An fMRI Feature Selection Method Based on a Minimum Spanning Tree for Identifying Patients with Autism
Abstract
1. Introduction
- (1)
- We constructed a minimum spanning tree from the original dataset and fully consider the effect of the feature context on classification to select the optimal feature subset.
- (2)
- In combination with the feature selection method we proposed to identify patients with ASD, the calculation of the model is simplified, and the recognition accuracy is improved.
- (3)
- We identified abnormal brain regions in patients with ASD by counting the regions with more frequent occurrences in the optimal feature subset. These abnormal brain regions provide important reference information for clinical decision-making.
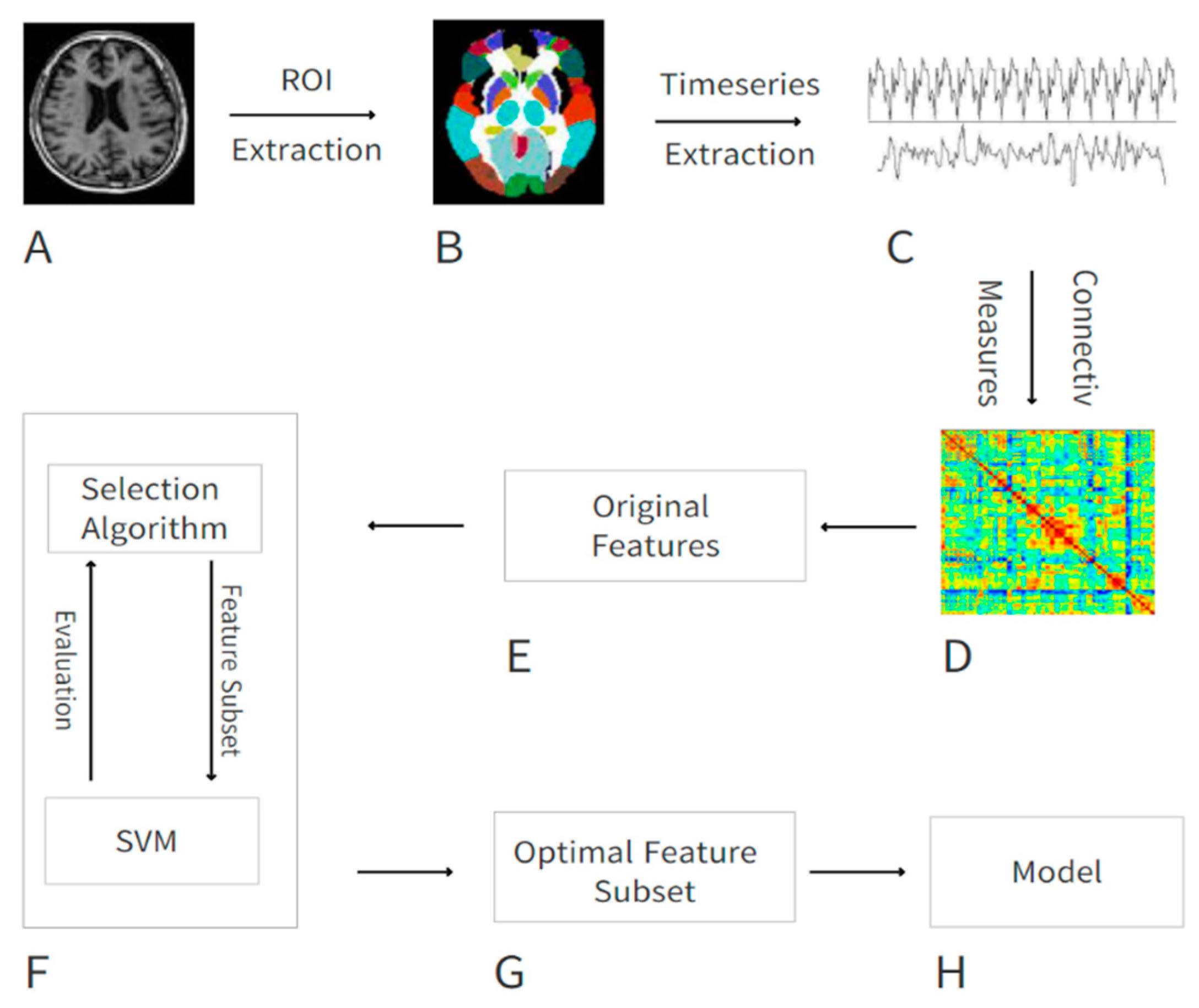
2. Materials and Methods
2.1. Demographic Information
2.2. Data Preprocessing
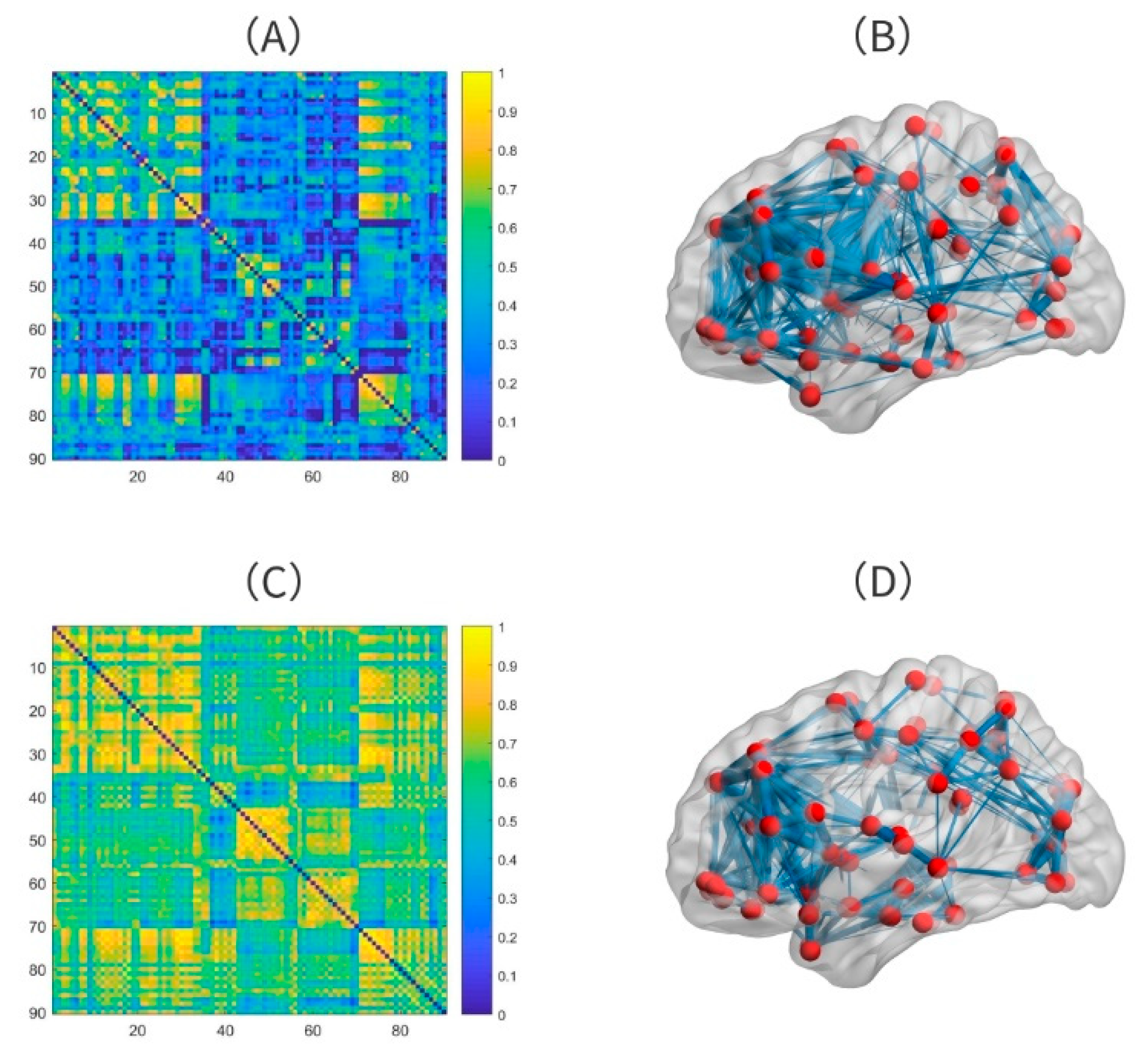
2.3. Features from Function Connectivity
2.4. Feature Selection
- (1)
- Construct an undirected weight feature graph
- (2)
- Build the minimum spanning tree
| Algorithm 1 Prim algorithm for MST |
| Input: The feature graph . Output: The edge set W of the minimum spanning tree. 1. BEGIN 2. Initialize: Set an empty set S and an edge set W. Assign a key value as INFINITE to all vertices in the input graph. 3. Assign key value as 0 for the first vertex. 4. while S doesn’t include all vertices do 5. (1) Pick a vertex u which is not there in S and has minimum key value. 6. (2) Add an edge (u,j) to W which j is the vertex with the smallest weight connected with u in S. 7. (3) Include u to S. 8. (4) Update key value of all adjacent vertices of u. 9. end while. 10. END BEGIN |
- (3)
- Select features
2.5. Classification Method and Evaluation of Performance
3. Results
3.1. Performance of the Model
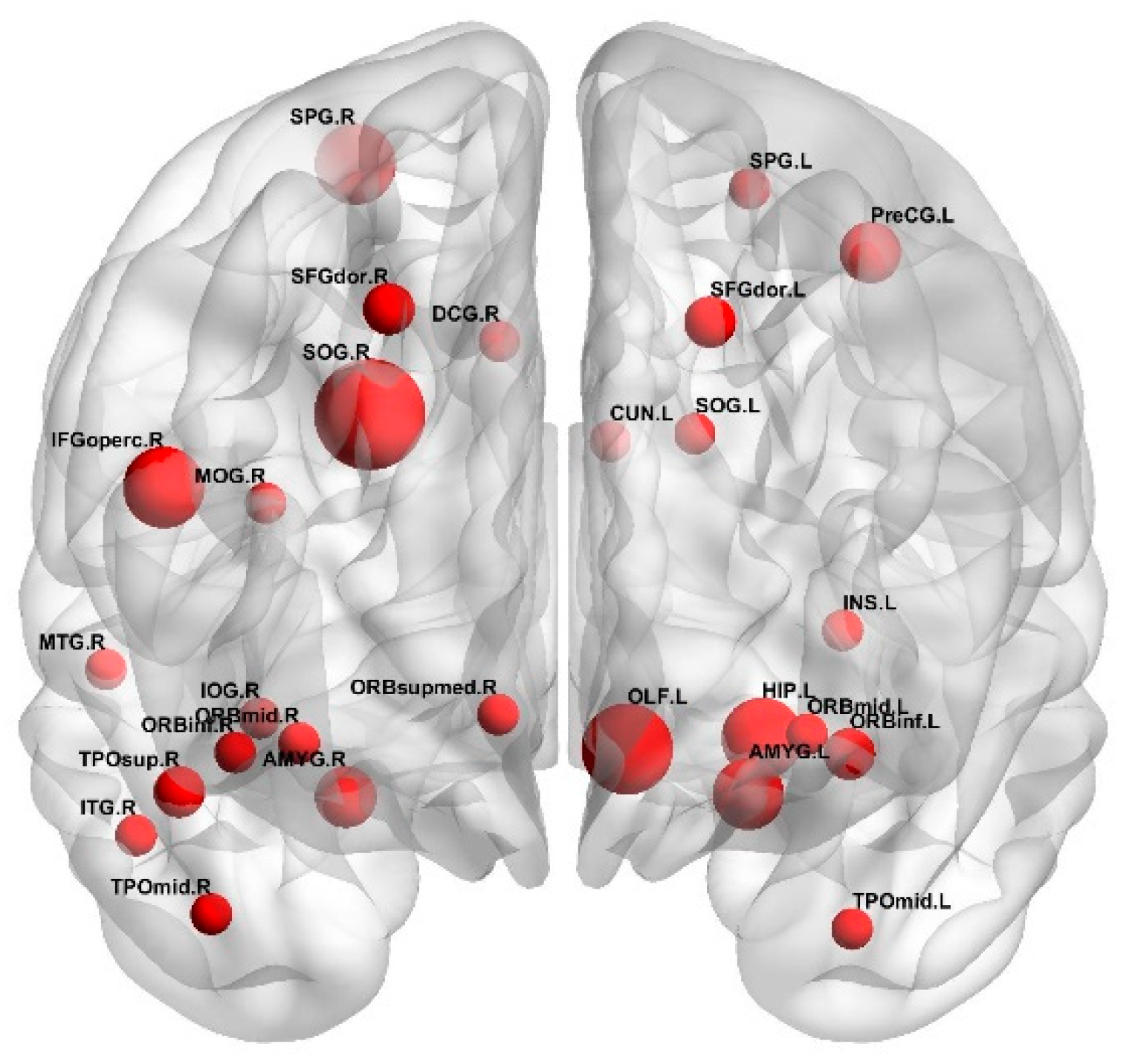
3.2. The Optimal Feature Set and the Abnormal Brain Regions
4. Discussion
4.1. Classification Effect
4.2. Comparison with Other Feature Selection Methods
4.3. Analysis of the Brain Regions with Greater Weight
- Superior occipital gyrus
- Olfactory cortex
- Inferior frontal gyrus
- Hippocampus
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Frith, U.; Happé, F. Autism spectrum disorder. Curr. Biol. 2005, 15, R786–R790. [Google Scholar] [CrossRef]
- Kong, Y.; Gao, J.; Xu, Y.; Pan, Y.; Wang, J.; Liu, J. Classification of autism spectrum disorder by combining brain connectivity and deep neural network classifier. Neurocomputing 2019, 324, 63–68. [Google Scholar] [CrossRef]
