A Modified Stein Variational Inference Algorithm with Bayesian and Gradient Descent Techniques
Abstract
:1. Introduction
- (1)
- The modified Stein variational gradient descent method (MSVGD) algorithm is proposed, in which an improved Stein method is used in a gradient increment calculation of KL divergence. A set of particles are used to approximate target distribution by minimizing the KL divergence;
- (2)
- The SVGD algorithm can keep the KL value reduced in the gradient descent theory. is only in the unit ball of a reproducing kernel Hilbert space (RKHS). The SVGD algorithm will become slow in searching for the parameter distribution because of the limitation of local optimization. It is quite hard to jump out of the local optimum using the SVGD algorithm. In the referece [31], Stein’s operator is based on . Considering in the design of the operator can increase the chance to jump out of the local optimum, especially in the case of complex distribution.
2. Model Formulation and Preliminaries
2.1. Stein Method
2.2. Variational Inference
3. Modified Stein Variational Inference Using KL Minimizing
3.1. Stein Operators Selection
3.2. Stein Transform for Differential Computing of KL
3.3. Modified Stein Variational Gradient Descent Method with Particle Swarm Optimization
| Algorithm 1 Modified Stein Variational Gradient Descent Method (MSVGD) |
Input: A group of random particles and target pdf Set the initial state of particles , constant parameter , step size .
Output: The particles that tries to match the goal distribution . |
3.4. MSVGD Algorithm and Its Computational Difficulty
4. Numerical Examples
4.1. Experimental Setups
4.2. Comparison with Different VI Models in Five Data Sets
4.3. Comparison with Different Non-VI Classification Models
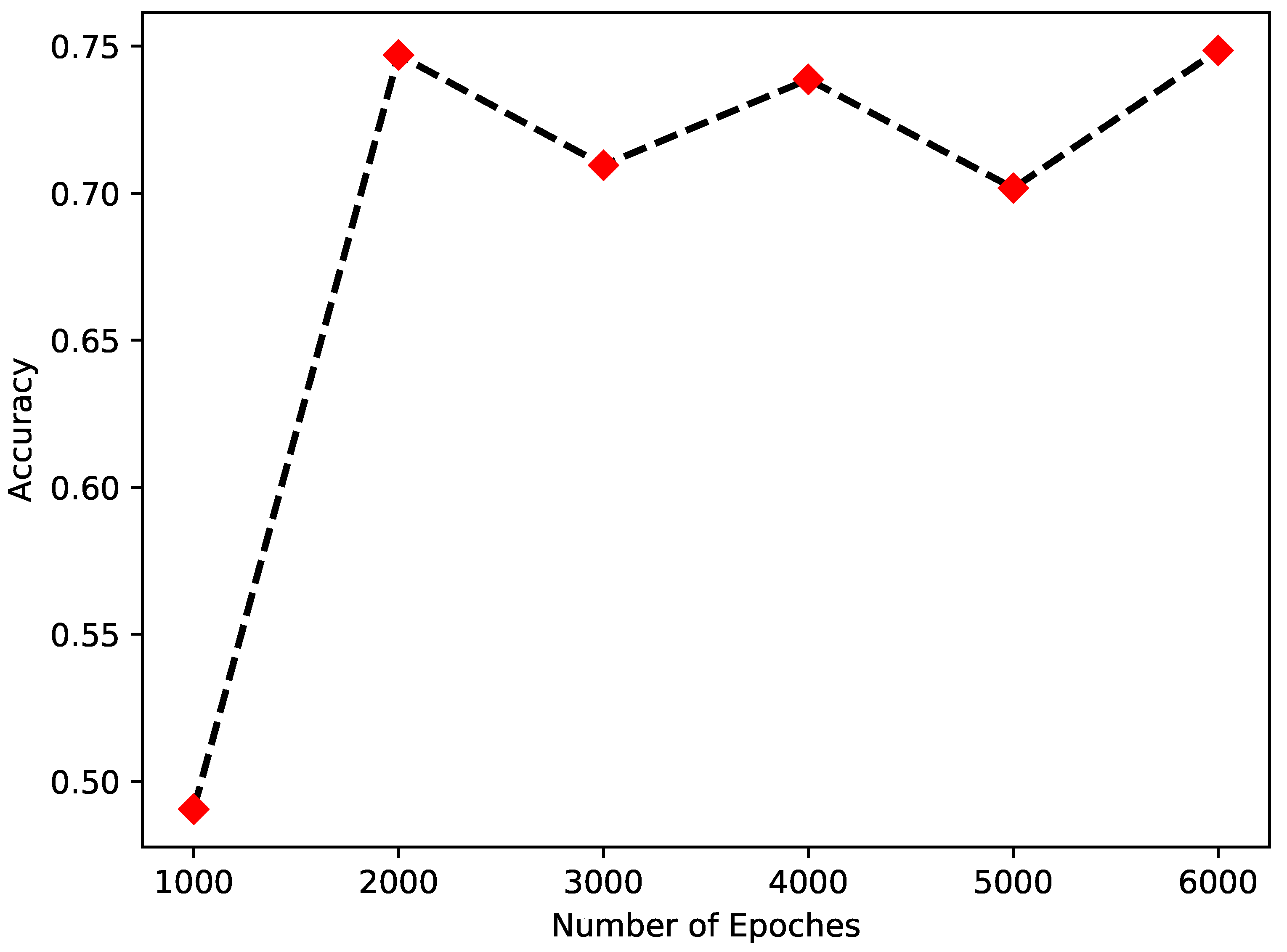
4.4. Analysis of Parameters and Function Q(x) in MSVGD Algorithm
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Attias, H. A variational baysian framework for graphical models. Adv. Neural Inf. Process. Syst. 2000, 12, 209–215. [Google Scholar]
- Puggard, W.; Niwitpong, S.A.; Niwitpong, S. Bayesian Estimation for the Coefficients of Variation of Birnbaum–Saunders Distributions. Symmetry 2021, 13, 2130. [Google Scholar] [CrossRef]
