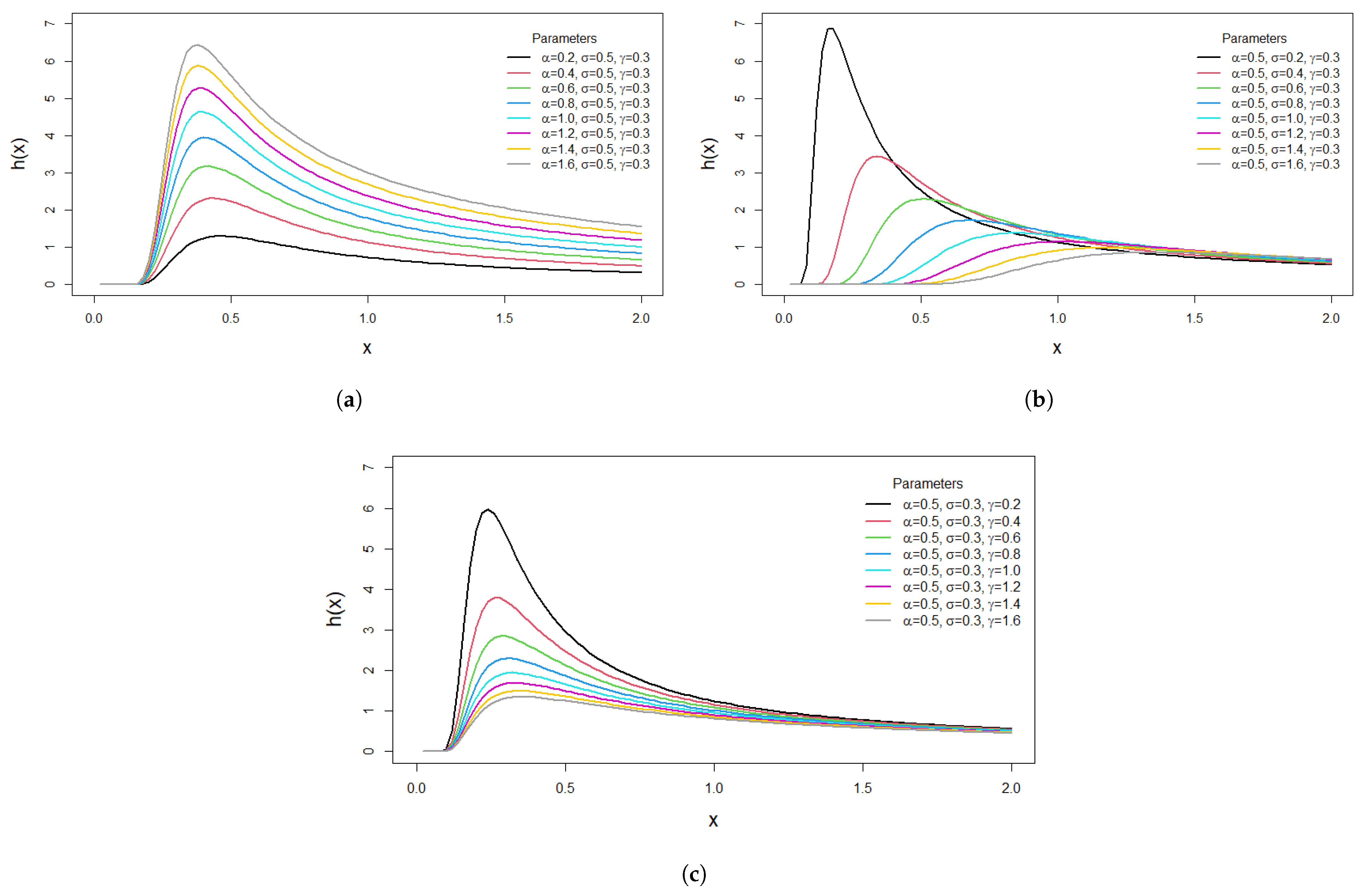
Figure 2a–c demonstrate the behavior of the HRF for the M-O-EIR distribution. It is observed that the hazard rate function increases and then decreases with time, which is highly beneficial in survival analysis.