- Zecavati, N.; Spence, S.J. Neurometabolic disorders and dysfunction in autism spectrum disorders. Curr. Neurol. Neurosci. Rep. 2009, 9, 129–136. [Google Scholar] [CrossRef]
- Amaral, D.G.; Schumann, C.M.; Nordahl, C.W. Neuroanatomy of autism. Trends Neurosci. 2008, 31, 137–145. [Google Scholar] [CrossRef]
- Khundrakpam, B.S.; Lewis, J.D.; Kostopoulos, P.; Carbonell, F.; Evans, A.C. Cortical Thickness Abnormalities in Autism Spectrum Disorders Through Late Childhood, Adolescence, and Adulthood: A Large-Scale MRI Study. Cereb. Cortex 2017, 27, 1721–1731. [Google Scholar] [CrossRef]
- Zablotsky, B.; Black, L.I.; Maenner, M.J.; Schieve, L.A.; Blumberg, S.J. Estimated prevalence of autism and other developmental disabilities following questionnaire changes in the 2014 national health interview survey. Natl. Health Stat. Rep. 2015, 87, 1–20. [Google Scholar]
- Karten, A.; Hirsch, J. Brief report: Anomalous neural deactivations and functional connectivity during receptive language in autism spectrum disorder: A functional MRI study. J. Autism Dev. Disord. 2015, 45, 1905–1914. [Google Scholar] [CrossRef]
- Fernell, E.; Eriksson, M.A.; Gillberg, C. Early diagnosis of autism and impact on prognosis: A narrative review. Clin. Epidemiol. 2013, 5, 33–43. [Google Scholar] [CrossRef] [PubMed]
- Mwangi, B.; Ebmeier, K.P.; Matthews, K.; Steele, J.D. Multi-centre diagnostic classification of individual structural neuroimaging scans from patients with major depressive disorder. Brain J. Neurol. 2012, 135, 1508–1521. [Google Scholar] [CrossRef] [PubMed]
- Plitt, M.; Barnes, K.A.; Martin, A. Functional connectivity classification of autism identifies highly predictive brain features but falls short of biomarker standards. NeuroImage Clin. 2015, 7, 359–366. [Google Scholar] [CrossRef] [PubMed]
- Bosl, W.J.; Tager-Flusberg, H.; Nelson, C.A. EEG Analytics for Early Detection of Autism Spectrum Disorder: A data-driven approach. Sci. Rep. 2018, 8, 6828. [Google Scholar] [CrossRef] [PubMed]
- Pereira, A.M.; Campos, B.M.; Coan, A.C.; Pegoraro, L.F.; de Rezende, T.J.R.; Obeso, I.; Dalgalarrondo, P.; da Costa, J.C.; Dreher, J.; Cendes, F. Differences in Cortical Structure and Functional MRI Connectivity in High Functioning Autism. Front. Neurol. 2018, 9, 539. [Google Scholar] [CrossRef]
