1. Introduction
For all proteolysis patterns, it is important to know how the fragmentation of the polypeptide chain occurs over time—that is, the kinetics of proteolysis. Enzymatic hydrolysis of proteins (proteolysis) occurs under the action of proteolytic enzymes (proteases), which are natural chiral catalysts. As a result of the hydrolysis of peptide bonds, these enzymes hydrolyze proteins into shorter polypeptides, short peptides, and even amino acids. This process plays an important role in various fields of biology, biotechnology, and proteomics. Proteolysis is involved in biological processes at the level of cells and biological systems, including digestion, autolysis, activation of enzymes, cell regulation, etc. The wide application of protein hydrolysis in the food industry includes the processing of milk proteins by various proteolytic enzymes [
1]. For all cases of proteolysis, it is important to know how the fragmentation of the polypeptide chain occurs over time—that is, the kinetics of proteolysis.
The mechanism of action and specificity of proteolytic enzymes has been intensively studied for synthetic substrates with ester or amide bonds and small peptides containing amino acid residues specific for a given enzyme [
2]. Trypsin (EC 3.4.21.4) is a serine protease found in the digestive system, where it breaks up food proteins into peptides [
3,
4]. Its optimal catalyst conditions are pH 7.8 and a temperature of 37 °C [
5]. Trypsin has a catalytic triad within its active site that involves Ser
195, His
57, and Asp
102 residues. Positively charged side chains of Arg and Lys residues in protein substrates participate in electrostatic interactions with Asp
189 at the bottom of the trypsin active site [
6]. Therefore, trypsin cleaves predominantly peptide bonds at the carboxyl side of lysine and arginine (Arg-X, Lys-X bonds) unless they are followed by proline [
4,
5,
6]. In addition, the rate of hydrolysis of these bonds depends also on other neighboring amino acid residues providing so-called secondary specificity. For small substrates with one bond to be hydrolyzed, the hydrolysis kinetics is of the Michaelis-Menten type.
Unlike the hydrolysis of small substrates, proteolysis is a polysubstrate process that involves the cleavage of peptide bonds of various specificities. Hydrolysis is complicated by unequal and often limited access of peptide bonds to the action of the enzyme since different sites in protein substrate are differently exposed to the surrounding solvent [
7]. This steric effect, known as masking, was proposed to be a significant limiting factor of hydrolysis [
7]. The degradation of the protein substrate and the corresponding demasking of the peptide bonds in the course of protein degradation are difficult to describe because of the complex nature of unfolding, dissociation, and solubilization of a large number of polypeptide fragments. Thus, the kinetic regularities of the total hydrolysis of peptide bonds are very complex and they do not obey the Michaelis-Menten law.
The main bovine whey protein is a small globular protein β-lactoglobulin (β-LG) [
8]. In vivo, it has the ability to bind and transport small biomolecules, such as fatty acids and vitamin A. The monomeric β-LG consists of 162 amino acid residues (18.3 kDa), and its compact globule is stabilized by two disulfide bonds (Cys