- Wilson, A.G.; Hu, Z.; Salakhutdinov, R.R.; Xing, E.P. Stochastic variational deep kernel learning. Adv. Neural Inf. Process. Syst. 2016, 29, 2586–2594. [Google Scholar]
- Chen, H.; Jiang, B.; Ding, S.X.; Huang, B. Data-driven fault diagnosis for traction systems in high-speed trains: A survey, challenges, and perspectives. IEEE Trans. Intell. Transp. Syst. 2020, 23, 1700–1716. [Google Scholar] [CrossRef]
- Gershman, S.; Hoffman, M.; Blei, D. Nonparametric variational inference. arXiv 2012, arXiv:1206.4665. [Google Scholar]
- Rezende, D.; Mohamed, S. Variational Inference with Normalizing Flows. Int. Conf. Mach. Learn. 2015, 37, 1530–1538. [Google Scholar]
- Liu, Q.; Wang, D. Stein variational gradient descent: A general purpose bayesian inference algorithm. Adv. Neural Inf. Process. Syst. 2016, 29, 2378–2386. [Google Scholar]
- Anderson, J.R.; Peterson, C. A mean field theory learning algorithm for neural networks. Complex Syst. 1987, 1, 995–1019. [Google Scholar]
- Tian, Q.; Wang, W.; Xie, Y.; Wu, H.; Jiao, P.; Pan, L. A Unified Bayesian Model for Generalized Community Detection in Attribute Networks. Complexity 2020, 2020, 5712815. [Google Scholar] [CrossRef]
- Jaakkola, T.; Saul, L.K.; Jordan, M.I. Fast learning by bounding likelihoods in sigmoid type belief networks. Adv. Neural Inf. Process. Syst. 1996, 8, 528–534. [Google Scholar]
- Kullback, S.; Leibler, R.A. On information and sufficiency. Ann. Math. Stat. 1951, 22, 79–86. [Google Scholar] [CrossRef]
- Lopez Quintero, F.O.; Contreras-Reyes, J.E.; Wiff, R.; Arellano-Valle, R.B. Flexible Bayesian analysis of the von Bertalanffy growth function with the use of a log-skew-t distribution. Fishery Bull. 2017, 115, 13–26. [Google Scholar] [CrossRef]
- Murphy, K.; Weiss, Y.; Jordan, M.I. Loopy belief propagation for approximate inference: An empirical study. arXiv 2013, arXiv:1301.6725. [Google Scholar]
- Minka, T.P. Expectation propagation for approximate Bayesian inference. In Proceedings of the Seventeenth Conference on Uncertainty in Artificial Intelligence, Seattle, WA, USA, 2–5 August 2001; pp. 362–369. [Google Scholar]
- Wainwright, M.J.; Jordan, M.I. Graphical Models, Exponential Families, and Variational Inference, Ser. Foundations and Trends in Machine Learning; NOW Publishers: Hanover, MA, USA, 2008; Volume 1. [Google Scholar]
