Abstract
In situations where several positive random variables cannot be described using symmetrical distributions, a positively asymmetric distribution which has garnered much attention for studying them is the Birnbaum-Saunders (BS) distribution. This distribution was originally proposed to study fatigue over time in materials and has become widely employed for reliability and fatigue studies. In statistics, the coefficient of variation (CV) is employed to measure relative variation. Furthermore, comparing the CVs of several samples from BS distributions is an important approach to assess the variation among them. Herein, we propose estimation methods for the simultaneous confidence intervals (SCIs) for all pairwise differences between the CVs of multiple BS distributions based on the percentile bootstrap, the generalized confidence interval (GCI), the method of variance estimates recovery (MOVER) based on the asymptotic confidence interval (ACI) and GCI, Bayesian credible interval, and the highest posterior density (HPD) interval. The coverage probabilities and average lengths of the proposed methods were examined via a simulation study to determine their performance. The results demonstrate that GCI and the MOVER based on the GCI method provided satisfactory performances in almost every case studied. Particulate matter ≤ 2.5 m (PM2.5) concentration datasets from three areas in northern Thailand were used to illustrate the effectiveness of the proposed methods.
1. Introduction
In practice, several random variables typically regarded as being continuous, non-negative, and random can be represented using a probability distribution. However, the density functions associated with these random variables are frequently asymmetrical and positively skewed, and so neither the normal nor symmetrical distributions can be used to describe them. One positively skewed distribution that has received much attention for describing these random variables is the Birnbaum–Saunders (BS) distribution. This distribution was initially proposed by Birnbaum and Saunders [1] as a failure-time distribution for describing the total amount of time until the damage caused by the development and growth of a dominant crack reaches the threshold at which the component fails. The BS distribution has two positive parameters, which are the shape and scale parameters . A more general derivation of the BS distribution was presented by Desmond [2] based on a biological model. By relaxing the assumptions provided by Birnbaum and Saunders, the author also improved the physical justification for the use of this distribution. Furthermore, the relation of the BS distribution and the inverse Gaussian distribution can be seen in [3].
The BS distribution has been utilized in a variety of applications, including environmental science [4], finance [5], agriculture [6], engineering [2,7], and air pollution [8]. Due to this extensive range of applications, several researchers have provided methods to estimate the parameters of the BS distribution. Birnbaum and Saunders [7] first introduced maximum likelihood estimations (MLEs) for and . Subsequently, Engelhardt et al. [9] determined the asymptotic joint distribution of and and obtained asymptotic confidence intervals (ACIs) for them. Achcar [10] applied Bayesian estimation by approximating the marginal posterior distributions of and . Lu and Chang [11] used the bootstrap approach to construct prediction intervals for future realizations of the BS distribution. Ng et al. [12] proposed modified moment estimators (MMEs) for and and then reduced the bias of their MLEs and MMEs by applying a bias-reduction approach. Wang [13] constructed the generalized confidence interval (GCI) for , the mean, quantiles, and a reliability function of the BS distribution. Wang et al. [14] developed two robust estimators, which are the percentile and the repeated median estimators, for the BS parameters. Wang et al. [15] investigated Bayesian credible intervals for and using the inverse-gamma prior.
The statistical measure of relative dispersion that is the most widely used is the coefficient of variation (CV). Since it is a unit-free measure of variation, it can be used to compare the relative variability of several variables measured in different units. It is calculated as the ratio of the standard deviation to the mean , which is denoted by the formula . The CV has been widely applied in several areas such as medicine, science, economics, and engineering. In practice, the population CV is unknown, and estimating it is usually accomplished using a sample estimate of the CV. In statistics, point estimation and the confidence interval can be used to estimate the characteristic of interest of a population. The confidence interval is more useful than a point estimator because it provides a range of predicted values that most probably contain the unknown population parameter of interest and can also be utilized for hypothesis testing. Therefore, estimating the confidence interval for the difference between the CVs of two independent populations can be extended to compare the dispersion between them. There is no significant difference between them when the confidence interval includes 0. Confidence-interval estimation for the CV and the difference between the CVs has been applied to various distributions. Wong and Wu [16] developed third-order accurate confidence-interval estimation for the CV of non-normal and normal distributions using likelihood-based methods. Pang et al. [17] applied a simulation-based Bayesian approach to obtain confidence-interval estimation for the CV of a three-parameter Weibull distribution. The method of variance estimates recovery (MOVER), GCI, and ACI were introduced by Sangnawakij and Niwitpong [18] to estimate the confidence intervals for the CV and the difference between the CVs of exponential distributions. Recently, Puggard et al. [19] developed confidence-interval estimators for the CV and the difference between the CVs of BS distributions using GCI, bootstrap confidence interval (BCI), BayCrI, and the HPD interval.