66-Cys
160 and Cys
106-Cys
119) [
9,
10]. β-LG contains 15 lysine and three arginine residues, which are potentially specific for trypsin, and two tryptophan residues at positions 19 and 61, which mostly provide intrinsic fluorescence. β-LG is able to form dimers at physiological environment conditions and exists in monomeric form at high temperatures and pH values higher than eight [
11].
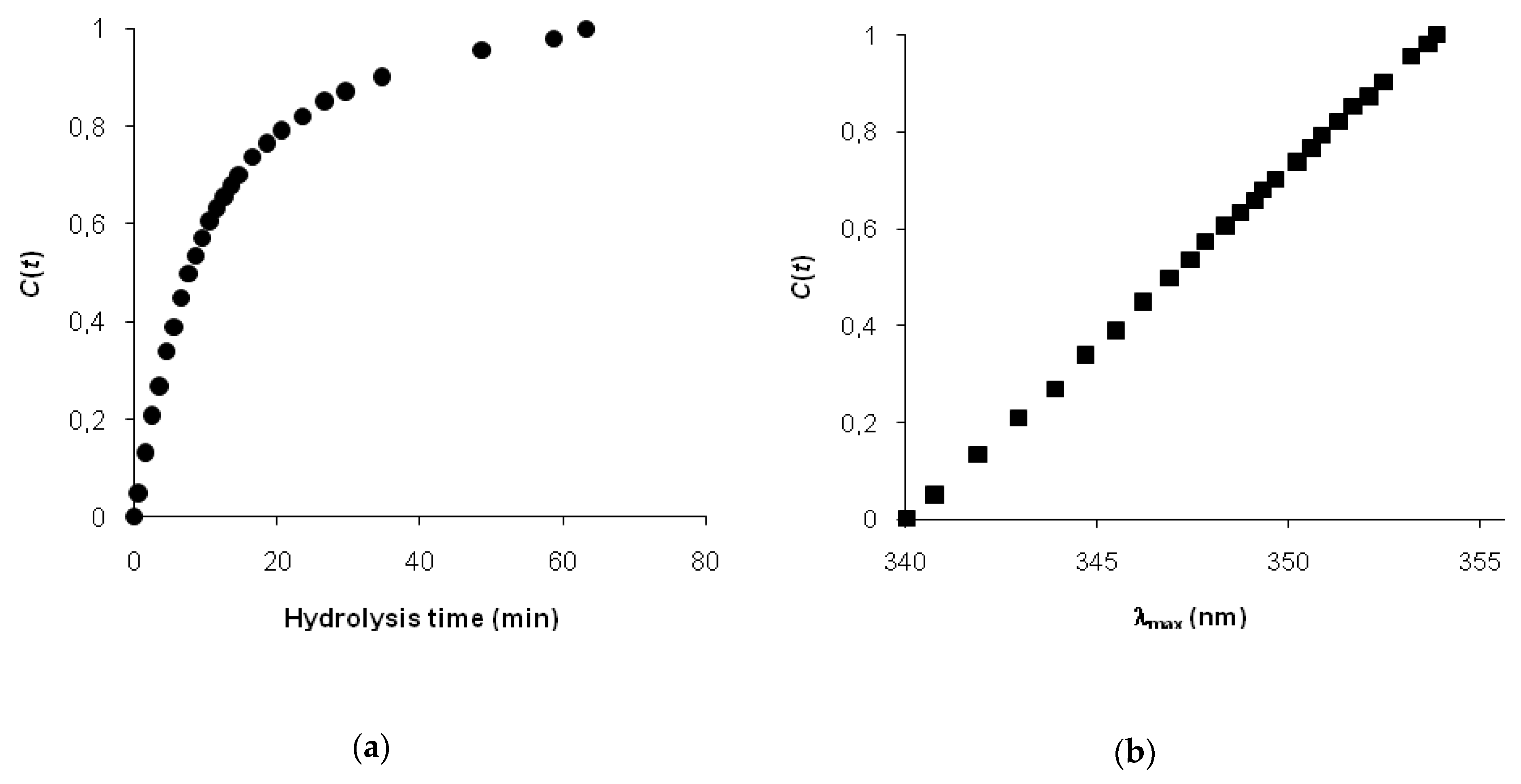
Since Adler-Nissen’s first presentation—the degree of hydrolysis—the percentage of hydrolyzed peptide bonds has been widely used as a universal parameter for controlling proteolysis and optimizing the process [
1]. An alternative to the degree of hydrolysis could be the degree of changes in a secondary structure and the conformation of protein substrate, as measured by an appropriate spectral technique. In recent years, some progress has been made in comparing spectral data obtained at different times of hydrolysis using fluorescence [
12,
13] and infrared spectroscopies [
14,
15,
16,
17]. It was demonstrated that the degradation of protein substrate can be quantified by a shift in tryptophan fluorescence [
12,
13]. This shift is caused by an increase in the polarity of the medium around Trp residues [
18,
19], being a result of the proteolytically induced degradation of the protein structure.
Proteolysis of β-LG by various proteases was intensively studied in connection with the practical importance of this process for food processing [
20,
21,
22,
23]. In addition to kinetic aspects, these studies revealed the main intermediate and final proteolysis products. For tryptic proteolysis of β-LG, the identification and quantification of peptides were accurately performed, opening the way for establishing the sequence of steps for protein fragmentation [
10,
22,
23]. Using the simple fragmentation pathway, the hydrolysis kinetics was successfully described by the limited number of differential equations [
23].
In proteolysis, a general tendency is the continuous degradation of the protein structure and thereby an increase in accessibility of the remaining peptide bonds to the enzyme. This process provides the peptide bond demasking [
7,
24], giving a local increase in the rate of hydrolysis at a certain period when most of the initially masked bonds become demasked. Owing to the demasking effect, the rate of accumulation of amino nitrogen (rate of hydrolysis) may be a non-monotonous function of the time of hydrolysis [
7]. To reveal the subtle features of the hydrolysis mechanism, one can differentiate the hydrolysis degree by time and study the kinetics in terms of the rate of hydrolysis and the degree of hydrolysis [
7]. Another possibility is to analyze the proteolysis kinetics in terms of the degree of hydrolysis and the degree of masking (or degree of demasking) using the two-step proteolysis model [
7,
12,
13]. In this model, proteolysis is considered as a simple two-step process with consecutive demasking and hydrolysis steps [
7]. The demasking step was quantified previously for the hydrolysis of β-casein (β-CN) by trypsin under the assumption that the degree of demasking is proportional to the fluorescence shift [
12,
13]. To analyze proteolysis in the framework of a two-step model, it is necessary to determine the numerical values of the rate constants included in the model [
7,
25].