6. Bayesian Estimation
Bayesian estimation is a statistical method that uses prior distribution for estimating the value of an unknown parameter. Many authors have contributed in studying Bayesian estimation: see Aboraya et al. [
22], Eliwa et al. [
23], Elbatal et al. [
24], and Alotaibi et al. [
25]. Assume that the independent prior distributions of the parameters
,
, and
are gamma distributions, that is,
,
, and
. Therefore, the joint prior density can be expressed as
where
and
are the hyper-parameters of the prior distribution, all of which are positive constants.
The joint posterior density function of
,
, and
is obtained by multiplying the likelihood function and the joint prior density of the parameters, as follows:
Therefore, the conditional posterior density functions of the parameters
,
, and
are obtained from Equation (
31), as follows:
In most cases, Bayesian estimation is employed under the squared-error loss function (SELF), which is the mean of the marginal of the parameter. The Bayesian estimators of the parameters
,
, and
regarding SELF are given as follows:
However, it is not possible to apply Equation (
35) directly. Hence, numerical approximations will be applied using the Gibbs sampler algorithm 1 to obtain random samples of conditional posterior density functions in Equations (
32)–(
34). This algorithm describes the implementation of Gibbs sampling to estimate parameters
,
, and
of a statistical model as follows:
| Algorithm 1 Gibbs Sampler Algorithm |
- 1:
Prior Distribution Initialization: - 2:
Define hyper-parameters for gamma prior distributions: - 3:
for , for , and for . - 4:
Density Function : - 5:
- 6:
Quantile Function : - 7:
- 8:
Distribution Function : - 9:
Numerically find q for given . - 10:
Random Generation : - 11:
Generate n random variables. - 12:
Gibbs Sampling Steps: - 13:
Define the conditional posterior distributions in Equations ( 32)–( 34) - 14:
Initialize the parameters , , using prior information. - 15:
for to do - 16:
Draw a sample from the conditional posterior - 17:
Update given current , and hyper-parameters . - 18:
Draw a sample from the conditional posterior - 19:
Update given current , and hyper-parameters . - 20:
Draw a sample from the conditional posterior - 21:
Update given current , and hyper-parameters . - 22:
end for - 23:
Output: Estimated parameters , , .
|