For several independent populations, CVs can be compared by constructing simultaneous confidence intervals (SCIs) to examine the differences between them, which simultaneously determines not only which of the CVs are equal but also which differences are significant. Various methods have been developed to construct SCIs for all pairwise differences between the CVs of several populations from various distributions. Thangjai and Niwitpong [20] examined SCIs for all pairwise differences between the CVs of two-parameter exponential distributions utilizing parametric bootstrapping, GCI, and MOVER. Recently, Yosboonruang et al. [21] introduced SCIs for all pairwise differences between the CVs of delta-lognormal distributions using three different methods: fiducial generalized confidence interval (FGCI), the Bayesian approach, and MOVER. However, to the best of our knowledge, there have not been any publications about the statistical comparison of all pairwise differences between the CVs of several BS distributions. Hence, the purpose of this study is to apply the concepts of the percentile bootstrap (PB), GCI, MOVER based on ACI and GCI, BayCrI, and HPD interval approaches to construct SCIs for all pairwise differences between the CVs of several BS distributions.
The rest of the paper is organized as follows. The theoretical background for the BS distribution and the methods for constructing SCIs for all pairwise differences between the CVs of several BS distributions are presented in Section 2. The details of a simulation study and the numerical results are included in Section 3. In Section 4, we use particulate matter ≤ 2.5 m (PM2.5) data from several locations in northern Thailand to illustrate the efficacies of the methods with real data. Finally, the conclusions of the study are presented in the last section.
2. Methods
Let ; be a positive random sample from k independent BS distributions with shape and scale parameters and , respectively, denoted as . Its probability density function (PDF) and cumulative distribution function (CDF) are, respectively, given by
and
with , , where is the CDF of a standard normal distribution. Some important properties associated with are (a) , (b) , and (c) , where refers to a standard normal distribution. Using property (a), the expected value and variance of can be easily obtained as
and
respectively. Similarly, using property (c), it then follows that
and
Following this, the CV for is given by
Therefore, all pairwise differences between the CVs of several samples can be derived as
for and . The procedures for constructing the SCIs for are described in further detail in the following subsection.
2.1. The PB Approach
The application of the bootstrap approach introduced by Efron [22] consists of obtaining a large number of bootstrap samples from original sample and then using the information extracted to improve the inference. There are many bootstrap approaches for estimating the confidence interval of parameters, such as the standard bootstrap, PB, the biased-corrected PB, and the biased-corrected and accelerated bootstrap methods. In this study, the PB method was applied to calculate the SCIs for . Since in Equation (8) is a function of , an estimator for the latter is considered. Let ; be a sample of size taken from a BS distribution. The MMEs for provided by Ng et al. [12] are
Bootstrap samples denoted by of size are drawn with replacement from the initial sample. The corresponding bootstrap (denoted as ) is then calculated using the bootstrap samples. Hence, the bootstrap estimator for can be expressed as
Suppose that B bootstrap samples are available, then B values for can be derived and arranged from the smallest to the largest (denoted by ), which provides the empirical bootstrap distribution of . From the ordered collection of —for —the SCIs for based on the PB approach can be written as , where stands for the th percentiles of . Moreover, these SCIs have the asymptotic coverage property [23]. Algorithm 1 demonstrates the computation of SCIs based on the PB approach.
| Algorithm 1 The PB approach |
|
2.2. The GCI Approach
GCI was initially introduced by Weerahandi [24]. Let Y be an observable random variable and the PDF of Y be , where y is the observed value of random variable Y, is the parameter of interest, and is a nuisance parameter. Suppose that we are interested in constructing the confidence interval for , then let be a function of . is the generalized pivotal quantity (GPQ) for if satisfies the following two conditions:
- The observed value of denoted as is free of nuisance parameter .