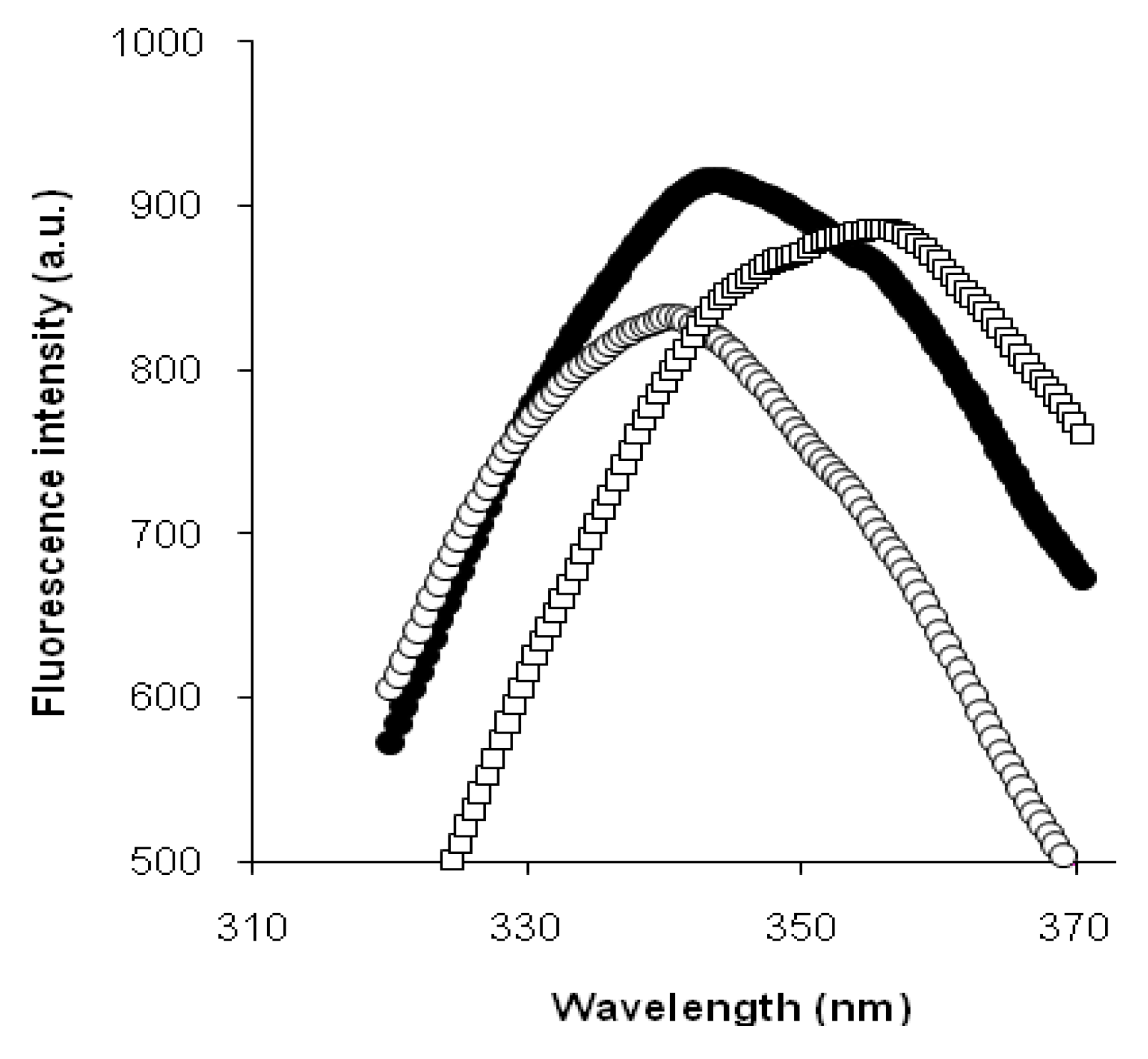
We assumed that the period of intensive demasking, caused by the structural opening of protein, could be clearly identified by the red shift in tryptophan fluorescence [
12]. Additionally, aggregation of proteolysis products or refolding of polypeptide chains could cause a decrease in protein openness, consequently slowing the shift in fluorescence. This would give different sets of periods of fast and slow shifts in tryptophan fluorescence during the hydrolysis of various protein substrates.
Here, we present a kinetic analysis of fluorescence data obtained for the hydrolysis of globular β-LG by trypsin and also compare this process with the tryptic hydrolysis of β-casein (β-CN), which is an example of an amphiphilic structure-unordered protein. This illustrates a new algorithm for distinguishing differences in proteolysis of various proteins by fluorescent analysis. Fluorescence data were used to extract the important modeling parameter, the degree of masking as a function of hydrolysis time. After adding data on the degrees of hydrolysis, the analysis of β-LG proteolysis by trypsin was performed using the two-step proteolysis model. Thus, we present the benefits of using fluorescence data along with the degrees of hydrolysis for quantitative analysis of proteolysis.
