Abstract
In biomedicine, the critical task is to decode Drug–Drug Interactions (DDIs) from complex biomedical texts. The scientific community employs Knowledge Graph Embedding (KGE) methods, enhanced with advanced neural network technologies, including capsule networks. However, existing methodologies primarily focus on the structural details of individual entities or relations within Biomedical Knowledge Graphs (BioKGs), overlooking the overall structural context of BioKGs, molecular structures, positional features of drug pairs, and their critical Relational Mapping Properties. To tackle the challenges identified, this study presents HSTrHouse an innovative hierarchical self-attention BioKGs embedding framework. This architecture integrates self-attention mechanisms with advanced neural network technologies, including Convolutional Neural Network (CNN) and Graph Neural Network (GNN), for enhanced computational modeling in biomedical contexts. The model bifurcates the BioKGs into entity and relation layers for structural analysis. It employs self-attention across these layers, utilizing PubMedBERT and CNN for position feature extraction, and a GNN for drug pair molecular structure analysis. Then, we connect the position and molecular structure features to integrate them into the self-attention calculation of entity and relation. After that, the output of the self-attention layer is combined with the connected vectors of the position feature and molecular structure feature to obtain the final representation vector, and finally, to model the Relational Mapping Properties (RMPs), the representation vector is embedded into the complex vector space using Householder projections to obtain the BioKGs model. The paper validates HSTrHouse’s efficacy by comparing it with advanced models on three standard BioKGs for DDIs research.
1. Introduction
Drug–Drug Interactions (DDIs), a consequence of concurrent multiple medication administration, may lead to severe or life-threatening Adverse Drug Reactions (ADRs). The recent surge in medical literature has led to an extensive yet unexplored corpus of potential DDIs, significantly influencing the medical co-medication landscape. This presents a substantial challenge in pharmaceutical management and development. Accurate DDI prediction, which enhances the structure of Biomedical Knowledge Graphs (BioKGs), is vital for improving patient welfare and advancing public health.
Amidst the dynamic evolution of computational paradigms, a varied array of methodologies have been utilized to tackle the complexities inherent in DDIs, leading to notable experimental successes. Sridhar et al. [1] ingeniously approached DDIs prediction by integrating a probabilistic planning framework with drug similarity analysis, identifying drug pairs with a heightened interaction likelihood. However, the predominant reliance on similarity-based methods, despite their commendable performance, is constrained by their limited capacity to address complex data structures like drug interaction networks and their inability to capture higher-order connectivity features, necessitating an alternative approach.
Additionally, the emergence of neural network paradigms, especially the Bidirectional Encoder Representations from Transformers (BERT) [2], has spurred research in DDI extraction. Key to this trend are pre-trained biomedical language models like BioBERT [3], SciBERT [4], BlueBERT [5], and PubMedBERT [6]. While these methods have advanced the field, they primarily focus on structural connectivities between nodes, often overlooking the subtleties within node attributes and edge typologies [7].
To mitigate limitations in existing algorithms, Knowledge Graph (KG)-based methods, as detailed in [8], have garnered attention for their ability to represent multiple potential relationships between two entities, thus addressing misclassification issues. However, these prevalent KG frameworks often overlook crucial factors such as the molecular structure and positional attributes of drug pairs, along with other significant elements that influence drug interactions. This oversight can impact the comprehensive modeling of drug relationships within these systems. Additionally, the complex dynamics of Relational Mapping Properties (RMPs) in the BioKG realm as depicted in Figure 1 pose further challenges. Most models, except for KG2ECapsule [9], fail to address these aspects. KG2ECapsule [9] uses a Bernoulli distribution-based sampling method, enhancing effectiveness, and employs capsule networks, leading to impressive predictive results. However, this approach increases training complexity and computational demands, limiting its use to smaller BioKG datasets.
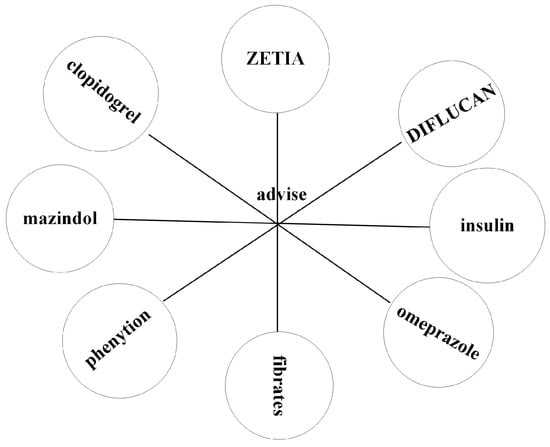
Figure 1.
RMPs (N-N) relationship in BioKG. The image clearly shows that there is an “advise” relationship pattern between “ZETIA” and “fibrates”, forming the triplet (ZETIA, advise, fibrates). Similarly, the relationship pattern “advise” also exists between “mazindol” and “insulin”, forming the triplet (mazindol, advise, insulin). Other triplets with the “advise” relationship can also be formed. These types of triplets possess RMP (N-N) relationships in the BioKG.
To address these challenges, this paper proposes an advanced hierarchical self-attention BioKGs embedding model, named HSTrHouse. This model innovatively combines the Transformer architecture with KG-based frameworks. It integrates self-attention mechanisms along with sophisticated neural network technologies like Convolutional Neural Network (CNN) and Graph Neural Network (GNN), thereby enhancing the model’s capability to encapsulate complex biomedical relationships and features effectively. As depicted in Figure 2, this study delineates the structural information of the entire BioKGs by segregating it into two distinct layers: the entity layer and the relation layer. Initially, within the entity layer, drug pairs acquire entity embeddings through an Encoder layer. Concurrently, drug pair-specific position and molecular structure features are extracted utilizing technologies such as PubMedBERT [6], CNN, and GNN. These features are then amalgamated. Subsequently, the integrated features, in conjunction with entity embeddings, are employed to construct a matrix space. The relation matrix is then mapped into this space, serving as the input for the relation layer. Through the Encoder layer, relation embeddings are obtained. Finally, to model the RMPs, both entity and relation embeddings are projected into a complex vector space using Householder projections. To sum up, our contributions are as follows:
- In this paper, we combine the self-attention mechanism and KG-based models to construct a model HSTrHouse that can consider the entire BioKG structure information by using Householder projections in complex vector space to model complex relation patterns, such as RMPs and hierarchies.