- Fitzgerald, W.J. Markov chain Monte Carlo methods with applications to signal processing. Signal Process. 2001, 81, 3–18. [Google Scholar] [CrossRef]
- Porteous, I.; Newman, D.; Ihler, A.; Asuncion, A.; Smyth, P.; Welling, M. Fast collapsed gibbs sampling for latent dirichlet allocation. In Proceedings of the 14th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, Las Vegas, NV, USA, 24–27 August 2008; pp. 569–577. [Google Scholar]
- Andrieu, C.; Thoms, J. A tutorial on adaptive MCMC. Stat. Comput. 2008, 18, 343–373. [Google Scholar] [CrossRef]
- Angelino, E.; Johnson, M.J.; Adams, R.P. Patterns of Scalable Bayesian Inference. Found. Trends Mach. Learn. 2016, 9, 119–247. [Google Scholar] [CrossRef] [Green Version]
- Blei, D.M.; Kucukelbir, A.; McAuliffe, J.D. Variational inference: A review for statisticians. J. Am. Stat. Assoc. 2017, 112, 859–877. [Google Scholar] [CrossRef] [Green Version]
- Martino, L. A review of multiple try MCMC algorithms for signal processing. Digit. Signal Process. 2018, 75, 134–152. [Google Scholar] [CrossRef] [Green Version]
- Salimans, T.; Kingma, D.; Welling, M. Markov chain monte carlo and variational inference: Bridging the gap. In Proceedings of the International Conference on Machine Learning, Lille, France, 6–11 July 2015; pp. 1218–1226. [Google Scholar]
- Mandt, S.; Hoffman, M.; Blei, D. A variational analysis of stochastic gradient algorithms. In Proceedings of the International Conference on Machine Learning, New York, NY, USA, 20–22 June 2016; pp. 354–363. [Google Scholar]
- Hoffman, M.D.; Blei, D.M.; Wang, C.; Paisley, J. Stochastic variational inference. J. Mach. Learn. Res. 2013, 14, 1303–1347. [Google Scholar]
- Dieng, A.B.; Tran, D.; Ranganath, R.; Paisley, J.; Blei, D. Variational Inference via χ Upper Bound Minimization. In Proceedings of the Advances in Neural Information Processing Systems, Long Beach, CA, USA, 4–9 December 2017; pp. 2732–2741. [Google Scholar]
- Dai, Z.; Damianou, A.; González, J.; Lawrence, N. Variational auto-encoded deep Gaussian processes. arXiv 2015, arXiv:1511.06455. [Google Scholar]
- Jang, E.; Gu, S.; Poole, B. Categorical reparameterization with gumbel-softmax. arXiv 2016, arXiv:1611.01144. [Google Scholar]
- Maddison, C.J.; Mnih, A.; Teh, Y.W. The Concrete Distribution: A Continuous Relaxation of Discrete Random Variables. arXiv 2016, arXiv:1611.00712. [Google Scholar]