3. Discussion
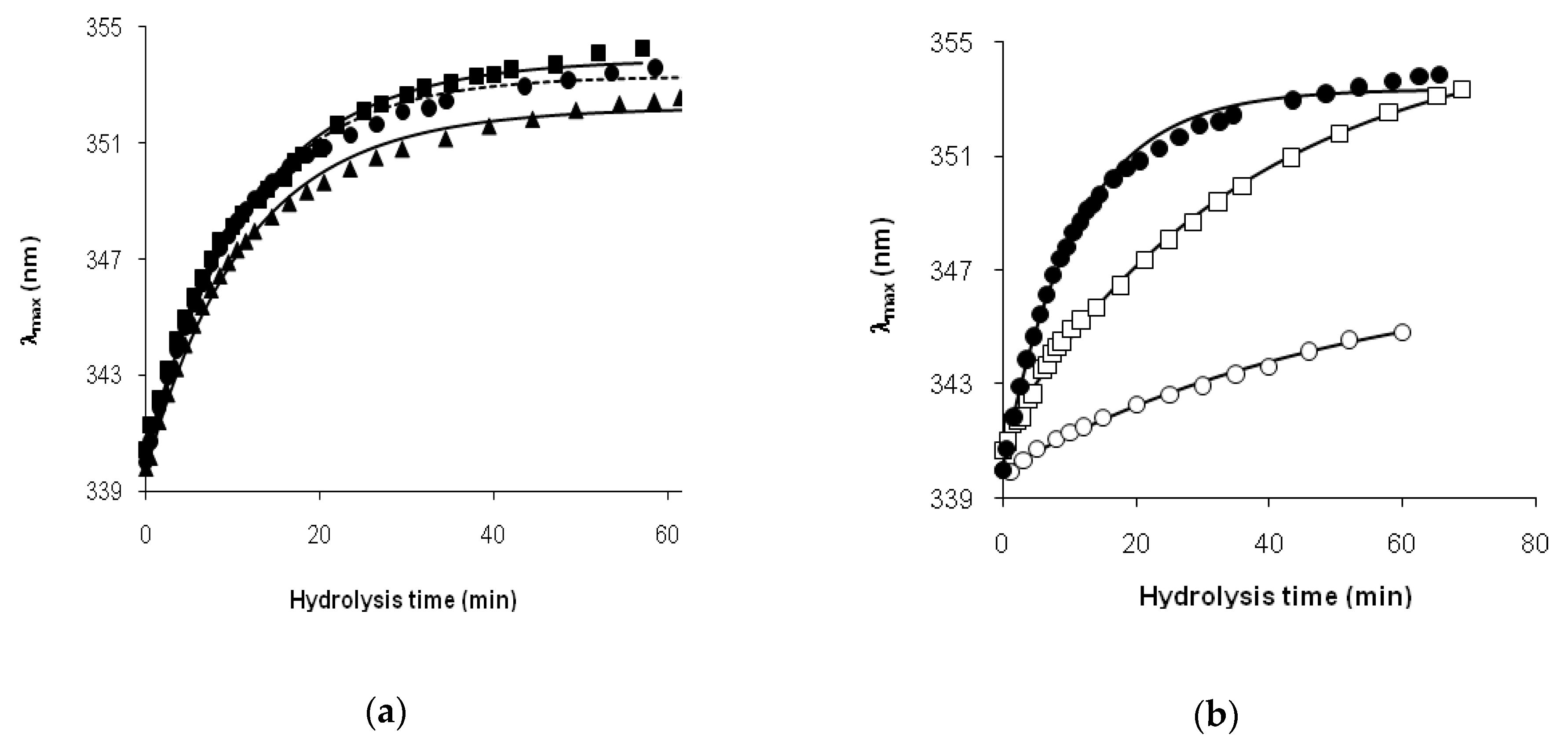
The hydrolysis of β-LG by trypsin was the case when the intact protein and relatively short peptide fragments [f(1–8), f(9–14), f(15–40), f(41–70), f(71–75), f(76–138), f(139–141), f(142–148), f(149–162)] were present in the reaction mixture, whereas longer peptides, combinations of these peptide fragments, did not accumulate [
23]. The conversion of β-LG into its fragments could be satisfactorily described by a limited number of differential equations without taking into account the accumulation and consumption of long intermediate fragments [
23]. This picture is quite realistic for the “one-by-one” mechanism, which we used to process the fluorescence spectra. In this mechanism of proteolysis, the degradation of the protein globule limits the demasking process. The complication of the “one-by-one” mechanism may be in considering the incomplete disruption of the globule with the conservation of a certain core [
31] that is more resistant to hydrolysis. In this case, the demasking kinetics can be bimodal, consisting of two processes with different rates. A good approximation of the demasking kinetics by Equation (3) with fast and slow exponential terms supports this idea.