- Our model integrates PubMedBERT [6], CNN, and GNN to capture the position feature and molecular structure features in the abstract description, increasing the interpretability of the properties and relations of the entities.
- We have conducted extensive experiments on three BioKGs to demonstrate the effectiveness of HSTrHouse in predicting DDIs and their interpretability.
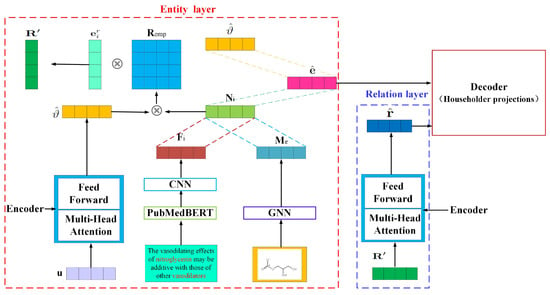
Figure 2.
Overview of our method. The illustration demonstrates the sentence “The vasodilating effects of nitroglycerin may be additive with those of other vasodilators”, identifying “nitroglycerin” and “vasodilators” as drug entities. Initially, within the entity layer, drug pairs acquire entity embeddings through an Encoder layer. Concurrently, drug pair-specific position and molecular structure features are extracted utilizing technologies such as PubMedBERT, CNN, and GNN. Subsequently, the position feature and the molecular structure are concatenated, resulting in the formation of . Subsequently, the integrated features , in conjunction with entity embeddings, are employed to construct a matrix space . The relation matrix is then mapped into this space, serving as the input for the relation layer. Through the Encoder layer, relation embeddings are obtained. Finally, to model the RMPs, both entity and relation embeddings are projected into a complex vector space using Householder projections.
2. Related Work
The work of Li et al. [10] predicted potential DDIs based on the pharmacokinetics (PK) model, one of the first computational DDIs prediction approaches. After that, the earliest techniques for DDIs were based on traditional feature-based machine learning methods; Chowdhury et al. [11] used kernel methods to extract DDIs, and Thomas et al. [12] used majority voting methods to extract DDIs. After this, many researchers improved the results by using more feature types, such as relative position features, syntactic structure features, and phrasal auxiliary verb features [13,14,15]. However, these methods usually rely heavily on manually performed feature engineering and feature selection, which limits their effectiveness and leads to some possible patterns that need to be noticed. Since then, deep neural network-based models have been widely used, which can mine knowledge in high-dimensional spaces. They can be broadly classified into three categories: (I) matrix factorization (MF)-based [16,17], (II) random walk (RW)-based [18,19], and (III) neural network (NN)-based [20,21]. They are comparable to traditional machine learning-based models, and achieved better performance compared to traditional machine learning-based models.
Since the powerful BERT [2] achieved the best results on many natural language processing tasks, many researchers have applied it to the task of extracting DDIs from biomedical language models, such as BioBERT [3], and SciBERT [4]. Zhu et al. proposed a BioBERT-based model [22] incorporating external entity information to provide a new baseline for DDIs extraction. Asada et al. used drug description and molecular structure information to improve the SciBERT [23] model’s performance for DDIs extraction. EGFI [24] combined BioBERT and Bio-GPT2 to construct the model’s classification part and generation part and used a multi-headed self-focus mechanism to fuse multiple semantic information to achieve strict contextual modeling and improve the performance of DDIs extraction. However, this class of models can only represent a few potential relationships between two entities and only focuses on the connections between nodes while ignoring node attributes and edge types.
With the development of KGE techniques, KGEs have been applied to the DDIs prediction task, enabling the automatic drug-to-entity capture of features required for inference. KGE methods have been shown to provide competitive performance in the DDIs prediction task. Among others, Tiresias [25] first integrated various drug-related variables into a BioKG, which was then used to compute several similarity measures among all drugs and predict potential DDIs using a logistic regression classifier. Celebi et al. [26] applied several classical KGE models, such as TransE [27] and TransD [28], to predict potential interactions between drugs, and BERTKG-DDIs [29] based on the classical KGE models, which combines the interactions of drug embeddings with other biomedical entities and the domain-specific BioBERT embedding-based Relation Classification (RC) architecture in combination. However, the above methods use addition, subtraction, or simple multiplication operators. Thus, they can only capture linear relationships between entities. Ma et al. [30] used a multi-view graph self-encoder to integrate multiple types of drug-related information. They added an attention mechanism to calculate the weights corresponding to each view for better interpretability.
With KGNN [31], in order to extract the higher-order structure and semantic relations of KG, the idea of the neural network is used to learn the neighborhood of each entity in KG as their local receptive domain. Then, the neighborhood information is integrated with the deviation of the current entity representation. Xin [15] integrated the neural network and knowledge graph, considered different levels of text features in the neural network part, used RotatE [32] in the KG part, and achieved excellent performance. However, most of the existing network-based methods for this class of models usually consider the prediction of DDIs as a binary classification problem, which does not correspond to the actual problem, where there are more relational correspondence properties (1-N, N-1, N-N) between the drug pairs problem. For this reason, KG2ECapsule [9] combines the idea of the Bernoulli distribution with a capsule network to model the relationship pattern between drug pairs. However, the high complexity of the Bernoulli distribution method model effect needs to be improved. Meanwhile, the model uses a capsule network. It achieves good results but only applies to small BioKG datasets due to its complex training process, significant computational overhead, and large data requirements for capsule network applications. For this reason, this paper combines the self-attention mechanism with a KG-based model. It constructs HSTrHouse, a hierarchical self-attentive embedding model based on a self-attentive mechanism and Convolutional Neural Network.
3. Model Building
As illustrated in Figure 2, we derive word-level position features using PubMedBERT [6] combined with a convolutional neural network, and molecular structure features through a graph neural network (GNN). These features are amalgamated into a composite feature vector. Simultaneously, vectors representing drug pairs are processed through an Encoder to obtain embeddings for these pairs. These embeddings and the composite feature are used to establish a matrix space where a relation matrix is introduced as the input to the relation layer. This layer utilizes the Encoder to generate relation embeddings. Ultimately, for modeling relational metabolic pathways (RMPs), both entity and relation embeddings are transformed into a complex vector space using Householder projections.