- Van den Heuvel, M.P.; Hulshoff Pol, H.E. Exploring the brain network: A review on resting-state fMRI functional connectivity. Eur. Neuropsychopharmacol. 2010, 20, 519–534. [Google Scholar] [CrossRef]
- Bi, X.; Wang, Y.; Shu, Q.; Sun, Q.; Xu, Q. Classification of Autism Spectrum Disorder Using Random Support Vector Machine Cluster. Front. Genet. 2018, 9, 18. [Google Scholar] [CrossRef]
- Travers, B.G.; Adluru, N.; Ennis, C.; Tromp, D.P.M.; Destiche, D.; Doran, S.; Bigler, E.D.; Lange, N.; Lainhart, J.E.; Alexander, A.L. Diffusion tensor imaging in autism spectrum disorder: A review. Autism Res. 2012, 5, 289–313. [Google Scholar] [CrossRef]
- Ardekani, B.A.; Tabesh, A.; Sevy, S.; Robinson, D.G.; Bilder, R.M.; Szeszko, P.R. Diffusion tensor imaging reliably differentiates patients with schizophrenia from healthy volunteers. Hum. Brain Mapp. 2011, 32, 1–9. [Google Scholar] [CrossRef]
- Kaufmann, T.; Skåtun, K.C.; Alnæs, D.; Brandt, C.L.; Doan, N.T.; Agartz, I.; Melle, I.S.; Andreassen, O.A.; Westlye, L.T. Disintegration of sensorimotor brain networks in schizophrenia. Eur. Psychiatry 2016, 33, S33–S34. [Google Scholar] [CrossRef][Green Version]
- Zhao, F.; Zhang, H.; Rekik, I.; An, Z.; Shen, D. Diagnosis of Autism Spectrum Disorders Using Multi-Level High-Order Functional Networks Derived From Resting-State Functional MRI. Front. Hum. Neurosci. 2018, 12, 184. [Google Scholar] [CrossRef]
- Jin, B.; Strasburger, A.; Laken, S.J.; Kozel, F.A.; Johnson, K.A.; George, M.S.; Lu, X. Feature selection for fMRI-based deception detection. BMC Bioinform. 2009, 10 (Suppl. 9), S15. [Google Scholar] [CrossRef]
- Guo, X.; Dominick, K.C.; Minai, A.A.; Li, H.; Erickson, C.A.; Lu, L.J. Diagnosing Autism Spectrum Disorder from Brain Resting-State Functional Connectivity Patterns Using a Deep Neural Network with a Novel Feature Selection Method. Front. Neurosci. 2017, 11, 460. [Google Scholar] [CrossRef]
- Dyrba, M.; Grothe, M.; Kirste, T.; Teipel, S.J. Multimodal analysis of functional and structural disconnection in Alzheimer’s disease using multiple kernel SVM. Hum. Brain Mapp. 2015, 36, 2118–2131. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Cheng, C.; Cao, X.; Xiang, J.; Chen, J.; Zhang, K. Resting-state functional connectivity abnormalities in first-onset unmedicated depression. Neural Regen. Res. 2014, 9, 153–163. [Google Scholar] [PubMed]