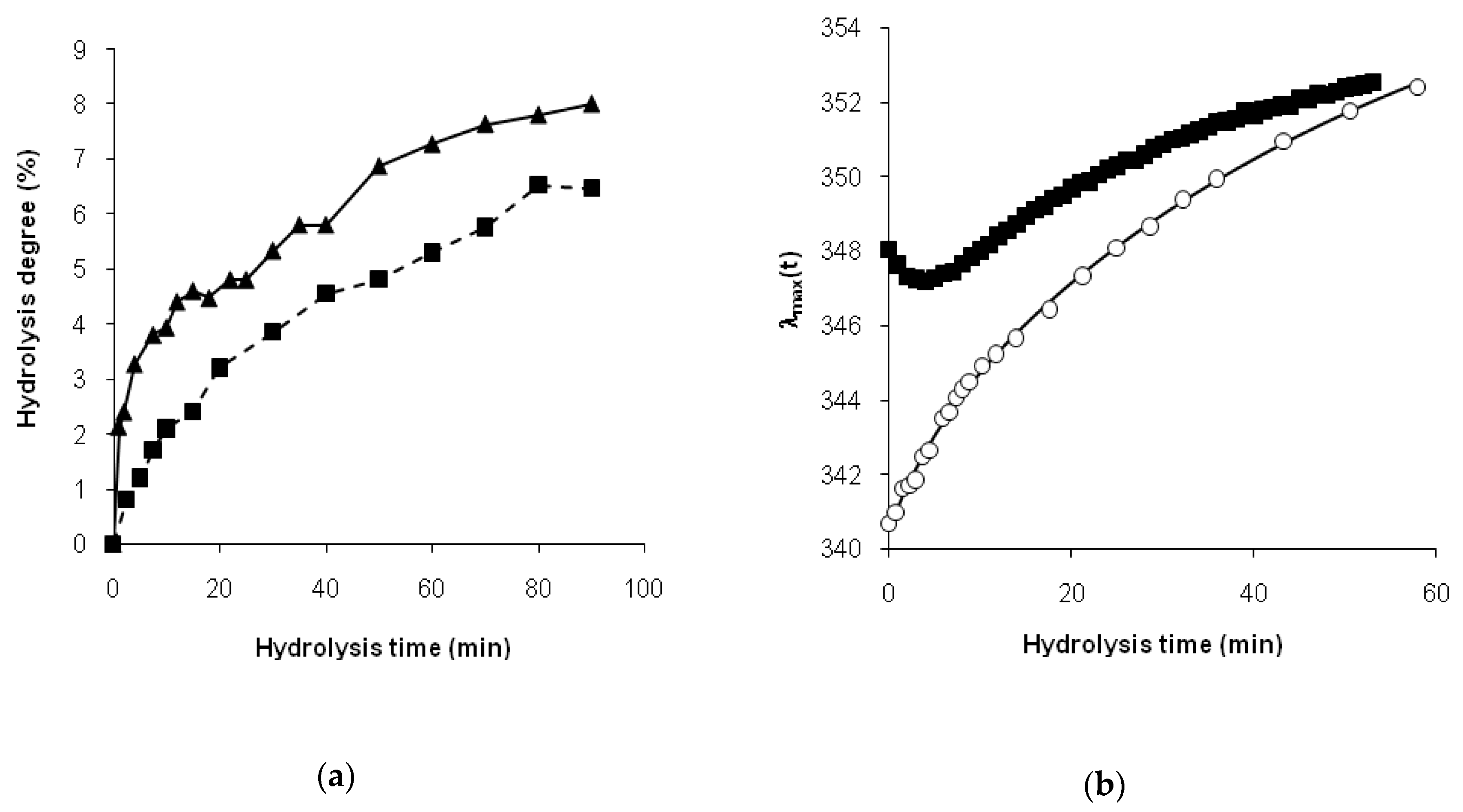
For the hydrolysis of β-CN, the relatively short period of the reduction in
λmax(
t) could be explained by the aggregation of the rests of β-casein molecules after the loss of its charged N-terminal moieties as a result of the relatively rapid hydrolysis of the Arg
25-Ile
26 bond [
13,
30]. This remarkable trypsinolysis of β-CN process of aggregation in combination with the degradation of the initial casein micelles was monitored in detail by the static light scattering [
32]. It was shown that aggregates were formed when the concentration of trypsin was low enough to provide an overall low rate of hydrolysis in order to obtain time for the formation of aggregates [
32]. Casein micelles and aggregates—as structural elements with limited openness—may have retarded proteolysis, limiting the accessibility of peptide bonds. In the result of degradation of aggregates, the peptide bonds became accessible for enzyme attack, passing from a masked state to a demasked one. Our previous FTIR absorption data at 1632 cm
−1 demonstrated a non-monotonic change in the concentration of β-sheets during trypsinolysis of β-CN, i.e., an increase and a subsequent decrease in β-structures [
15], which was consistent with the obtained fluorescence data.
Thus, we found that the read shift of tryptophan fluorescence in the course of proteolysis, estimated as the dependence of the wavelength of fluorescence maximum on the hydrolysis time, was significantly different for tryptic proteolysis of β-LG and β-CN. This difference was explained by different demasking scenarios for these substrates.
The difference in proteolysis mechanisms for different protein substrates should be especially noticeable at the initial stage of proteolysis, where spectral methods are able to monitor conformational changes. We demonstrated that fluorescence spectroscopy is quite appropriate for this purpose, both on a quantitative and a qualitative level. At a qualitative level, we can compare λmax(t) functions using empirical equations, such as Equation (3), as was demonstrated by comparing the proteolysis processes for β-LG and β-CN. At a quantitative level, the best way to use fluorescence data is to calculate the parameter x by Equation (4) and use the two-step proteolysis model.
The hydrolysis of each specific bond of the protein substrate was determined for the proteolysis of the whey protein isolate by
Bacillus licheniformis protease [
33]. It was determined precisely by mass-spectroscopy technique using a set of kinetic curves for all peptides arising from the cleavage of the analyzed bond. A significant difference in the selectivity parameters for different peptide bonds was reported [
33]. In the present study, we took into account three categories of the hydrolysis rate constants, as in the previous publication [
13], whereas the Equation (5) could be used for more types of peptide bonds. Measuring selectivity parameters can help determine the hydrolysis rate constants used in the two-step model. However, the relationship between the selectivity values and the hydrolysis rate constants is not simple in the presence of masking effect [
34].