3.1. Position Feature
In this study, textual descriptions of drug pairs are parsed using the WordPiece algorithm [33]. This algorithm splits the text into word pieces, creating a sequence , with n as the count of word pieces. Each word piece is processed through PubMedBERT [6] to achieve its contextualized vector . Additionally, we generate paired relative position embeddings for each word piece, represented as and , which are then merged with the word embeddings and supplied to a CNN layer. The CNN layer’s computation is defined by:
where is the Gaussian Error Linear Unit activation function [34], indicates the convolutional weights, k the window size, and ⊙ denotes element-wise multiplication, with as the bias vector. Different window sizes are utilized to provide multiple perspectives. The resultant position feature consolidates the output from each CNN filter into a standardized vector, achieved by the following max-pooling step:
3.2. Molecular Structure
The analysis of drug molecular structures is efficiently performed using a Graph Neural Network (GNN) that adeptly handles the complex graph structures of molecules. In this graph-based model, atoms are represented as nodes and bonds as edges within the molecular graph . According to the framework by Tsubaki et al. [35], we adopt a neural GNN approach that utilizes extensive molecular fragments, or r-radius subgraphs, which are akin to molecular fingerprints. These subgraphs effectively encapsulate the atomic details and their relational context within the molecule.
In this method, the fingerprint vectors act as stand-ins for atomic representations and are initially randomized. These vectors undergo iterative refinement influenced by the graph structure of the molecule. For example, the vector for the i-th atom in a drug molecule is represented as , with indicating the set of adjacent atoms. The refinement of vector during the ℓ-th iteration is captured by:
where is the ReLU activation function, and , represent the weight and bias parameters, respectively. Following this, the atomic vectors are consolidated to form a molecular representation, which then passes through a linear transformation as follows:
where m denotes the total number of atomic fingerprints, and and are the weight and bias of the output layer, respectively. The combined structure and position features are then merged to yield:
3.3. Encoder Module
In this research, we incorporate a self-attention mechanism within our model to robustly capture the sequential dependencies and interactions, thus producing enriched feature representations. The embedding of a drug entity is fed into the Entity Self-Attention layer, where it is divided into three components for the transformer architecture as detailed in [14]: the query , the key , and the value . Each of has dimensions , and the weight matrices are dimensioned respectively. Using a single-head attention model for simplicity, we calculate the attention coefficients via the softmax normalization as shown:
The value vectors are then aggregated using the calculated attention weights to produce the attention mechanism’s output:
For the entity-level self-attention schema, the outputs from the attention mechanism are concatenated to generate the final vector:
with ⨁ representing concatenation, and K the count of distinct attention heads. This concatenated vector is then passed into the Encoder’s subsequent stage, the Feed-Forward Network (FFN). This network includes a two-layer setup with ReLU and linear activations respectively:
where indicates the Rectified Linear Unit function, , , , and serve as the trainable parameters.
Following this, a spatial model is constructed that integrates the entity features with relation vectors:
where ⊙ denotes the Hadamard product, facilitating the combination of entity and relational features. This vector is further processed through a relation-level self-attention layer and an FFN to produce the final integrated output . Ultimately, the output vector from the Entity Self-Attention layer is amalgamated with its position feature and molecular structure as follows:
3.4. Decoder Module
3.4.1. Knowledge Graph
In the realm of KGE models, substantial advancements have been made, with numerous models being developed to enhance our understanding and representation of complex relationships. The pioneering model in this domain, TransE [36], introduced a translational distance-based approach, embedding entities and , along with the relation , and employed the functional mapping . Despite its groundbreaking nature, TransE [36] encountered limitations in accurately learning 1-n relations, leading to the development of several extensions [37,38] to address these deficiencies. Further expanding on the concept, RotatE [39] proposed an adaptation of TransE [36] into the complex vector space, enabling the representation of asymmetric relation patterns. Similarly, QuatE [40] advanced the model into quaternion vectors to facilitate 3D space rotation. DualE [41] innovated by incorporating dual quaternions into the embedding of relational entities, successfully capturing non-combinatorial and multiple relation patterns. Additionally, some scholars have explored embedding vectors in hyperbolic geometric spaces to articulate hierarchical relations as demonstrated in models like [42,43]. To express deeper interactions between entities and relations, others have integrated deep learning into KGE model construction. This includes the ConvKB [44], which utilizes one-dimensional convolution, and the ConvR [45], which employs adaptive convolution. CapsE [46] leverages the capabilities of capsule neural networks, while RSN [47] applies Recurrent Neural Network (RNN) methodologies.
In a recent innovative approach, BiQUE [48] introduced the use of biquaternions in KGE models, combining rotation and hyperbolic geometry to model a variety of relation patterns. HousE [49], based on dual Householder transformations, models chain and RMPs relationship patterns. In this paper, we primarily employ HousE’s Householder Projections to implement transformations within the complex vector space, aiming to model the relational pattern of the BioKG dataset.
3.4.2. Householder Projections
In the realm of vector transformations, the adjusted Householder matrix is central. This matrix, associated with a unit vector and a scalar , is defined for a dimension as:
where represents the identity matrix of size , and the condition confirms p as a unit vector. The eigenvalues of mostly equal 1, except for one which is , making invertible when .
The function of the adjusted Householder matrix in transforming a vector x into a modified form is demonstrated geometrically as follows:
where adjusts the transformation magnitude along the axis defined by vector p.
Further extending this framework, with a collection of real scalars and matching unit vectors , where m denotes the number of transformations and each , the overall mapping process can be depicted as:
The resultant matrix is inherently invertible, stemming from the principle that the product of invertible matrices remains invertible. Such a composite of m modified Householder reflections is termed as Householder projections. Unlike the conventional Householder rotations, which strictly preserve distances, Householder projections introduce reversible alterations in the relative distances between points. This characteristic renders them especially suitable for temporal modeling, without compromising their capability to represent complex relational patterns.
In our model architecture, the Decoder Module plays a crucial role in handling complexRMPs. It accomplishes this by performing Householder projections between drug–entity pairs () obtained from the Encoder Module. These drug–entity vectors are then rotated in the complex vector space to establish the relationship vectors, thereby modeling the intricate relational patterns between drug entities. For each temporal aspect t, we define two parameter types, the axes and the scalars , with m being a positive integer. Each row consists of m k-dimensional unit vectors (projection axes), i.e., satisfying . Similarly, each row comprises m real values (projection scalars). The embedding of relation r is denoted as , where . Each row is composed of k-dimensional unit vectors, with and .