- The probability distribution of is free of unknown parameters.
Therefore, the GCI for is given by , where stands for the th percentiles of Q.
Now, we develop the SCIs for using the concept of GCI. Let be a random sample from a BS distribution with and . According to Sun [25] and Wang [13], the respective GPQs for and are given by
and
where are the observed values of , has a t-distribution with degrees of freedom (), , , and has a Chi-squared distribution with degrees of freedom (). In Equation (11), and are the two solutions of the following equation:
where , , and . Following this, the GPQs for can be derived as
Subsequently, the GPQs for can be expressed as
for and . Hence, the SCIs for based on the GCI approach can be written as , where stands for the th percentiles of . The asymptotic coverage probability of the SCIs based on the GCI approach was proven by Thangjai and Niwitpong [20]. The steps to compute SCIs based on the GCI approach are shown in Algorithm 2.
| Algorithm 2 The GCI approach |
|
2.3. The MOVER Approach
MOVER approach was used by Zou and Donner [26] to construct a closed-form approximation of the confidence interval for the differences between parameters ; for and based on the confidence intervals of the individual parameters. To consider the differences between parameters , let and be independent unbiased estimates of and , respectively. Furthermore, let and denote the confidence interval for and . Therefore, the MOVER confidence interval for can be expressed as
and
2.3.1. The MOVER Based on ACI Approach
From Equation (7), the estimates for are
where is provided by Equation (9). According to Puggard et al. [27], the estimates of the variances for can be written as
Following this, the ACI for becomes
Therefore, the SCIs for using MOVER based on ACI can be defined by
Algorithm 3 describes the procedures for constructing SCIs using MOVER based on ACI approach.
| Algorithm 3 The MOVER based on ACI approach |
2.3.2. The MOVER Based on GCI Approach
The GPQ for is provided by Equation (14). Subsequently, the GCI for can be expressed as
where stands for the th percentiles of . Therefore, the the SCIs for using MOVER based on GCI becomes
Algorithm 4 summarizes the steps for constructing SCIs using MOVER based on GCI approach. Note that the asymptotic coverage property of the SCIs based on the MOVER based on ACI or GCI approaches is provided in [20,21,23].
| Algorithm 4 The MOVER based on GCI approach |
2.4. The BayCrI Approach
The Bayesian method permits statistical inference on a parameter based on data obtained from two different sources: experimentally (via the likelihood function) and expert knowledge (via a prior distribution). Recently, the application of Bayesian method can be seen in [28,29]. For the BS distribution, Xu and Tang [30] proved that the reference prior, which is also a type of independent Jeffreys prior, is not suitable for Bayesian estimation because it results in an improper posterior distribution. To ensure the propriety of the posterior, Wang et al. [15] proposed using proper priors with known hyperparameters to estimate the confidence intervals of the parameters from a BS distribution.
Suppose that the prior distribution for is an inverse-gamma distribution with parameters and , denoted as . Furthermore, an inverse-gamma distribution with parameters and is assumed to be the prior distribution for , denoted as ). According to Equation (1), the likelihood function is given by
By combining the likelihood function in Equation (24) and the prior distribution, the joint posterior distribution of the parameters is given by
From Equation (25), the marginal posterior distribution of given the data is derived as
Subsequently, the posterior conditional distribution of given is
A Markov-chain Monte Carlo was employed to obtain posterior samples using Equations (26) and (27). Since the marginal posterior distribution of (26) is mathematically intractable, the generalized ratio-of-uniforms method of Wakefield et al. [31] was used to generate the posterior sample for (denoted as ) [15]. The details of this method are explained as follows.
Suppose the pair of variables is uniformly distributed over
where is given by Equation (26) and is a constant. Subsequently, has PDF . The accept-reject technique using convenient 1D enveloping rectangle is often applied to draw random points uniformly distributed in , where
and
and are finite, while [15]. Therefore, the generalized ratio-of-uniforms method for generating the posterior sample from Equation (26) consists of the following three steps:
- (1)
- Calculate and .