The complexity of the modeling of proteolysis lies in the large number of elementary acts that make up proteolysis. Even if it were possible to determine the full set of kinetic parameters for the main peptide bonds in the main peptide fragments, this would not be enough since we need to know at which point of proteolysis these fragments can be attacked by the enzyme. Therefore, information on the demasking of peptide bonds is absolutely necessary. An alternative to the complete kinetic description of proteolysis is the use of simple empirical models containing only a few parameters, such as the exponential model [
29]. These parameters can be accurately measured, although they do not make precise sense. Representing the two-step proteolysis model, we propose a compromise between complete kinetic analysis and empirical modeling. The parameters of the two-step model have a certain chemical meaning and can be measured using fluorescence spectroscopy, as shown in this study.
4. Materials and Methods
4.1. Materials
β-LG (L3908), β-CN (C6905) from bovine milk, and trypsin from bovine pancreas (T1426) treated with N-tosyl-L-phenylalanine chloromethyl ketone (TPCK) were purchased from Sigma–Aldrich and used without further treatment. TPCK was used to inhibit the contaminating chymotrypsin activity without affecting the activity of trypsin. Phosphate buffer solution was prepared with doubly distilled water and stored at 4 °C before use. Trypsin solutions were freshly prepared by diluting trypsin in phosphate buffer. All other reagents were of analytical grade and obtained from commercial sources.
4.2. Proteolysis Reaction
The protein substrate (β-lactoglobulin or β-casein) was dissolved in 20 mM phosphate buffer (pH 7.9) at 37 °C by stirring. The enzymatic hydrolysis was carried out in a volume of 2 mL for the determination of amino nitrogen in a bath stirred reactor or in a 1 cm quartz cuvette for fluorescence measurements at various concentrations of the substrate and enzyme. For β-LG, for example, enzymatic hydrolysis with a substrate concentration of 0.25 g/L was initiated by adding 10 µL of trypsin solution (1 g/L) to provide a final enzyme concentration in the reaction mixture of 5.0 mg/L. These conditions were accepted as standard for the proteolysis modeling by the two-step model or for comparing the dependences of the degree of hydrolysis on the time of hydrolysis. To determine the kinetics of demasking at a constant concentration of trypsin, the concentration of β-LG was 1, 0.5, and 0.25 g/L at trypsin concentration of 15 mg/L. To determine the demasking kinetics at a constant concentration of β-LG, the concentration of trypsin was 0.9, 5, and 15 mg/L at β-LG concentration of 0.25 g/L.
4.3. Determination of Hydrolysis Degree by OPA Method
The
o-phthalaldehyde (OPA) method for the determination of amino nitrogen is based on the reaction of
o-phthalaldehyde and 2-mercaptoethanol with amino groups released during proteolysis [
35]. A stock OPA solution was prepared by mixing 25 mL of 100 mM sodium tetrahydroborate, 2.5 mL of 20% sodium dodecyl sulfate (SDS) solution (w/w), 40 mg of OPA, and 100 μL of β-mercaptoethanol. The final volume of OPA solution (50 mL) was adjusted with distilled water.
For the termination of proteolysis after a required time and to monitor the reaction by the OPA-method, 20 μL samples from the proteolysis reactor were placed in the test-tubes with 1 mL of OPA solution, incubated for 5 min at room temperature, and then the absorption values at 340 nm were determined against a blank sample. The latter was prepared in the same way except for the absence of the enzyme. As a result, the dependence of amino nitrogen N(t) on time of hydrolysis was determined. For the determination of ΔN, a complete hydrolysis of proteins in 6N HCl at 110 °C under Argon during 24 h was performed.
4.4. Fluorescence Spectroscopy and Determination of Demasking Kinetics by Fluorescence Measurements
Fluorescence emission during proteolysis was measured using a Perkin-Elmer LS 55 Luminescence Spectrometer (Waltham, MA, USA) at 90° relative to the excitation beam at excitation wavelengths of 280 nm. The spectral bandwidth of the excitation and emission light was set to 10 and 5 nm. A thermostated cuvette holder was used to keep the sample at 37 °C. The emission spectra were recorded at a scanning speed of 150 nm/min.
To determine the wavelength of fluorescence maximum, we used a parabolic function
I(
λ) =
aλ2 +
bλ + const for the approximation of fluorescence spectrum, taking only a small area with a bandwidth of 30 nm around the parabola center at
λmax = −
b/2
a [
12].
4.5. Kinetic Analysis by Two-Step Proteolysis Model
We used the two-step proteolysis model, which was originally formulated for Arg-X and Lys-X peptide bonds (X is an arbitrary amino acid residue) in protein substrates [
7,
12]. These bonds include masked peptide bonds, demasked peptide bonds, the peptide bond that are already hydrolyzed for the current moment, and the unhydrolyzable (resistant) bonds.