For each drug–entity pair, HSTrHouse transforms the head entity and tail entity with r-specific Householder projections:
Subsequently, HSTrHouse applies r-specific Householder rotations to the projected head point , formulated as:
To optimize distance-based models effectively, a loss function akin to the negative sampling loss [27] is employed, described as follows:
where is the sigmoid function, denotes a fixed margin, and () represents the i-th negative quadruple, with n being the number of negative samples.
Employing self-adversarial negative sampling as per [32], negative quadruples are sampled from the following distribution:
where is the temperature parameter. Thus, the final loss function is formulated as:
4. Experiments
4.1. Datasets
To rigorously evaluate the effectiveness of our proposed framework, we conduct a detailed link prediction analysis using three principal BioKG benchmark datasets. These datasets, specifically OGB-Biokg [50], DrugBank [51], and KEGG [52], are recognized for their academic and scientific merit and are instrumental in benchmarking computational approaches within the intricate field of biomedical knowledge. Detailed characteristics of these datasets, encompassing their structural complexities and unique features, are provided in Table 1.

Table 1.
Statistics for the various experimental datasets.
When depicting a drug molecule graphically, the primary input comes from SMILES string encoding, which is gathered from various datasets. We apply preprocessing scripts to derive molecular fingerprints from these graphical representations, following the methods described by Tsubaki et al. [35].
The OGB-Biokg, carefully assembled by Stanford University, stands as an extensive Biomedical Knowledge Graph. This graph is structured with five unique categories of entities and is linked by 51 directed relationships, showcasing the complex interconnections present in biomedical data.
The DrugBank database is an essential hub that compiles comprehensive data on drugs and their biological targets. It integrates bioinformatics and cheminformatics, providing exhaustive details on drugs including their chemical, pharmacological, and pharmaceutical attributes alongside rich target data that includes sequence, structure, and pathway details.
KEGG is recognized as a pivotal resource for understanding complex functions and interactions in biological systems, extending from the cellular level to entire ecosystems. It offers molecular-level details, encompassing interactions, reactions, and pathways across diverse areas such as metabolism, genetic and environmental information processing, cellular processes, organismal systems, and human diseases, thus delivering a holistic perspective on life sciences.
4.2. Baselines and Metrics
In our study, we conducted a thorough comparative analysis of our models against a diverse array of baseline methodologies. To assess the efficacy of traditional network representation learning techniques, we incorporated several benchmark models: matrix factorization-based (Laplacian [17]), random walk-based (DeepWalk [19]), and neural network-based (LINE [21]). Furthermore, our evaluation also encompasses a variety of knowledge graph-based models, specifically KGNN [31], KGAT [53], R-GCN [54], BERTKG-DDIs [29], Xin [15], and KG2ECapsule [9].
To rigorously validate the effectiveness of the proposed Householder projections within our framework, we designed two variants of HSTrHouse. These variants involve substituting the Householder projections with the earlier non-reversible prediction methodologies utilized in TransH and TransR, resulting in two distinct models named HSTrTH and HSTrTR, respectively.
The effectiveness of the model was quantified using a diverse array of metrics, each designed to shed light on different aspects of its predictive capabilities and reliability. These measures are detailed as follows:
- Accuracy (Acc.): This essential metric assesses the overall rate of correct predictions made by the model.where TP stands for True Positives, TN for True Negatives, FP for False Positives, and FN for False Negatives. It provides a snapshot of how well the model performs across all categories.
- Precision (Pre.): This metric measures the accuracy of the model in identifying only relevant instances as positive.It gains importance in situations where the implications of false positives are severe.
- Recall (Rec.): Also known as Sensitivity, this metric assesses the model’s ability to detect all actual positives.It is essential in applications where failing to identify a positive instance could be detrimental.
- F1 Score (F1): Balances Precision and Recall, providing a single score that gauges the accuracy of the model’s positive predictions and its thoroughness in capturing positive instances.This metric is particularly valuable in situations where classes are imbalanced.
- Area Under the ROC Curve (AUC): This metric measures the area beneath the Receiver Operating Characteristic (ROC) curve, which plots the True Positive Rate (TPR, or Recall) against the False Positive Rate (FPR). An AUC near 1 signifies superior model performance. The ROC curve’s area is usually calculated through a graphical method rather than a direct formula.
- Area Under the Precision–Recall Curve (AUPR): The AUPR metric quantifies the area under the Precision–Recall curve, which is crucial for evaluating models on imbalanced datasets. Like the AUC, the precise value of AUPR is typically derived through graphical analysis rather than a straightforward mathematical formula.
4.3. Implementation Details
In the training phase of our model, we meticulously define and optimize several hyperparameters. The batch size is set at 512 for uniformity across all datasets. We explore a range of embedding dimensions , specifically tuning it within the set . The learning rate is varied between 0.01 and 1 to identify the optimal rate for convergence. Additionally, we select a fixed margin from the set , crucial for the stability and performance of the model. The dropout rate, an important factor in preventing overfitting, is experimented with among the values 0.0, 0.2, and 0.4. For the number of modified Householder reflections m used in the Householder projections, we consider values within . The regularization coefficient , an integral component for model generalization, is tuned between the values . In the context of the attention mechanism, the number of attention heads is optimized across , while the dimensions of keys and values are varied within and the dimension of heads is chosen from .
4.4. Experimental Results and Analysis
The empirical results obtained from three distinguished BioKG datasets, presented in Table 2, emphatically affirm the exceptional effectiveness of our novel HSTrHouse model. This model demonstrates superior performance across a range of evaluative metrics, positioning it as a leading contender in the field. The results distinctly illustrate the enhanced capabilities of our biomedical model, which is predicated upon the KG structure. Additionally, our research illuminates the impressive efficacy of the HSTrTH and HSTrTR variants. While these models exhibit slightly lower performance metrics compared to those based on the Bernoulli distribution, such as KG2ECapsule [9], and our newly introduced HSTrHouse model, their results are nonetheless notable. Specifically, the approach focusing on projection for modeling RMPs demonstrates tangible outcomes. However, it is important to recognize that, despite their promising nature, the results of the HSTrTH and HSTrTR models do not quite reach the level of those achieved by methodologies incorporating the Bernoulli distribution and Householder projection techniques. These findings underscore the potential for further refinement and optimization in projection-based modeling approaches within the realm of BioKGs.