For more details, see Mohamed et al. [
17], Aboraya et al. [
22], and Elbatal et al. [
24].
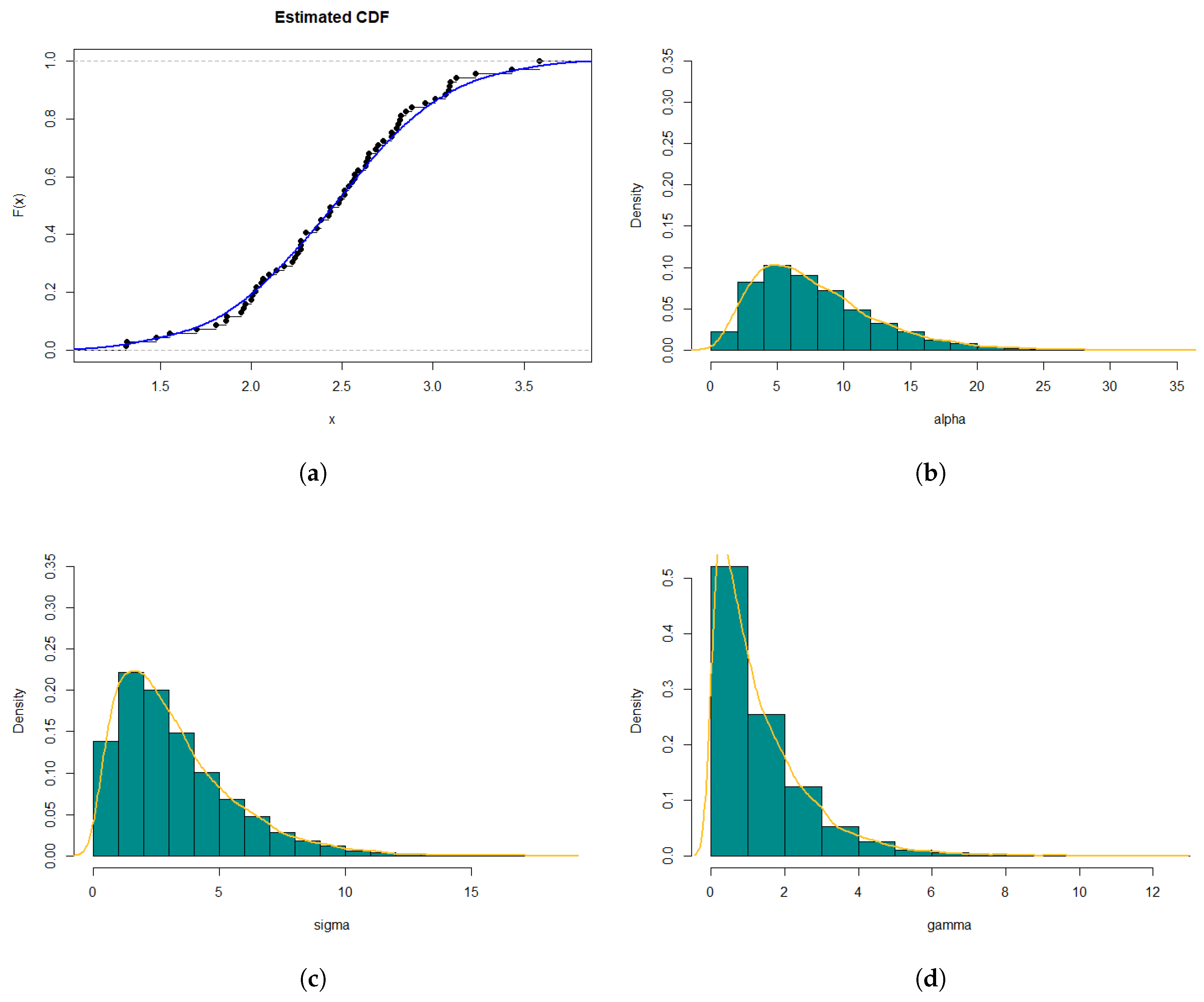
7. Simulation Studies
In this section, simulation studies were conducted using the R programming language to evaluate the theoretical results of the estimation process. Tables
Table 4,
Table 6, and
Table 8 present the maximum likelihood (ML) estimation results for parameters
,
, and
for different sample sizes. The initial values of the parameters were chosen randomly in three cases:
Case 1: (
Table 1)
,
, and
.
Case 2: (
Table 3)
,
, and
.
Case 3: (
Table 5)
,
, and
.
The tables provide the ML estimates, bias, mean squared error (MSE), 95% confidence interval (lower limit, upper limit), and the length of the confidence interval for each parameter.
For example, in
Table 4 and for
, the ML estimates are
,
, and
. The bias represents the difference between the ML estimate and the true value of the parameter, so for
, the bias is approximately
. The MSE provides a measure of the accuracy of the estimation, with lower values indicating better estimation performance. The 95% confidence interval gives a range of plausible values for the estimate of parameters, where the lower limit and upper limit represent the lower and upper bounds of the interval, respectively. The length of the confidence interval measures the width of the interval.
Table 5,
Table 7, and
Table 9 present the Gibbs estimation results for the initial guess of the parameters in Cases 1, 2, and 3, along with the sample sizes (
). Additionally, they provide the Gibbs estimates, bias, MSE, 95% confidence interval (lower limit, upper limit), and the length of the confidence interval.
For example, in
Table 5 and for
, the Gibbs estimates are
,
, and
. The bias is the difference between the Gibbs estimate and the true value of the parameter. The MSE measures the accuracy of the Gibbs estimation. The 95% confidence interval provides a range of plausible values for the estimate, and the length of the interval indicates its width.
The tables given allow us to assess how the ML estimation performs in relation to the Gibbs estimation method for different sample sizes. It is observed that the Gibbs method performs better since it provides smaller biases, smaller MSEs, and narrower confidence intervals.
Table 4.
ML estimates of the parameters , , and for sample sizes .
Table 4.