- Fekete, T.; Wilf, M.; Rubin, D.; Edelman, S.; Malach, R.; Mujica-Parodi, L.R. Combining classification with fMRI-derived complex network measures for potential neurodiagnostics. PLoS ONE 2013, 8, e62867. [Google Scholar] [CrossRef] [PubMed]
- Bassett, D.S.; Nelson, B.G.; Mueller, B.A.; Camchong, J.; Lim, K.O. Altered resting state complexity in schizophrenia. Neuroimage 2012, 59, 2196–2207. [Google Scholar] [CrossRef] [PubMed]
- Arbabshirani, M.R.; Plis, S.; Sui, J.; Calhoun, V.D. Single subject prediction of brain disorders in neuroimaging: Promises and pitfalls. Neuroimage 2017, 145, 137–165. [Google Scholar] [CrossRef]
- Rakić, M.; Cabezas, M.; Kushibar, K.; Oliver, A.; Lladó, X. Improving the detection of autism spectrum disorder by combining structural and functional MRI information. NeuroImage Clin. 2020, 25, 102181. [Google Scholar] [CrossRef]
- Khazaee, A.; Ebrahimzadeh, A.; Babajani-Feremi, A. Identifying patients with Alzheimer’s disease using resting-state fMRI and graph theory. Clin. Neurophysiol. 2015, 126, 2132–2141. [Google Scholar] [CrossRef]
- Kang, J.; Han, X.; Song, J.; Niu, Z.; Li, X. The identification of children with autism spectrum disorder by SVM approach on EEG and eye-tracking data. Comput. Biol. Med. 2020, 120, 103722. [Google Scholar] [CrossRef]
- Fredo, A.J.; Jahedi, A.; Reiter, M.; Müller, R.A. Diagnostic classification of autism using resting-state fMRI data and conditional random forest. Age 2018, 12, 6–41. [Google Scholar]
- Eslami, T.; Mirjalili, V.; Fong, A.; Laird, A.R.; Saeed, F. ASD-DiagNet: A Hybrid Learning Approach for Detection of Autism Spectrum Disorder Using fMRI Data. Front. Neuroinform. 2019, 13, 70. [Google Scholar] [CrossRef]
- Li, G.; Liu, M.; Sun, Q.; Shen, D.; Wang, L. Early Diagnosis of Autism Disease by Multi-channel CNNs. Machine learning in medical imaging. MLMI (Workshop) 2018, 11046, 303–309. [Google Scholar]
- Di Martino, A.; Yan, C.; Li, Q.; Denio, E.; Castellanos, F.X.; Alaerts, K.; Anderson, J.S.; Assaf, M.; Bookheimer, S.Y.; Dapretto, M.; et al. The autism brain imaging data exchange: Towards a large-scale evaluation of the intrinsic brain architecture in autism. Mol. Psychiatry 2014, 19, 659–667. [Google Scholar] [CrossRef] [PubMed]
- Chao-Gan, Y.; Yu-Feng, Z. DPARSF: A MATLAB Toolbox for “Pipeline” Data Analysis of Resting-State fMRI. Front. Syst. Neurosci. 2010, 4, 13. [Google Scholar] [PubMed]