Table 2.
Experimental results of HSTrHouse and baseline models on three datasets.
4.4.1. Different Features
To meticulously investigate the salient attributes of the position feature and the discernible influence exerted by the molecular structure features on the performance dynamics of the model, we conducted a structured study. Informed by the foundational model, we iteratively undertook an exclusion process, explicitly targeting the position and molecular structure features. This yielded two distinct model instances, denoted as HSTrHP and HSTrHM. These instances were subjected to an exhaustive array of experiments conducted on the BioKG dataset OGB-Biokg.
The outcomes of these systematic experiments are succinctly presented in Figure 3. The findings incontrovertibly manifest that both the omission of the position feature and the absence of the molecular structure features invariably contribute to a degradation in the performance metrics exhibited by the model. Notably, the discernible disparity between HSTrHP and HSTrHM across the three performance indicators is an insightful inference gleaned from the results. This incongruity serves as an indirect yet cogent validation, establishing that the position feature harbors a substantively more profound impact on the model’s performance than the molecular structure feature.
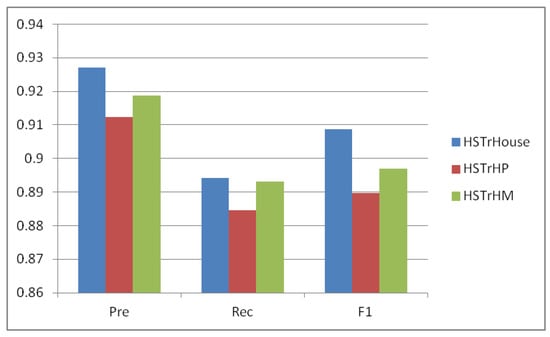
Figure 3.
Ablation experiments were performed on the OGB-Biokg dataset.
4.4.2. The Numbers of Modified Householder Matrices
We investigate the impact of m on the performance (F1) of HousE. The results are shown in Figure 4. With the increase in m on both datasets, the model’s performance increases first and then decreases. Moreover, the values of m for the best performance on the two datasets are different (m = 6 on OGB-Biokg, m = 3 on KEGG), mainly because it can be seen from Table 1 that the thoroughness of the OGB-Biokg dataset is significantly higher than that of the KEGG dataset.
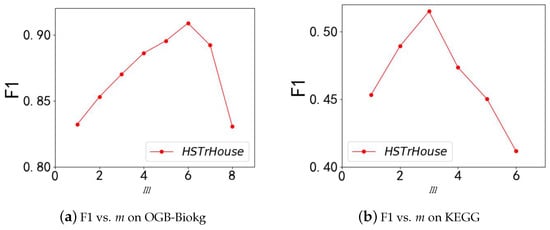
Figure 4.
(a,b) show the F1 results of HSTrHouse with different numbers of modified Householder matrices on OGB-Biokg and KEGG.
4.5. Extended Applications
Beyond routine Relation Extraction (RE) tasks, the purview of Biomedical Natural Language Processing (NLP) research encapsulates an array of intricate undertakings. These include, but are not limited to, Question Answering (QA), Named Entity Recognition (NER), Evidence-Based Medical Information Extraction (PICO), and Document Classification (DC), among other distinctive tasks.
In order to rigorously evaluate the robustness and adaptability of the HSTrHouse model within the complex domain of Biomedical NLP, an extensive validation process was conducted utilizing three distinct datasets: BC5-Chemical, PubMedQA, and BioASQ. The empirical results, detailed in Figure 5, unequivocally demonstrate the model’s superior performance compared to existing approaches, particularly in Question Answering (QA) tasks across these varied datasets. This comprehensive assessment not only confirms the effectiveness of HSTrHouse but also highlights its broad applicability and utility in the extensive field of Biomedical NLP research.
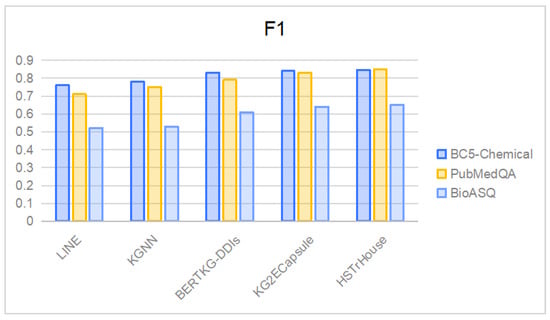
Figure 5.
Comparison of the proposed model with previous methods on the test datasets of BC5-Chemical, PubMedQA, and BioASQ.
However, it is pertinent to note that while the model exhibits performance improvements, the magnitude of these enhancements warrants a closer examination. This observation opens avenues for future research, indicating a fertile ground for continued academic investigation and enhancement of the model. Such scrutiny is vital for advancing the field of Biomedical NLP and unlocking further potential of models like HSTrHouse.
5. Conclusions
This study presents a hierarchical self-attention embedding model that integrates a self-attention mechanism with a Convolutional Neural Network architecture. The proposed model employs PubMedBERT for contextual embeddings, coupled with CNN and GNN layers to extract positional features and molecular structure characteristics of drug entities, respectively. Furthermore, the model leverages the rotation of Householder projections in complex vector space to effectively capture and model the interaction patterns between pairs of drugs. The utility and efficacy of the developed model are substantiated through comprehensive correlation and ablation studies performed on three benchmark BioKG. These experiments confirm the model’s robustness and its ability to provide significant insights into drug interactions, thereby highlighting its potential applications in the field of drug discovery and development.
Author Contributions
L.J.: investigation, methodology, software, and writing—original draft; S.Z.: investigation, data curation, visualization, attestation, and writing—original draft. All authors have read and agreed to the published version of the manuscript.