- (2)
- Simulate and from and , where refer to a uniform distribution with parameters s and t, then compute .
- (3)
- If , set , otherwise go back to step (2).
From Equation (27), the values are obtained by applying the package from the R software suite. Since the posterior sample for (denoted as ) is the square root of (), the Bayesian estimators for can be written as
Therefore, the SCIs for based on the BayCrI approach can be written as , where stands for the th percentiles of . The asymptotic coverage property of the SCIs based on the BayCrI approach was proven by Yosboonruang et al. [21]. Furthermore, we also applied the package (version 0.2.2) from the R software suite to calculate the SCIs based on the HPD interval for . The HPD interval has the property that the density of every point within the interval is higher than that of every point outside of it, and so provides the shortest length for a given probability level [32]. Algorithm 5 was used to obtain the SCIs based on the BayCrI and HPD approaches.
| Algorithm 5 The BayCrI and HPD approaches |
|
3. Simulation Study Settings and Results
The performances of the proposed methods were evaluated via a Monte Carlo simulation study under the criteria of the average length and the coverage probability of the SCI based on 3000 replications with 3000 pivotal quantities for GCI, for PB, and for BayCrI and the HPD interval. The method that performs the best for a given scenario is the one with a coverage probability higher than or close to 0.95 and the shortest average length. We compare the effectiveness of the proposed methods in various circumstances. Therefore, we considered k = 3, 5 and 10 sample cases. The settings for parameters and sample sizes are reported in Table 1. Meanwhile, the values of the scale parameters were kept fixed at 1.0 without any loss of generality. In addition, and were chosen for constructing BayCrI and the HPD interval [15]. In Table 2, Table 3 and Table 4 and Figure 1, Figure 2 and Figure 3, PB is the percentile bootstrap approach, GCI is the generalized confidence interval approach, MOVER1 is the MOVER based on ACI approach, MOVER2 is the MOVER based on GCI approach, BayCrI is the Bayesian credible interval approach, and HPD is the highest posterior density interval approach.

Table 1.
Parameter settings for sample cases , or 10.

Table 2.
The coverage probabilities and the average lengths of the 95% SCIs for all pairwise differences between the CVs of three BS distributions ().

Table 3.
The coverage probabilities and the average lengths of the 95% SCIs for all pairwise differences between the CVs of five BS distributions .

Table 4.
The coverage probabilities and the average lengths of the 95% SCIs for all pairwise differences between the CVs of 10 BS distributions .
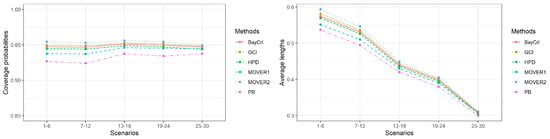
Figure 1.
A summary of the coverage probabilities and the average lengths of the methods in Table 2.
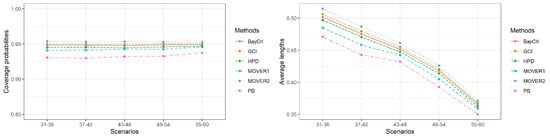
Figure 2.
A summary of the coverage probabilities and the average lengths of the methods in Table 3.
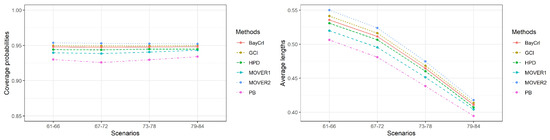
Figure 3.
A summary of the coverage probabilities and the average lengths of the methods in Table 4.