- Tzourio-Mazoyer, N.; Landeau, B.; Papathanassiou, D.; Crivello, F.; Etard, O.; Delcroix, N.; Mazoyer, B.; Joliot, M. Automated Anatomical Labeling of Activations in SPM Using a Macroscopic Anatomical Parcellation of the MNI MRI Single-Subject Brain. Neuroimage 2002, 15, 273–289. [Google Scholar] [CrossRef] [PubMed]
- Khazaee, A.; Ebrahimzadeh, A.; Babajani-Feremi, A. Application of advanced machine learning methods on resting-state fMRI network for identification of mild cognitive impairment and Alzheimer’s disease. Brain Imaging Behav. 2016, 10, 799–817. [Google Scholar] [CrossRef]
- You, Y.; Liang, D.; Wei, R.; Li, M.; Li, Y.; Wang, J.; Wang, X.; Jia, W.; Chen, T. Evaluation of metabolite-microbe correlation detection methods. Anal. Biochem. 2019, 567, 106–111. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Dong, Z.; Phillips, P.; Wang, S.; Ji, G.; Yang, J.; Yuan, T. Detection of subjects and brain regions related to Alzheimer’s disease using 3D MRI scans based on eigenbrain and machine learning. Front. Comput. Neurosci. 2015, 9, 66. [Google Scholar] [CrossRef]
- Blennow, K.; Zetterberg, H. Cerebrospinal fluid biomarkers for Alzheimer’s disease. J. Alzheimer’s Dis. 2009, 18, 413–417. [Google Scholar] [CrossRef]
- Holtzman, D.M.; Morris, J.C.; Goate, A.M. Alzheimer’s disease: The challenge of the second century. Sci. Transl. Med. 2011, 3, 71s–77s. [Google Scholar] [CrossRef]
- Wang, C.; Xiao, Z.; Wu, J. Functional connectivity-based classification of autism and control using SVM-RFECV on rs-fMRI data. Physica Med. 2019, 65, 99–105. [Google Scholar] [CrossRef]
- Chen, C.P.; Keown, C.L.; Jahedi, A.; Nair, A.; Pflieger, M.E.; Bailey, B.A.; Müller, R. Diagnostic classification of intrinsic functional connectivity highlights somatosensory, default mode, and visual regions in autism. NeuroImage Clin. 2015, 8, 238–245. [Google Scholar] [CrossRef] [PubMed]
- DSouza, A.M.; Abidin, A.Z.; Wismüller, A. Classification of autism spectrum disorder from resting-state fMRI with mutual connectivity analysis. In Proceedings of the Medical Imaging 2019: Biomedical Applications in Molecular, Structural, and Functional Imaging, San Diego, CA, USA, 19–21 February 2019. [Google Scholar]
- Heinsfeld, A.S.; Franco, A.R.; Craddock, R.C.; Buchweitz, A.; Meneguzzi, F. Identification of autism spectrum disorder using deep learning and the ABIDE dataset. NeuroImage Clin. 2017, 17, 16–23. [Google Scholar] [CrossRef] [PubMed]