- Stein, C. A bound for the error in the normal approximation to the distribution of a sum of dependent random variables. In Proceedings of the Sixth Berkeley Symposium on Mathematical Statistics and Probability, Volume 2: Probability Theory, The Regents of the University of California, Oakland, CA, USA, 1 January 1972. [Google Scholar]
- Wang, Y.; Chen, J.; Liu, C.; Kang, L. Particle-based energetic variational inference. Stat. Comput. 2021, 31, 1–17. [Google Scholar] [CrossRef]
- Liu, Q.; Lee, J.; Jordan, M. A kernelized Stein discrepancy for goodness-of-fit tests. In Proceedings of the International Conference on Machine Learning, New York, NY, USA, 20–22 June 2016; pp. 276–284. [Google Scholar]
- Liu, Y.; Ramachandran, P.; Liu, Q.; Peng, J. Stein variational policy gradient. arXiv 2017, arXiv:1704.02399. [Google Scholar]
- Ranganath, R.; Tran, D.; Altosaar, J.; Blei, D. Operator variational inference. In Proceedings of the Advances in Neural Information Processing Systems, Barcelona, Spain, 5–10 December 2016; pp. 496–504. [Google Scholar]
- Paisley, J.; Blei, D.; Jordan, M. Variational Bayesian inference with stochastic search. arXiv 2012, arXiv:1206.6430. [Google Scholar]
- Jaakkola, T.S.; Jordan, M.I. Bayesian parameter estimation via variational methods. Stat. Comput. 2000, 10, 25–37. [Google Scholar] [CrossRef]
- Tanveer, M.; Tiwari, A.; Choudhary, R.; Jalan, S. Sparse pinball twin support vector machines. Appl. Soft Comput. 2019, 78, 164–175. [Google Scholar] [CrossRef]
- Haque, M.E.; Sudhakar, K.V. ANN back-propagation prediction model for fracture toughness in microalloy steel. Int. J. Fatigue 2002, 24, 1003–1010. [Google Scholar] [CrossRef]
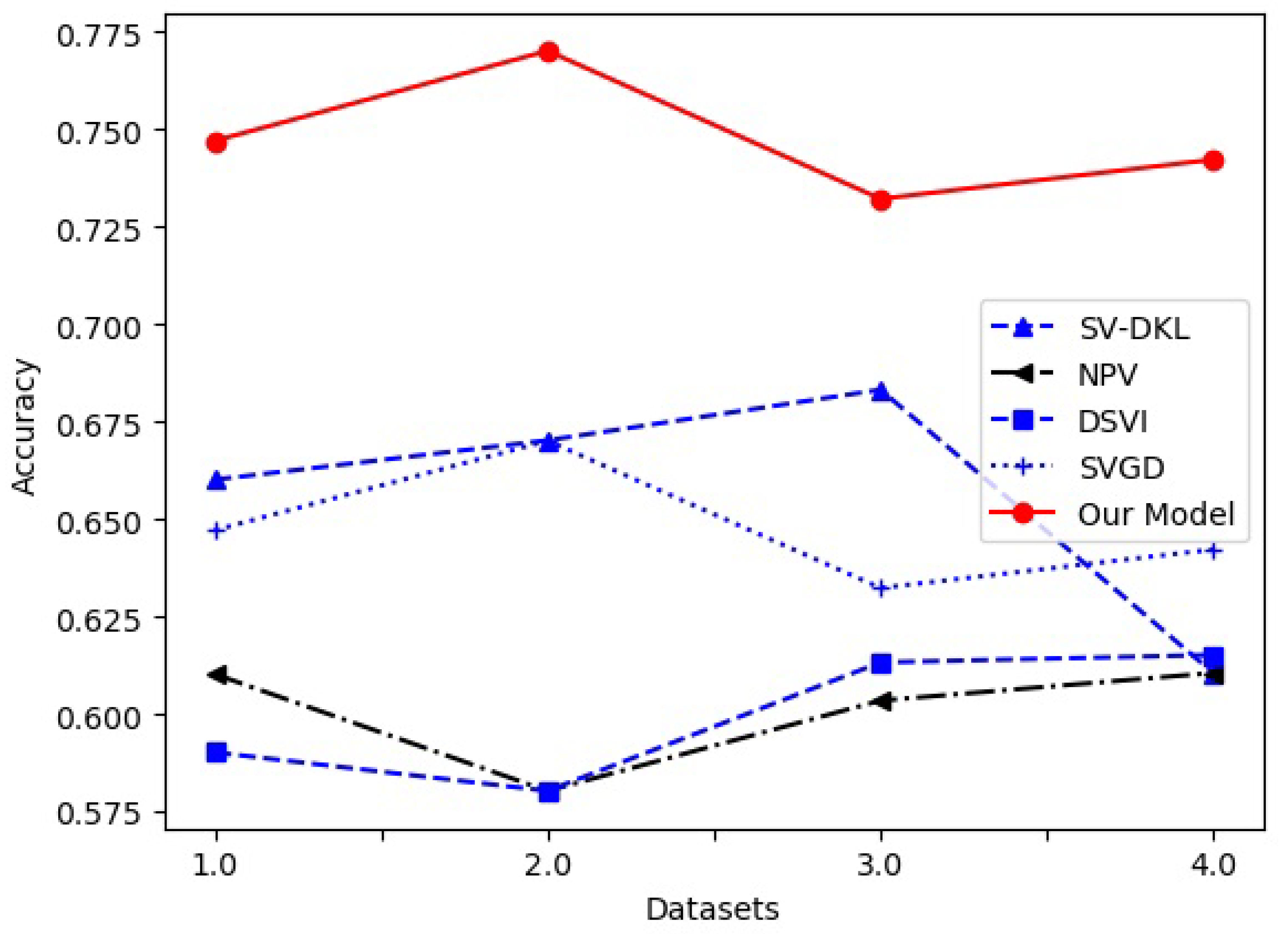
| Model Data | Iris | Pima | Covertype | Heart Disease | p-Value | |
|---|---|---|---|---|---|---|
| SV-DKL [3] | Acc | 0.6601 | 0.6702 | 0.6832 | 0.6104 | 0.010 |
| 0.2662 | 0.2134 | 0.2361 | 0.2415 | |||
| 0.6234 | 0.5915 | 0.6183 | 0.6453 | |||
| Average runtime (s) | 29 | 28 | 72 | 34 | ||
| NPV [5] | Acc | 0.6102 | 0.5802 | 0.6034 | 0.6105 | 0.000 |
| 0.3562 | 0.2536 | 0.2824 | 0.2713 | |||
| 0.5235 | 0.5115 | 0.6355 | 0.5425 | |||
| Average runtime (s) | 30 | 32 | 70 | 30 | ||