The results for , or 10 are presented in Table 2, Table 3 and Table 4, respectively. In addition, Figure 1, Figure 2 and Figure 3 summarize the coverage probabilities and the average lengths of the methods in Table 2, Table 3 and Table 4, respectively. As the simulation results of these three scenarios were similar, we can draw the following conclusions. Overall, the average lengths of all of the methods tended to decrease when the sample sizes were increased, and, in contrast, when shape parameter was increased. The coverage probabilities of PB, MOVER based on ACI, BayCrI, and the HPD interval were slightly below 0.95 in almost of the situations whereas those for GCI and MOVER based on GCI were above or close to 0.95 under all circumstances, even for a high value. In addition, the coverage probabilities of these two methods were very stable at 0.95. The average lengths reported in Table 2, Table 3 and Table 4 indicated that although those of PB, MOVER based on ACI, BayCrI, and the HPD interval were shorter than GCI and MOVER based on GCI, their coverage probabilities were slightly lower than 0.95 in almost every case, and, as a result, they did not perform satisfactorily. Among the remaining methods, GCI achieved the shortest average lengths in almost every situation. However, the differences in average length between GCI and MOVER based on GCI were very small.
4. Empirical Application of the Methods with Three Real Datasets
The average daily PM2.5 concentrations datasets from Lamphun, Mae Hong Son, and Nan in northern Thailand were used to evaluate the performances of the proposed methods for constructing the SCIs for all pairwise differences between the CVs of three BS distributions (Table 5). These data were monitored in March 2019 by the Thailand Pollution Control Department [33]. Since the PM2.5 concentration data consist of positive values, they could be modeled using a lognormal, gamma, exponential, Weibull, or BS distribution. Therefore, it is important to determine the best-fitting distribution of these data, for which the minimum Akaike information criterion (AIC) and Bayesian information criterion (BIC) were utilized. The results in Table 6 and Table 7 indicate that the best-fitting distribution for all three PM2.5 concentration datasets is the BS distribution. The summary statistics of the PM2.5 concentration datasets from three different areas in northern Thailand are reported in Table 8. We considered and ; for constructing the SCIs based on BayCrI and the HPD interval.

Table 5.
PM2.5 concentration data from three different areas in northern Thailand.

Table 6.
AIC values from the fitting of five candidate distributions.

Table 7.
BIC values for the fitting of five candidate distributions.

Table 8.
Summary statistics for the PM2.5 concentration data from three different areas in northern Thailand.
The 95% SCIs based on PB, GCI, MOVER based on ACI or GCI, BayCrI, and the HPD interval for all pairwise differences between the CVs are summarized in Table 9. The average lengths of PB, GCI, MOVER based on ACI or GCI, BayCrI, and the HPD interval were 0.2695, 0.3940, 0.3533, 0.3998, 0.3919, and 0.3644, respectively. According to these results, PB provided the shortest average lengths, followed by MOVER based on ACI, whereas MOVER based on GCI provided the longest ones, followed by GCI, which is in good agreement with the results from the simulation study for = (30, 30, 30) and = (0.5, 0.5, 0.5). However, one should be aware that GCI and MOVER based on GCI provided satisfactory coverage probabilities whereas PB, MOVER based on ACI, BayCrI, and the HPD interval did not. Therefore, when considering the coverage probability and average length together, GCI and MOVER based on GCI are recommended for constructing the SCIs for all pairwise differences between the CVs of PM2.5 concentration datasets from the three areas in northern Thailand.

Table 9.
The 95% SCIs for all pairwise differences between the CVs of PM2.5 concentration data from three different areas in northern Thailand.
5. Conclusions
We developed SCIs for all pairwise differences between the CVs of multiple BS distributions using PB, GCI, MOVER based on ACI or GCI, BayCrI, and the HPD interval. Their efficacies were assessed using a Monte Carlo simulation based on their coverage probabilities and average lengths under various scenarios. Based on the findings of this, GCI and MOVER based on GCI provided satisfactory performance in most situations studied. Moreover, the proposed methods were applied to construct the SCIs for all pairwise differences between the CVs of PM2.5 concentration datasets from three different areas in northern Thailand, the results of which were similar to those obtained from the simulation study. As a result, GCI and MOVER based on GCI are recommended when considering the coverage probability and average length together. In practice, several random variables are not independent of each other. Therefore, correlated data that follows the BS distributions is of interest for future research.