- Dvornek, N.C.; Ventola, P.; Duncan, J.S. Combining phenotypic and resting-state fmri data for autism classification with recurrent neural networks. In Proceedings of the IEEE International Symposium on Biomedical Imaging, Washington, DC, USA, 4–7 April 2018. [Google Scholar]
- Simard, I.; Luck, D.; Mottron, L.; Zeffiro, T.A.; Soulières, I. Autistic fluid intelligence: Increased reliance on visual functional connectivity with diminished modulation of coupling by task difficulty. NeuroImage Clin. 2015, 9, 467–478. [Google Scholar] [CrossRef] [PubMed]
- Ha, S.; Sohn, I.; Kim, N.; Sim, H.J.; Cheon, K. Characteristics of Brains in Autism Spectrum Disorder: Structure, Function and Connectivity across the Lifespan. Exp. Neurobiol. 2015, 24, 273–284. [Google Scholar] [CrossRef] [PubMed]
- Keown, C.L.; Shih, P.; Nair, A.; Peterson, N.; Mulvey, M.E.; Müller, R. Local functional overconnectivity in posterior brain regions is associated with symptom severity in autism spectrum disorders. Cell Rep. 2013, 5, 567–572. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Li, M.; Yang, C.; Fang, X.; Ye, M.; Wei, L.; Liu, J.; Li, B.; Gan, Y.; Yang, B.; et al. Altered Functional Connectivity in Children With Low-Function Autism Spectrum Disorders. Front. Neurosci. 2019, 13, 806. [Google Scholar] [CrossRef]
- Ecker, C.; Suckling, J.; Deoni, S.C.; Lombardo, M.V.; Bullmore, E.T.; Baron-Cohen, S.; Catani, M.; Jezzard, P.; Barnes, A.; Bailey, A.J.; et al. Brain anatomy and its relationship to behavior in adults with autism spectrum disorder: A multicenter magnetic resonance imaging study. Arch. Gen. Psychiatry 2012, 69, 195–209. [Google Scholar] [CrossRef]
- Turetsky, B.I.; Hahn, C.; Borgmann-Winter, K.; Moberg, P.J. Scents and nonsense: Olfactory dysfunction in schizophrenia. Schizophr. Bull. 2009, 35, 1117–1131. [Google Scholar] [CrossRef][Green Version]
- Moberg, P.J.; Agrin, R.; Gur, R.E.; Gur, R.C.; Turetsky, B.I.; Doty, R.L. Olfactory dysfunction in schizophrenia: A qualitative and quantitative review. Neuropsychopharmacology 1999, 21, 325–340. [Google Scholar] [CrossRef]
- Soudry, Y.; Lemogne, C.; Malinvaud, D.; Consoli, S.; Bonfils, P. Olfactory system and emotion: Common substrates. Eur. Ann. Otorhinolaryngol. Head Neck Dis. 2011, 128, 18–23. [Google Scholar] [CrossRef]
- Menassa, D.A.; Sloan, C.; Chance, S.A. Primary olfactory cortex in autism and epilepsy: Increased glial cells in autism. Brain Pathol. 2017, 27, 437–448. [Google Scholar] [CrossRef] [PubMed]