ML estimates of the parameters , , and for sample sizes .
| n | Parameter | 95% Confidence Interval | Length |
|---|
| ML Estimate | Bias | MSE | Lower Limit | Upper Limit |
|---|
| |
| 0.5771750 | −0.02282498 | 0.004838136 | 0.4690902 | 0.6852599 | 0.2161697 |
| 150 |
| 0.9234943 | 0.02349434 | 0.012781062 | 0.7415818 | 1.1054069 | 0.3638252 |
| |
| 1.5719552 | 0.07195516 | 0.393476985 | 0.5468952 | 2.5970152 | 2.0501200 |
| |
| 0.6213724 | 0.02137245 | 0.005566749 | 0.5037812 | 0.7389637 | 0.2351825 |
| 200 |
| 0.8760288 | −0.02397124 | 0.007869966 | 0.7355247 | 1.0165328 | 0.2810081 |
| |
| 1.8002179 | 0.30021791 | 0.432649746 | 0.8374797 | 2.7629561 | 1.9254764 |
| |
| 0.5802084 | −0.019791619 | 0.005113568 | 0.4671709 | 0.6932459 | 0.2260749 |
| 300 |
| 0.9064734 | 0.006473443 | 0.005329572 | 0.7868551 | 1.0260918 | 0.2392368 |
| |
| 1.4749322 | −0.025067831 | 0.264474681 | 0.6299620 | 2.3199024 | 1.6899404 |
| |
| 0.6304493 | 0.030449308 | 0.003509457 | 0.5468565 | 0.7140421 | 0.1671856 |
| 400 |
| 0.9073363 | 0.007336349 | 0.001354344 | 0.8480131 | 0.9666596 | 0.1186464 |
| |
| 1.5793075 | 0.079307471 | 0.106944222 | 1.0574131 | 2.1012018 | 1.0437887 |
Table 5.
Gibbs estimates of the parameters , , and for sample sizes .
Table 5.
Gibbs estimates of the parameters , , and for sample sizes .
| n | Parameter | 95% Confidence Interval | Length |
|---|
| Gibbs Estimate | Bias | MSE | Lower Limit | Upper Limit |
|---|
| 150 |
| 0.6145842 | 0.009843254 | 0.0009402291 | 0.5668129 | 0.6623555 | 0.09554261 |
|
| 0.8986206 | 0.023575332 | 0.0017916399 | 0.8407913 | 0.9564499 | 0.11565853 |
|
| 1.5177550 | 0.115959169 | 0.0167852140 | 1.4227046 | 1.6128053 | 0.19010066 |
| 200 |
| 0.6142425 | 0.04944098 | 0.003039235 | 0.5741225 | 0.6543624 | 0.08023986 |
|
| 0.9436900 | 0.01171312 | 0.001074259 | 0.8933342 | 0.9940459 | 0.10071170 |
|
| 1.5629566 | −0.01958582 | 0.006698013 | 1.4322395 | 1.6936736 | 0.26143411 |
| 300 |
| 0.6102177 | 0.01796004 | 0.0007921347 | 0.5745712 | 0.6458642 | 0.07129299 |
|
| 0.9287885 | −0.04775314 | 0.0035509068 | 0.8701530 | 0.9874240 | 0.11727103 |
|
| 1.5054043 | 0.02681624 | 0.0038531711 | 1.4133128 | 1.5974958 | 0.18418302 |
| 400 |
| 0.6224760 | 0.0035316688 | 0.0005690846 | 0.5836662 | 0.6612859 | 0.07761973 |
|
| 0.9026814 | 0.0003655791 | 0.0030841192 | 0.8113286 | 0.9940343 | 0.18270569 |
|
| 1.5033342 | −0.0227326067 | 0.0051544409 | 1.3913090 | 1.6153594 | 0.22405044 |
Table 6.
ML estimates of the parameters , , and for sample sizes .
Table 6.