Funding
This work has received support from the 2023 Guangxi Higher Education Young Teachers’ Research Basic Ability Enhancement Project. The project is titled “Research on Interactive Display of Digital Guilin Museum Based on Virtual Reality” and its project number is 2023KY0806.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Acknowledgments
We extend our heartfelt appreciation for the support received from the 2023 Guangxi Higher Education Young Teachers’ Research Basic Ability Enhancement Project. Our project, entitled ‘Research on Interactive Display of Digital Guilin Museum Based on Virtual Reality’, designated with the project number 2023KY0806, has significantly contributed to the advancement of our research.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Sridhar, D.; Fakhraei, S.; Getoor, L. A probabilistic approach for collective similarity-based drug–drug interaction prediction. Bioinformatics 2016, 32, 3175–3182. [Google Scholar] [CrossRef] [PubMed]
- Devlin, J.; Chang, M.; Lee, K.; Toutanova, K. BERT: Pre-training of Deep Bidirectional Transformers for Language Understanding. In Proceedings of the 2019 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, NAACL-HLT 2019, Minneapolis, MN, USA, 2–7 June 2019; Burstein, J., Doran, C., Solorio, T., Eds.; Association for Computational Linguistics: Stroudsburg, PA, USA, 2019; Volume 1, pp. 4171–4186. [Google Scholar] [CrossRef]
- Lee, J.; Yoon, W.; Kim, S.; Kim, D.; Kim, S.; So, C.H.; Kang, J. BioBERT: A pre-trained biomedical language representation model for biomedical text mining. Bioinformatics 2020, 36, 1234–1240. [Google Scholar] [CrossRef] [PubMed]
- Beltagy, I.; Lo, K.; Cohan, A. SciBERT: A Pretrained Language Model for Scientific Text. In Proceedings of the 2019 Conference on Empirical Methods in Natural Language Processing and the 9th International Joint Conference on Natural Language Processing, EMNLP-IJCNLP 2019, Hong Kong, China, 3–7 November 2019; Inui, K., Jiang, J., Ng, V., Wan, X., Eds.; Association for Computational Linguistics: Stroudsburg, PA, USA, 2019; pp. 3613–3618. [Google Scholar] [CrossRef]
- Peng, Y.; Chen, Q.; Lu, Z. An Empirical Study of Multi-Task Learning on BERT for Biomedical Text Mining. In Proceedings of the 19th SIGBioMed Workshop on Biomedical Language Processing, BioNLP 2020, Online, 9 July 2020; Demner-Fushman, D., Cohen, K.B., Ananiadou, S., Tsujii, J., Eds.; Association for Computational Linguistics: Stroudsburg, PA, USA, 2020; pp. 205–214. [Google Scholar] [CrossRef]
- Gu, Y.; Tinn, R.; Cheng, H.; Lucas, M.; Usuyama, N.; Liu, X.; Naumann, T.; Gao, J.; Poon, H. Domain-Specific Language Model Pretraining for Biomedical Natural Language Processing. ACM Trans. Comput. Healthc. 2022, 3, 2:1–2:23. [Google Scholar] [CrossRef]
- Nickel, M.; Murphy, K.; Tresp, V.; Gabrilovich, E. A Review of Relational Machine Learning for Knowledge Graphs. Proc. IEEE 2016, 104, 11–33. [Google Scholar] [CrossRef]
- Guan, N.; Song, D.; Liao, L. Knowledge graph embedding with concepts. Knowl. Based Syst. 2019, 164, 38–44. [Google Scholar] [CrossRef]
- Su, X.; You, Z.; Huang, D.; Wang, L.; Wong, L.; Ji, B.; Zhao, B. Biomedical Knowledge Graph Embedding with Capsule Network for Multi-Label Drug-Drug Interaction Prediction. IEEE Trans. Knowl. Data Eng. 2023, 35, 5640–5651. [Google Scholar] [CrossRef]
- Li, L.; Yu, M.; Chin, R.; Lucksiri, A.; Flockhart, D.A.; Hall, S.D. Drug–drug interaction prediction: A Bayesian meta-analysis approach. Stat. Med. 2007, 26, 3700–3721. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, M.F.M.; Lavelli, A. FBK-irst: A Multi-Phase Kernel Based Approach for Drug-Drug Interaction Detection and Classification that Exploits Linguistic Information. In Proceedings of the 7th International Workshop on Semantic Evaluation, SemEval@NAACL-HLT 2013, Atlanta, GA, USA, 14–15 June 2013; Diab, M.T., Baldwin, T., Baroni, M., Eds.; The Association for Computer Linguistics: Stroudsburg, PA, USA, 2013; pp. 351–355. [Google Scholar]
- Thomas, P.; Neves, M.L.; Rocktäschel, T.; Leser, U. WBI-DDI: Drug-Drug Interaction Extraction using Majority Voting. In Proceedings of the 7th International Workshop on Semantic Evaluation, SemEval@NAACL-HLT 2013, Atlanta, GA, USA, 14–15 June 2013; Diab, M.T., Baldwin, T., Baroni, M., Eds.; The Association for Computer Linguistics: Stroudsburg, PA, USA, 2013; pp. 628–635. [Google Scholar]
- Bokharaeian, B.; Díaz, A. NIL_UCM: Extracting Drug-Drug interactions from text through combination of sequence and tree kernels. In Proceedings of the International Workshop on Semantic Evaluation, SemEval@NAACL-HLT 2013, Atlanta, GA, USA, 14–15 June 2013; Diab, M.T., Baldwin, T., Baroni, M., Eds.; The Association for Computer Linguistics: Stroudsburg, PA, USA, 2013; pp. 644–650. [Google Scholar]
- Kim, S.; Liu, H.; Yeganova, L.; Wilbur, W.J. Extracting drug-drug interactions from literature using a rich feature-based linear kernel approach. J. Biomed. Inform. 2015, 55, 23–30. [Google Scholar] [CrossRef] [PubMed]
- Jin, X.; Sun, X.; Chen, J.; Sutcliffe, R.F.E. Extracting Drug-Drug Interactions from Biomedical Texts using Knowledge Graph Embeddings and Multi-focal Loss. In Proceedings of the 31st ACM International Conference on Information & Knowledge Management, Atlanta, GA, USA, 17–21 October 2022; Hasan, M.A., Xiong, L., Eds.; ACM: New York, NY, USA, 2022; pp. 884–893. [Google Scholar] [CrossRef]