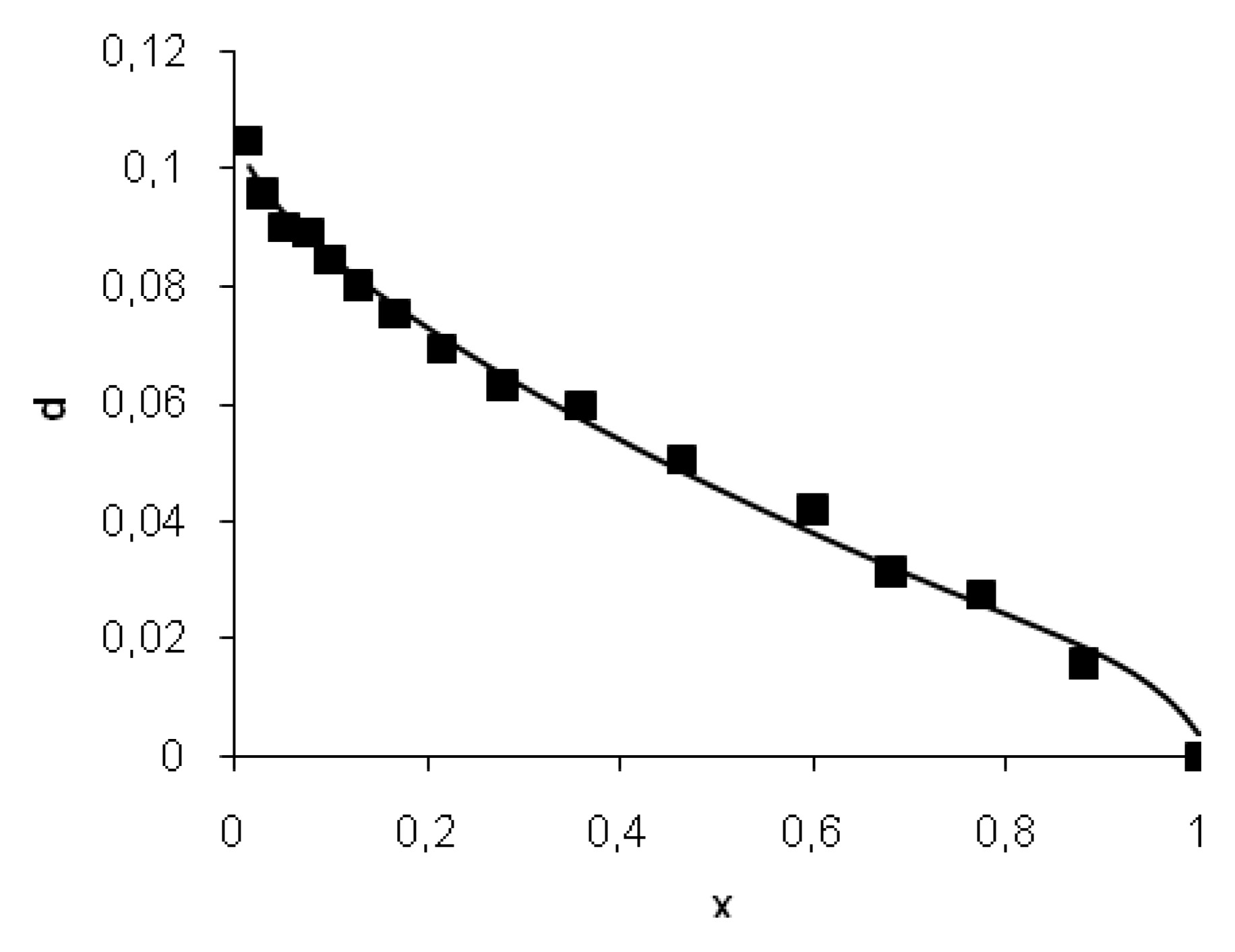
In the framework of the two-step proteolysis model [
7,
12], the degree of peptide bond hydrolysis (
d = DH/100) and the degree of peptide bond masking (
x) are connected by the following equation:
where
is the initial concentration of masked peptide bonds of the jth type,
X0 is the initial concentration of all masked peptide bonds,
m is the initial degree of masking (portion of initially masked peptide bonds), and
n is the portion of bonds specific but resistant to hydrolysis.
aj and
bj are the parameters of the two-step model:
where
kh is the maximum hydrolysis constant,
kj/
kh is the relative hydrolysis rate constant for
j-type bonds, and
kd/
kh is the relative rate constant of demasking, which is proposed to be the same for any type of peptide bonds
j.
The model includes independent variables d and x, which were determined using measurements of amino nitrogen and maximum fluorescence wavelength. The model parameters kj/kh and are the constant values. In the result of modeling, the parameters n, m, and kd/kh were determined by the fitting of experimental data with Equation (5).
For trypsin, specific amino acid residues are Arg and Lys, for which the hydrolysis rate constants
kj are not equal to 0. We assumed that the hydrolysis rate constants for Arg-X and Lys-X bonds roughly fall into the categories of high, intermediate, and low specific bonds among 161 peptide bonds of β-LG (3 Arg-X and 15 Lys-X bonds). For highly specific bonds,
k1/
kh = 1 (1/3 part of specific bonds,
= 0.0373). For intermediate specific bonds,
k2/
kh = 0.1 (1/3 part of specific bonds,
= 0.0373), and for weakly specific bonds,
k3/
kh = 0.02 (1/3 part of specific bonds,
= 0.0373). For non-specific bonds (other 143 bonds in β-LG),
= 0.8881 and
k4/
kh = 0. Estimates of the hydrolysis rate constants were taken from our previous study for proteolysis of β-CN [
12].