- Koehler, L.; Fournel, A.; Albertowski, K.; Roessner, V.; Gerber, J.; Hummel, C.; Hummel, T.; Bensafi, M. Impaired Odor Perception in Autism Spectrum Disorder Is Associated with Decreased Activity in Olfactory Cortex. Chem. Senses 2018, 43, 627–634. [Google Scholar] [CrossRef] [PubMed]
- Grèzes, J.; Wicker, B.; Berthoz, S.; de Gelder, B. A failure to grasp the affective meaning of actions in autism spectrum disorder subjects. Neuropsychologia 2009, 47, 1816–1825. [Google Scholar] [CrossRef] [PubMed]
- Philip, R.C.M.; Whalley, H.C.; Stanfield, A.C.; Sprengelmeyer, R.; Santos, I.M.; Young, A.W.; Atkinson, A.P.; Calder, A.J.; Johnstone, E.C.; Lawrie, S.M.; et al. Deficits in facial, body movement and vocal emotional processing in autism spectrum disorders. Psychol. Med. 2010, 40, 1919–1929. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Choi, U.; Park, S.; Oh, S.; Yoon, H.; Koh, Y.; Im, W.; Park, J.; Song, D.; Cheon, K.; et al. Abnormal activation of the social brain network in children with autism spectrum disorder: An FMRI study. Psychiatry Investig. 2015, 12, 37–45. [Google Scholar] [CrossRef]
- Bastiaansen, J.A.; Thioux, M.; Nanetti, L.; van der Gaag, C.; Ketelaars, C.; Minderaa, R.; Keysers, C. Age-related increase in inferior frontal gyrus activity and social functioning in autism spectrum disorder. Biol. Psychiatry 2011, 69, 832–838. [Google Scholar] [CrossRef]
- Watanabe, T.; Yahata, N.; Abe, O.; Kuwabara, H.; Inoue, H.; Takano, Y.; Iwashiro, N.; Natsubori, T.; Aoki, Y.; Takao, H.; et al. Diminished medial prefrontal activity behind autistic social judgments of incongruent information. PLoS ONE 2012, 7, e39561. [Google Scholar] [CrossRef]
- Cooper, R.A.; Richter, F.R.; Bays, P.M.; Plaisted-Grant, K.C.; Baron-Cohen, S.; Simons, J.S. Reduced Hippocampal Functional Connectivity during Episodic Memory Retrieval in Autism. Cereb. Cortex 2017, 27, 888–902. [Google Scholar] [CrossRef]
- Mackiewicz, K.L.; Sarinopoulos, I.; Cleven, K.L.; Nitschke, J.B. The effect of anticipation and the specificity of sex differences for amygdala and hippocampus function in emotional memory. Proc. Natl. Acad. Sci. USA 2006, 103, 14200–14205. [Google Scholar] [CrossRef]
- Mailo, J.; Tang-Wai, R. Insight into the precuneus: A novel seizure semiology in a child with epilepsy arising from the right posterior precuneus. Epileptic Disord. 2015, 17, 321–327. [Google Scholar] [CrossRef]