| DSVI [6] | Acc | 0.5901 | 0.5802 | 0.6132 | 0.6151 | 0.000 |
| 0.2634 | 0.2456 | 0.2631 | 0.2514 | |||
| 0.7235 | 0.6415 | 0.6883 | 0.7456 | |||
| Average runtime (s) | 26 | 32 | 67 | 30 | ||
| SVGD [31] | Acc | 0.6471 | 0.6701 | 0.6323 | 0.6422 | 0.001 |
| 0.4150 | 0.4456 | 0.4632 | 0.3815 | |||
| 0.7136 | 0.7416 | 0.6114 | 0.7324 | |||
| Average runtime (s) | 25 | 30 | 55 | 27 | ||
| our model | Acc | 0.7471 | 0.7702 | 0.7322 | 0.7423 | 0.000 |
| 0.5151 | 0.5452 | 0.5634 | 0.5814 | |||
| 0.6132 | 0.6414 | 0.6117 | 0.7345 | |||
| Average runtime (s) | 15 | 16 | 34 | 18 |
| Model Data | Iris | Pima | Covertype | Heart Disease |
|---|---|---|---|---|
| SVM [3] | 0.7212 | 0.7545 | 0.7221 | 0.7332 |
| BP [5] | 0.7061 | 0.7134 | 0.7124 | 0.7026 |
| our model | 0.7471 | 0.7702 | 0.7322 | 0.7423 |
| Accuracy | ||||
|---|---|---|---|---|
| Iris | Pima | Covertype | Heart Disease | |
| 0.7 | 0.5514 | 0.5644 | 0.5431 | 0.5322 |
| 0.8 | 0.5764 | 0.5631 | 0.5624 | 0.5521 |
| 0.9 | 0.6562 | 0.6531 | 0.6721 | 0.6620 |
| 1.0 | 0.7061 | 0.7134 | 0.7124 | 0.7026 |
| 1.1 | 0.6762 | 0.6920 | 0.6811 | 0.6825 |
| 1.2 | 0.5861 | 0.5833 | 0.5922 | 0.5924 |
| 1.3 | 0.5471 | 0.5732 | 0.5621 | 0.5470 |
| Q(x) | Accuracy | |||
|---|---|---|---|---|
| Iris | Pima | Covertype | Heart Disease | |
| 0.8 | 0.7212 | 0.7545 | 0.7421 | 0.7332 |
| 0.9 | 0.7061 | 0.7134 | 0.7124 | 0.7026 |
| 1 | 0.7471 | 0.7702 | 0.7322 | 0.7423 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, L.; Dong, J.; Zhang, J.; Yang, J. A Modified Stein Variational Inference Algorithm with Bayesian and Gradient Descent Techniques. Symmetry 2022, 14, 1188. https://doi.org/10.3390/sym14061188
Zhang L, Dong J, Zhang J, Yang J. A Modified Stein Variational Inference Algorithm with Bayesian and Gradient Descent Techniques. Symmetry. 2022; 14(6):1188. https://doi.org/10.3390/sym14061188
Chicago/Turabian StyleZhang, Limin, Jing Dong, Junfang Zhang, and Junzi Yang. 2022. "A Modified Stein Variational Inference Algorithm with Bayesian and Gradient Descent Techniques" Symmetry 14, no. 6: 1188. https://doi.org/10.3390/sym14061188
APA StyleZhang, L., Dong, J., Zhang, J., & Yang, J. (2022). A Modified Stein Variational Inference Algorithm with Bayesian and Gradient Descent Techniques. Symmetry, 14(6), 1188. https://doi.org/10.3390/sym14061188