4.6. Prediction of the Cleavage of Individual Peptide Bonds
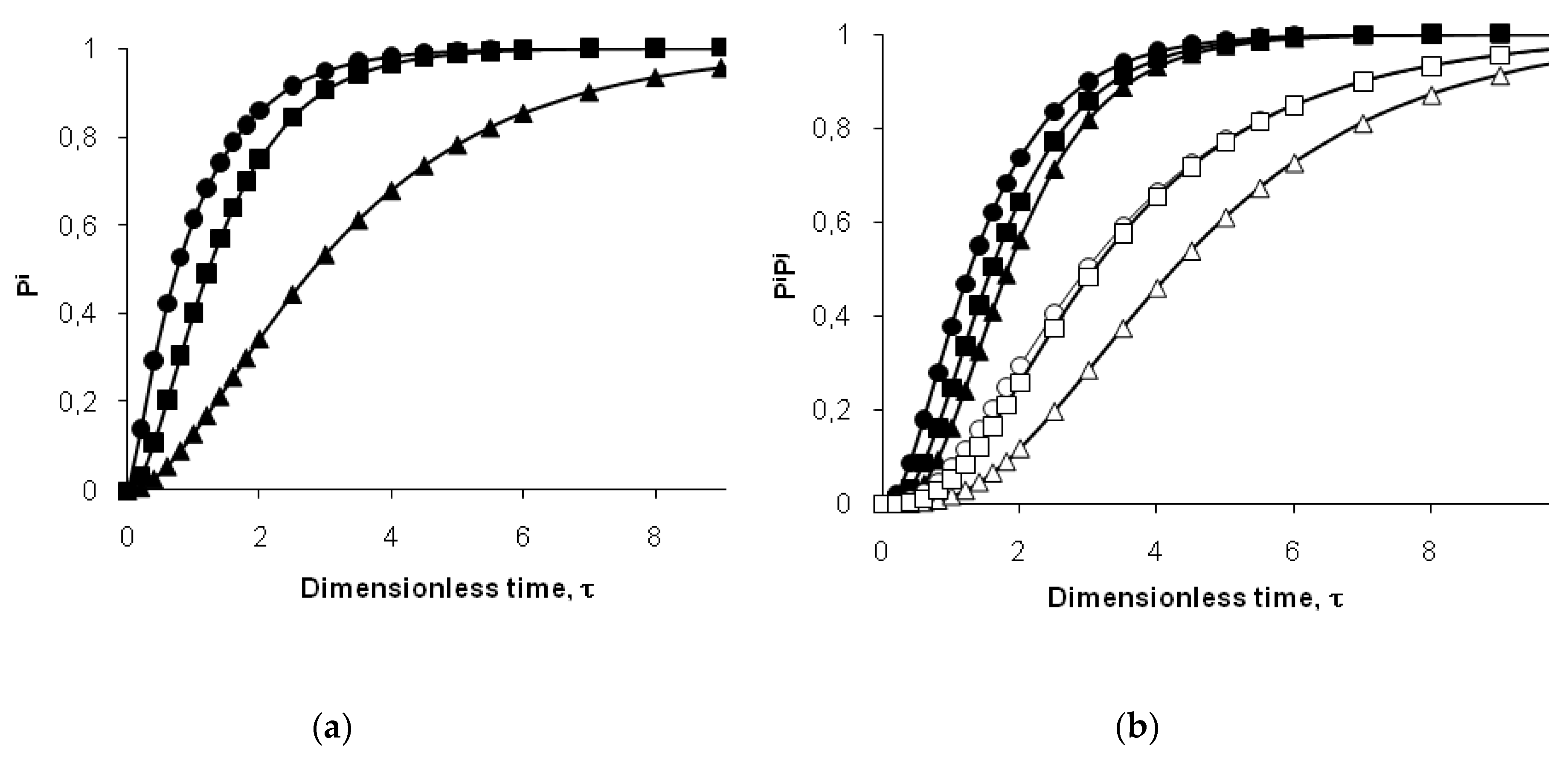
To predict the probability of cleavage (
Pi) for peptide bond
i, we used a version of the two-step model for individual bonds, which was previously tested for the hydrolysis of whey proteins by
Bacillus licheniformis protease [
34]. In this case,
ki is the rate constant of hydrolysis for the peptide bond
i in demasked state. The rate constant
kd is the rate constant of demasking for the site that masks the hydrolysis of bond
i. [
34]. It is assumed that all bonds of type
i are masked at the beginning of proteolysis. The evolution of the probability of cleavage with hydrolysis time
t is [
34]:
Using the dimensionless time of hydrolysis (
τ =
tkd),
Pi from Equation (8) can be represented as a function of the variable
τ and the parameter
kd/
ki:
The parameters of non-linear equations were calculated by the method of non-linear least squares fitting (Origin 5.0).