- Reinhardt, V.P.; Iosif, A.; Libero, L.; Heath, B.; Rogers, S.J.; Ferrer, E.; Nordahl, C.; Ghetti, S.; Amaral, D.; Solomon, M. Understanding Hippocampal Development in Young Children With Autism Spectrum Disorder. J. Am. Acad. Child Adolesc. Psychiatry 2020, 59, 1069–1079. [Google Scholar] [CrossRef] [PubMed]
- Via, E.; Radua, J.; Cardoner, N.; Happé, F.; Mataix-Cols, D. Meta-analysis of gray matter abnormalities in autism spectrum disorder: Should Asperger disorder be subsumed under a broader umbrella of autistic spectrum disorder? Arch. Gen. Psychiatry 2011, 68, 409–418. [Google Scholar] [CrossRef] [PubMed]
- Chaddad, A.; Desrosiers, C.; Hassan, L.; Tanougast, C. Hippocampus and amygdala radiomic biomarkers for the study of autism spectrum disorder. BMC Neurosci. 2017, 18, 52. [Google Scholar] [CrossRef] [PubMed]

| Autism (n = 59) | NC (n = 46) | p Value | |
|---|---|---|---|
| Gender(M/F) | 51/8 | 39/7 | 0.81 |
| Age(years) | 12.472.3 | 12.351.9 | 0.77 |
| Feature Set | Accuracy (%) | Sensitivity (%) | Specificity (%) |
|---|---|---|---|
| Optimal feature subset | 86.7 | 87.5 | 85.7 |
| Original feature set | 57.1 | 61.5 | 50 |
| Weight | Region |
|---|---|
| 8 | SOG.R |
| 6 | OLF.L |
| 5 | IFGoperc.R HIP.L |
| 4 | AMYG.L |
| 3 | PreCG.L |
| Methods | Accuracy (%) |
|---|---|
| Chen et al. [39] | 66 |
| Adora et al. [40] | 70–81 |
| Anibal et al. [41] | 70 |
| Nicha et al. [42] | 70.1 |
| Our method | 86.7 |
| Feature Set | Accuracy (%) | Sensitivity (%) | Specificity (%) |
|---|---|---|---|
| Fisher score | 64.3 | 87.5 | 66.7 |
| RFE | 68.8 | 83.3 | 60 |
| Our method | 86.7 | 87.5 | 85.7 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shi, C.; Zhang, J.; Wu, X. An fMRI Feature Selection Method Based on a Minimum Spanning Tree for Identifying Patients with Autism. Symmetry 2020, 12, 1995. https://doi.org/10.3390/sym12121995
Shi C, Zhang J, Wu X. An fMRI Feature Selection Method Based on a Minimum Spanning Tree for Identifying Patients with Autism. Symmetry. 2020; 12(12):1995. https://doi.org/10.3390/sym12121995
Chicago/Turabian StyleShi, Chunlei, Jiacai Zhang, and Xia Wu. 2020. "An fMRI Feature Selection Method Based on a Minimum Spanning Tree for Identifying Patients with Autism" Symmetry 12, no. 12: 1995. https://doi.org/10.3390/sym12121995
APA StyleShi, C., Zhang, J., & Wu, X. (2020). An fMRI Feature Selection Method Based on a Minimum Spanning Tree for Identifying Patients with Autism. Symmetry, 12(12), 1995. https://doi.org/10.3390/sym12121995