- Belkin, M.; Niyogi, P. Laplacian Eigenmaps for Dimensionality Reduction and Data Representation. Neural Comput. 2003, 15, 1373–1396. [Google Scholar] [CrossRef]
- Cao, S.; Lu, W.; Xu, Q. GraRep: Learning Graph Representations with Global Structural Information. In Proceedings of the 24th ACM International Conference on Information and Knowledge Management, CIKM 2015, Melbourne, VIC, Australia, 19–23 October 2015; Bailey, J., Moffat, A., Aggarwal, C.C., de Rijke, M., Kumar, R., Murdock, V., Sellis, T.K., Yu, J.X., Eds.; ACM: New York, NY, USA, 2015; pp. 891–900. [Google Scholar] [CrossRef]
- Perozzi, B.; Al-Rfou, R.; Skiena, S. DeepWalk: Online learning of social representations. In Proceedings of the 20th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, KDD ’14, New York, NY, USA, 24–27 August 2014; Macskassy, S.A., Perlich, C., Leskovec, J., Wang, W., Ghani, R., Eds.; ACM: New York, NY, USA, 2014; pp. 701–710. [Google Scholar] [CrossRef]
- Grover, A.; Leskovec, J. node2vec: Scalable Feature Learning for Networks. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; Krishnapuram, B., Shah, M., Smola, A.J., Aggarwal, C.C., Shen, D., Rastogi, R., Eds.; ACM: New York, NY, USA, 2016; pp. 855–864. [Google Scholar] [CrossRef]
- Tang, J.; Qu, M.; Wang, M.; Zhang, M.; Yan, J.; Mei, Q. LINE: Large-scale Information Network Embedding. In Proceedings of the International Conference on World Wide Web, WWW 2015, Florence, Italy, 18–22 May 2015; Gangemi, A., Leonardi, S., Panconesi, A., Eds.; ACM: New York, NY, USA, 2015; pp. 1067–1077. [Google Scholar] [CrossRef]
- Wang, D.; Cui, P.; Zhu, W. Structural Deep Network Embedding. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; Krishnapuram, B., Shah, M., Smola, A.J., Aggarwal, C.C., Shen, D., Rastogi, R., Eds.; ACM: New York, NY, USA, 2016; pp. 1225–1234. [Google Scholar] [CrossRef]
- Zhu, Y.; Li, L.; Lu, H.; Zhou, A.; Qin, X. Extracting drug-drug interactions from texts with BioBERT and multiple entity-aware attentions. J. Biomed. Inform. 2020, 106, 103451. [Google Scholar] [CrossRef]
- Asada, M.; Miwa, M.; Sasaki, Y. Using drug descriptions and molecular structures for drug-drug interaction extraction from literature. Bioinformatics 2021, 37, 1739–1746. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Lin, J.; Li, X.; Song, L.; Zheng, Z.; Wong, K. EGFI: Drug-drug interaction extraction and generation with fusion of enriched entity and sentence information. Briefings Bioinform. 2022, 23, bbab451. [Google Scholar] [CrossRef]
- Abdelaziz, I.; Fokoue, A.; Hassanzadeh, O.; Zhang, P.; Sadoghi, M. Large-scale structural and textual similarity-based mining of knowledge graph to predict drug-drug interactions. J. Web Semant. 2017, 44, 104–117. [Google Scholar] [CrossRef]
- Çelebi, R.; Yasar, E.; Uyar, H.; Gümüs, Ö.; Dikenelli, O.; Dumontier, M. Evaluation of knowledge graph embedding approaches for drug-drug interaction prediction using Linked Open Data. In Proceedings of the 11th International Conference Semantic Web Applications and Tools for Life Sciences, SWAT4LS 2018, Antwerp, Belgium, 3–6 December 2018; Volume 2275. [Google Scholar]
- Bordes, A.; Usunier, N.; García-Durán, A.; Weston, J.; Yakhnenko, O. Translating Embeddings for Modeling Multi-relational Data. In Proceedings of the Advances in Neural Information Processing Systems 26: 27th Annual Conference on Neural Information Processing Systems 2013, Lake Tahoe, NV, USA, 5–8 December 2013; pp. 2787–2795. [Google Scholar]
- Ji, G.; He, S.; Xu, L.; Liu, K.; Zhao, J. Knowledge Graph Embedding via Dynamic Mapping Matrix. In Proceedings of the 53rd Annual Meeting of the Association for Computational Linguistics and the 7th International Joint Conference on Natural Language Processing of the Asian Federation of Natural Language Processing, ACL 2015, Beijing, China, 26–31 July 2015; The Association for Computer Linguistics: Stroudsburg, PA, USA, 2015; Volume 1, pp. 687–696. [Google Scholar] [CrossRef]
- Mondal, I. BERTKG-DDI: Towards Incorporating Entity-Specific Knowledge Graph Information in Predicting Drug-Drug Interactions. In Proceedings of the Workshop on Scientific Document Understanding Co-Located with 35th AAAI Conference on Artificial Inteligence, SDU@AAAI 2021, Virtual Event, 9 February 2021; Veyseh, A.P.B., Dernoncourt, F., Nguyen, T.H., Chang, W., Celi, L.A., Eds.; CEUR Workshop Proceedings: 2021. Volume 2831. Available online: https://ceur-ws.org/Vol-2831/paper5.pdf (accessed on 21 December 2020).
- Ma, T.; Xiao, C.; Zhou, J.; Wang, F. Drug Similarity Integration through Attentive Multi-View Graph Auto-Encoders. In Proceedings of the Twenty-Seventh International Joint Conference on Artificial Intelligence, IJCAI 2018, Stockholm, Sweden, 13–19 July 2018; pp. 3477–3483. [Google Scholar] [CrossRef]
- Lin, X.; Quan, Z.; Wang, Z.; Ma, T.; Zeng, X. KGNN: Knowledge Graph Neural Network for Drug-Drug Interaction Prediction. In Proceedings of the Twenty-Ninth International Joint Conference on Artificial Intelligence, IJCAI 2020, Yokohama, Japan, 7–15 January 2021; pp. 2739–2745. [Google Scholar] [CrossRef]
- Sun, Z.; Deng, Z.; Nie, J.; Tang, J. RotatE: Knowledge Graph Embedding by Relational Rotation in Complex Space. In Proceedings of the 7th International Conference on Learning Representations, ICLR 2019, New Orleans, LA, USA, 6–9 May 2019; Available online: https://openreview.net/forum?id=HkgEQnRqYQ (accessed on 21 December 2018).