ML estimates of the parameters , , and for sample sizes .
| n | Parameter | 95% Confidence Interval | Length |
|---|
| ML Estimate | Bias | MSE | Lower Limit | Upper Limit |
|---|
| 100 |
| 2.153459 | 0.430914710 | 0.34059358 | 1.506018 | 2.800900 | 1.2948819 |
|
| 1.688962 | −0.001248683 | 0.02621104 | 1.422648 | 1.955277 | 0.5326293 |
|
| 1.786701 | 0.051803699 | 0.01235204 | 1.624951 | 1.948451 | 0.3234995 |
| 200 |
| 2.234282 | −0.464169284 | 0.249594891 | 1.930327 | 2.538236 | 0.6079094 |
|
| 1.734226 | −0.009221691 | 0.058223948 | 1.337584 | 2.130869 | 0.7932852 |
|
| 1.761486 | 0.018666838 | 0.005229557 | 1.646558 | 1.876413 | 0.2298556 |
| 300 |
| 2.218313 | −0.49967587 | 0.32090158 | 1.779294 | 2.657333 | 0.8780394 |
|
| 1.656387 | 0.09621738 | 0.02528184 | 1.448152 | 1.864621 | 0.4164684 |
|
| 1.874647 | −0.03478232 | 0.03313397 | 1.580730 | 2.168565 | 0.5878353 |
| 400 |
| 2.459155 | 0.01687739 | 0.49148448 | 1.306246 | 3.612063 | 2.3058174 |
|
| 1.706396 | 0.01827464 | 0.03103875 | 1.418146 | 1.994646 | 0.5764995 |
|
| 1.805303 | 0.01271099 | 0.03361914 | 1.504410 | 2.106197 | 0.6017874 |
Table 7.
Gibbs estimates of the parameters , , and for sample sizes .
Table 7.
Gibbs estimates of the parameters , , and for sample sizes .
| n | Parameter | 95% Confidence Interval | Length |
|---|
| Gibbs Estimate | Bias | MSE | Lower Limit | Upper Limit |
|---|
| 100 |
| 2.448847 | 0.2197759 | 0.25828077 | 1.695051 | 3.202644 | 1.5075932 |
|
| 1.634007 | −0.1146412 | 0.03801343 | 1.374582 | 1.893431 | 0.5188489 |
|
| 1.799770 | −0.1784849 | 0.03917636 | 1.659033 | 1.940506 | 0.2814732 |
| 200 |
| 2.273107 | −0.180399658 | 0.11908250 | 1.789191 | 2.757024 | 0.9678331 |
|
| 1.676353 | −0.006822676 | 0.02219400 | 1.431544 | 1.921162 | 0.4896185 |
|
| 1.823658 | −0.084395585 | 0.02146485 | 1.626654 | 2.020661 | 0.3940073 |
| 300 |
| 2.327360 | 0.406830338 | 0.226867665 | 1.919888 | 2.734831 | 0.8149426 |
|
| 1.636862 | −0.008154544 | 0.009547621 | 1.476687 | 1.797038 | 0.3203508 |
|
| 1.865932 | 0.100229560 | 0.049332036 | 1.539882 | 2.191983 | 0.6521015 |
| 400 |
| 2.301370 | 0.002194087 | 0.12992529 | 1.708438 | 2.894302 | 1.1858635 |
|
| 1.581303 | 0.100249401 | 0.02129398 | 1.406870 | 1.755735 | 0.3488647 |
|
| 1.803557 | 0.117629491 | 0.02659310 | 1.617764 | 1.989351 | 0.3715866 |
Table 8.
ML estimates of the parameters , , and for sample sizes .
Table 8.
ML estimates of the parameters , , and for sample sizes .