Author Contributions
Conceptualization, S.N.; Data curation, W.P.; Formal analysis, W.P. and S.N.; Funding acquisition, S.N.; Investigation, S.-A.N. and S.N.; Methodology, S.-A.N. and S.N.; Project administration, S.-A.N.; Resources, S.-A.N.; Software, W.P.; Supervision, S.-A.N. and S.N.; Visualization, S.-A.N.; Writing—original draft, W.P.; Writing—review and editing, S.-A.N. and S.N. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by National Science, Research and Innovation Fund (NSRF), and King Mongkut’s University of Technology North Bangkok with Contract no. KMUTNB–FF–65–22.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data of PM2.5 concentration were obtained from the Pollution Control Department [33].
Acknowledgments
The first author wishes to express gratitude for financial support provided by the Thailand Science Achievement Scholarship (SAST).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Birnbaum, Z.W.; Saunders, S.C. A new family of life distributions. J. Appl. Probab. 1969, 6, 319–327. [Google Scholar] [CrossRef]
- Desmond, A.F. Stochastic models of failure in random environments. Can. J. Stat. 1985, 13, 171–183. [Google Scholar] [CrossRef]
- Desmond, A.F. On the relationship between two fatigue-life model. IEEE Trans. Reliab. 1986, 35, 167–169. [Google Scholar] [CrossRef]
- Leiva, V.; Marchant, C.; Ruggeri, F.; Saulo, H. A criterion for environmental assessment using Birnbaum–Saunders attribute control charts. Environmetrics 2015, 26, 463–476. [Google Scholar] [CrossRef]
- Marchant, C.; Bertin, K.; Leiva, V.; Saulo, H. Generalized Birnbaum-Saunders kernel density estimators and an analysis of financial data. Comput. Stat. Data Anal. 2013, 63, 1–15. [Google Scholar] [CrossRef]
- Lio, Y.L.; Tsai, T.R.; Wu, S.J. Acceptance sampling plans from truncated life tests based on the Birnbaum–Saunders distribution for percentiles. Commun. Stat. Simul. Comput. 2010, 39, 119–136. [Google Scholar] [CrossRef]
- Birnbaum, Z.W.; Saunders, S.C. Estimation for a family of life distributions with applications to fatigue. J. Appl. Probab. 1969, 6, 328–347. [Google Scholar] [CrossRef]
- Leiva, V.; Barros, M.; Paula, G.A.; Sanhueza, A. Generalized Birnbaum–Saunders distributions applied to air pollutant concentration. Environmetrics 2008, 19, 235–249. [Google Scholar] [CrossRef]
- Engelhardt, M.; Bain, L.J.; Wright, F.T. Inference on the parameters of the Birnbaum–Saunders fatigue life distribution based on maximum likelihood estimation. Technometrics 1981, 23, 251–255. [Google Scholar] [CrossRef]
- Achcar, J. Inferences for the Birnbaum-Saunders fatigue life model using Bayesian methods. Comput. Stat. Data Anal. 1993, 15, 367–380. [Google Scholar] [CrossRef]
- Lu, M.; Chang, D.S. Bootstrap prediction intervals for the Birnbaum–Saunders distribution. Microelectron. Reliab. 1997, 37, 1213–1216. [Google Scholar] [CrossRef]
- Ng, H.K.T.; Kundu, D.; Balakrishnan, N. Modified moment estimation for the two-parameter Birnbaum–Saunders distribution. Comput. Stat. Data Anal. 2003, 43, 283–298. [Google Scholar] [CrossRef]
- Wang, B.X. Generalized interval estimation for the Birnbaum-Saunders distribution. Comput. Stat. Data Anal. 2012, 56, 4320–4326. [Google Scholar] [CrossRef]
- Wang, M.; Zhao, J.; Sun, X.; Park, C. Robust explicit estimation of the two-parameter Birnbaum–Saunders distribution. J. Appl. Stat. 2013, 40, 2259–2274. [Google Scholar] [CrossRef]
- Wang, M.; Sun, X.; Park, C. Bayesian analysis of Birnbaum-Saunders distribution via the generalized ratio-of-uniforms method. Comput. Stat. 2016, 31, 207–225. [Google Scholar] [CrossRef]