- Kudo, T.; Richardson, J. SentencePiece: A simple and language independent subword tokenizer and detokenizer for Neural Text Processing. In Proceedings of the 2018 Conference on Empirical Methods in Natural Language Processing, EMNLP 2018: System Demonstrations, Brussels, Belgium, 31 October–4 November 2018; Blanco, E., Lu, W., Eds.; Association for Computational Linguistics: Stroudsburg, PA, USA, 2018; pp. 66–71. [Google Scholar] [CrossRef]
- Hendrycks, D.; Gimpel, K. Bridging Nonlinearities and Stochastic Regularizers with Gaussian Error Linear Units. arXiv 2016, arXiv:1606.08415. [Google Scholar]
- Tsubaki, M.; Tomii, K.; Sese, J. Compound-protein interaction prediction with end-to-end learning of neural networks for graphs and sequences. Bioinformatics 2019, 35, 309–318. [Google Scholar] [CrossRef] [PubMed]
- Bordes, A.; Usunier, N.; Garcia-Duran, A.; Weston, J.; Yakhnenko, O. Translating embeddings for modeling multi-relational data. Adv. Neural Inf. Process. Syst. 2013, 26. [Google Scholar]
- Wang, Z.; Zhang, J.; Feng, J.; Chen, Z. Knowledge graph embedding by translating on hyperplanes. In Proceedings of the AAAI Conference on Artificial Intelligence, Quebec City, QC, Canada, 27–31 July 2014; Volume 35. [Google Scholar]
- Lin, Y.; Liu, Z.; Sun, M.; Liu, Y.; Zhu, X. Learning entity and relation embeddings for knowledge graph completion. In Proceedings of the Twenty-Ninth AAAI Conference on Artificial Intelligence, Austin, TX, USA, 25–30 January 2015. [Google Scholar]
- Sun, Z.; Deng, Z.H.; Nie, J.Y.; Tang, J. Rotate: Knowledge graph embedding by relational rotation in complex space. arXiv 2019, arXiv:1902.10197. [Google Scholar]
- Zhang, S.; Tay, Y.; Yao, L.; Liu, Q. Quaternion knowledge graph embeddings. Adv. Neural Inf. Process. Syst. 2019, 32. [Google Scholar]
- Cao, Z.; Xu, Q.; Yang, Z.; Cao, X.; Huang, Q. Dual quaternion knowledge graph embeddings. In Proceedings of the AAAI Conference on Artificial Intelligence, Virtual, 2–9 February 2021; Volume 35, pp. 6894–6902. [Google Scholar]
- Balazevic, I.; Allen, C.; Hospedales, T. Multi-relational poincaré graph embeddings. Adv. Neural Inf. Process. Syst. 2019, 32. [Google Scholar]
- Chami, I.; Wolf, A.; Juan, D.C.; Sala, F.; Ravi, S.; Ré, C. Low-dimensional hyperbolic knowledge graph embeddings. arXiv 2020, arXiv:2005.00545. [Google Scholar]
- Nguyen, D.Q.; Nguyen, T.D.; Nguyen, D.Q.; Phung, D. A novel embedding model for knowledge base completion based on convolutional neural network. arXiv 2017, arXiv:1712.02121. [Google Scholar]
- Jiang, X.; Wang, Q.; Wang, B. Adaptive convolution for multi-relational learning. In Proceedings of the 2019 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, Minneapolis, MN, USA, 2–7 June 2019; Volume 1, pp. 978–987. [Google Scholar]
- Nguyen, D.Q.; Vu, T.; Nguyen, T.D.; Nguyen, D.Q.; Phung, D. A capsule network-based embedding model for knowledge graph completion and search personalization. In Proceedings of the 2019 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, Minneapolis, MN, USA, 2–7 June 2019; Volume 1, pp. 2180–2189. [Google Scholar]
- Guo, L.; Sun, Z.; Hu, W. Learning to exploit long-term relational dependencies in knowledge graphs. In Proceedings of the International Conference on Machine Learning. PMLR, Long Beach, CA, USA, 9–15 June 2019; pp. 2505–2514. [Google Scholar]
- Guo, J.; Kok, S. BiQUE: Biquaternionic Embeddings of Knowledge Graphs. arXiv 2021, arXiv:2109.14401. [Google Scholar]
- Li, R.; Zhao, J.; Li, C.; He, D.; Wang, Y.; Liu, Y.; Sun, H.; Wang, S.; Deng, W.; Shen, Y.; et al. HousE: Knowledge Graph Embedding with Householder Parameterization. In Proceedings of the International Conference on Machine Learning, ICML 2022, Baltimore, MD, USA, 17–23 July 2022; Volume 162, pp. 13209–13224. [Google Scholar]
- Hu, W.; Fey, M.; Zitnik, M.; Dong, Y.; Ren, H.; Liu, B.; Catasta, M.; Leskovec, J. Open Graph Benchmark: Datasets for Machine Learning on Graphs. In Proceedings of the Advances in Neural Information Processing Systems 33: Annual Conference on Neural Information Processing Systems 2020, NeurIPS 2020, Virtual, 6–12 December 2020. [Google Scholar]
- Wishart, D.S.; Feunang, Y.D.; Guo, A.C.; Lo, E.J.; Marcu, A.; Grant, J.R.; Sajed, T.; Johnson, D.; Li, C.; Sayeeda, Z.; et al. DrugBank 5.0: A major update to the DrugBank database for 2018. Nucleic Acids Res. 2018, 46, D1074–D1082. [Google Scholar] [CrossRef]
- Kanehisa, M.; Furumichi, M.; Tanabe, M.; Sato, Y.; Morishima, K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017, 45, D353–D361. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; He, X.; Cao, Y.; Liu, M.; Chua, T. KGAT: Knowledge Graph Attention Network for Recommendation. In Proceedings of the 25th ACM SIGKDD International Conference on Knowledge Discovery & Data Mining, KDD 2019, Anchorage, AK, USA, 4–8 August 2019; Teredesai, A., Kumar, V., Li, Y., Rosales, R., Terzi, E., Karypis, G., Eds.; ACM: New York, NY, USA, 2019; pp. 950–958. [Google Scholar] [CrossRef]
- Schlichtkrull, M.S.; Kipf, T.N.; Bloem, P.; van den Berg, R.; Titov, I.; Welling, M. Modeling Relational Data with Graph Convolutional Networks. In Proceedings of the Semantic Web-15th International Conference, ESWC 2018, Heraklion, Crete, Greece, 3–7 June 2018; Gangemi, A., Navigli, R., Vidal, M., Hitzler, P., Troncy, R., Hollink, L., Tordai, A., Alam, M., Eds.; Springer: Berlin/Heidelberg, Germany, 2018; Volume 10843, pp. 593–607. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).