| n | Parameter | 95% Confidence Interval | Length |
|---|
| ML Estimate | Bias | MSE | Lower Limit | Upper Limit |
|---|
| 100 |
| 0.8316659 |
| 0.0011159390 | 0.7848806 | 0.8784513 | 0.09357065 |
|
| 0.8163681 |
| 0.0005374803 | 0.7782312 | 0.8545051 | 0.07627391 |
|
| 0.9579443 |
| 0.0018889469 | 0.9230478 | 0.9928409 | 0.06979311 |
| 200 |
| 0.8271646 | 0.0002249037 | 0.0011891715 | 0.7704390 | 0.8838902 | 0.11345115 |
|
| 0.8434598 | 0.0384664282 | 0.0030380032 | 0.7785221 | 0.9083975 | 0.12987531 |
|
| 0.9548462 | -0.0059776642 | 0.0003291505 | 0.9266683 | 0.9830242 | 0.05635589 |
| 300 |
| 0.8685745 | −0.086366396 | 0.0118576480 | 0.7594763 | 0.9776728 | 0.21819655 |
|
| 0.8265717 | −0.048953817 | 0.0032391426 | 0.7788195 | 0.8743239 | 0.09550447 |
|
| 0.9640693 | 0.006154031 | 0.0009376036 | 0.9147266 | 1.0134119 | 0.09868528 |
| 400 |
| 0.8709748 | −0.16175433 | 0.030191820 | 0.7665807 | 0.9753689 | 0.20878817 |
|
| 0.8352936 | 0.01690962 | 0.001155469 | 0.7867861 | 0.8838011 | 0.09701504 |
|
| 0.9497568 | −0.01820171 | 0.001278391 | 0.8991322 | 1.0003813 | 0.10124911 |
Table 9.
Gibbs estimates of the parameters , , and for sample sizes .
Table 9.
Gibbs estimates of the parameters , , and for sample sizes .
| n | Parameter | 95% Confidence Interval | Length |
|---|
| Gibbs Estimate | Bias | MSE | Lower Limit | Upper Limit |
|---|
| 100 |
| 0.7878036 | −0.014810786 | 0.0009424598 | 0.7435687 | 0.8320385 | 0.08846983 |
|
| 0.8284729 | 0.002281736 | 0.0020609117 | 0.7538888 | 0.9030570 | 0.14916823 |
|
| 0.9091723 | 0.025490909 | 0.0012062128 | 0.8703689 | 0.9479757 | 0.07760679 |
| 200 |
| 0.7603078 | 0.017708859 | 0.0007219984 | 0.7270644 | 0.7935513 | 0.06648688 |
|
| 0.8496209 | −0.055430117 | 0.0045450075 | 0.7864969 | 0.9127450 | 0.12624813 |
|
| 0.9062628 | −0.009306478 | 0.0004778868 | 0.8737236 | 0.9388021 | 0.06507852 |
| 300 |
| 0.7871234 | −0.00572992 | 0.001655168 | 0.7208657 | 0.8533810 | 0.13251538 |
|
| 0.8179235 | 0.01969022 | 0.001565852 | 0.7614602 | 0.8743867 | 0.11292647 |
|
| 0.9105627 | 0.03984153 | 0.002370133 | 0.8645384 | 0.9565870 | 0.09204865 |
| 400 |
| 0.7875281 | 0.040999324 | 0.0034598663 | 0.7181465 | 0.8569097 | 0.13876320 |
|
| 0.8315301 | −0.011180411 | 0.0008965500 | 0.7858373 | 0.8772229 | 0.09138554 |
|
| 0.9255909 | 0.005224829 | 0.0008809976 | 0.8775271 | 0.9736547 | 0.09612763 |
9. Conclusions
This paper introduces and examines a new generalized form of continuous distributions utilizing the Marshall–Olkin distribution as a foundation. Various significant mathematical properties are deduced, encompassing moments, the moment generating function, order statistics, entropy, and quantile function. Two distinct estimation approaches, namely, maximum likelihood estimation and Bayesian methods, are investigated. A Monte Carlo simulation is conducted to estimate parameters and scrutinize the behavior of the proposed distribution. Bayesian estimation is achieved through the Gibbs sampler and Metropolis–Hastings algorithm which show superb performance. Finally, the new distribution is applied to two real-world datasets, demonstrating the practical utility of the M-O-EIR distribution. It is determined that the M-O-EIR offers superior fits compared to other competitive distributions for these specific datasets. Furthermore, the effectiveness of the ML estimation is contrasted with the Bayesian method on the same dataset, revealing that the Bayesian method outperforms the ML estimation method. Future research could explore improving the proposed distribution by considering alternative baseline distributions within the Marshall–Olkin framework. Additionally, the model may be extended to incorporate other parameter-estimation techniques, such as L-moments and least squares estimation.