- Wong, A.C.M.; Wu, J. Small sample asymptotic inference for the coefficient of variation: Normal and nonnormal models. J. Stat. Plan. Inference 2002, 104, 73–82. [Google Scholar] [CrossRef]
- Pang, W.K.; Leung, P.K.; Huang, W.K.; Liu, W. On interval estimation of the coefficient of variation for the three-parameter weibull, lognormal and gamma distribution:a simulation-based approach. Eur. J. Oper. Res. 2005, 164, 367–377. [Google Scholar] [CrossRef]
- Sangnawakij, P.; Niwitpong, S.-A. Confidence intervals for coefficients of variation in two-parameter exponential distributions. Commun. Stat.-Simul. A Comput. 2017, 46, 6618–6630. [Google Scholar] [CrossRef]
- Puggard, W.; Niwitpong, S.A.; Niwitpong, S. Bayesian estimation for the coefficients of variation of Birnbaum–Saunders distributions. Symmetry 2021, 13, 2130. [Google Scholar] [CrossRef]
- Thangjai, W.; Niwitpong, S. Simultaneous confidence intervals for all differences of coefficients of variation of two-parameter exponential distributions. Thail. Stat. 2020, 18, 135–149. [Google Scholar]
- Yosboonruang, N.; Niwitpong, S.A.; Niwitpong, S. Simultaneous confidence intervals for all pairwise differences between the coefficients of variation of rainfall series in Thailand. PeerJ 2021, 9, e11651. [Google Scholar] [CrossRef] [PubMed]
- Efron, B. Bootstrap methods: Another look at the jackknife. Ann. Stat. 1979, 7, 1–26. [Google Scholar] [CrossRef]
- Thangjai, W.; Niwitpong, S.A.; Niwitpong, S. Simultaneous confidence intervals for all differences of coefficients of variation of log-normal distributions. Hacet. J. Math. Stat. 2019, 48, 1505–1521. [Google Scholar]
- Weerahandi, S. Generalized Inference in Repeated Measures: Exact Methods in MANOVA and Mixed Models; Wiley: Boston, MA, USA, 2004. [Google Scholar]
- Sun, Z.L. The confidence intervals for the scale parameter of the Birnbaum–Saunders fatigue life distribution. Acta Armamentarii. 2009, 30, 1558–1561. [Google Scholar]
- Zou, G.Y.; Donner, A. Construction of confidence limits about effect measures: A general approach. Stat. Med. 2008, 27, 1693–1702. [Google Scholar] [CrossRef]
- Puggard, W.; Niwitpong, S.A.; Niwitpong, S. Confidence intervals for common coefficient of variation of several Birnbaum–Saunders distributions. Symmetry 2022, 14, 2101. [Google Scholar] [CrossRef]
- Saunders, R.; Rawlings, A.; Birnbaum, A.; Iliopoulos, A.; Michopoulos, J.; Lagoudas, D.; Elwany, A. Additive Manufacturing Melt Pool Prediction and Classification via Multifidelity Gaussian Process Surrogates. Integr. Mater. Manuf. Innov. 2022, 11, 497–515. [Google Scholar] [CrossRef]
- Saunders, R.; Butler, C.; Michopoulos, J.; Lagoudas, D.; Elwany, A.; Bagchi, A. Mechanical behavior predictions of additively manufactured microstructures using functional Gaussian process surrogates. NPJ Comput. Mater. 2021, 7, 81. [Google Scholar] [CrossRef]
- Xu, A.; Tang, Y. Bayesian analysis of Birnbaum–Saunders distribution with partial information. Comput. Stat. Data Anal. 2011, 55, 2324–2333. [Google Scholar] [CrossRef]
- Wakefield, J.C.; Gelfand, A.E.; Smith, A.F.M. Efficient generation of random variates via the ratio-of-uniforms method. Stat. Comput. 1991, 1, 129–133. [Google Scholar] [CrossRef]
- Box, G.E.P.; Tiao, G.C. Bayesian Inference in Statistical Analysis; Wiley: New York, NY, USA, 1992. [Google Scholar]
- Reports on Smog Situation in the North Home Page. Available online: http://www.pcd.go.th/ (accessed on 20 October 2022). (In Thai).
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).