1. Introduction
Individual identification is a technique that is used to identify a user using behavioral or physical characteristics that are the sole characteristics of an individual. Currently, the range of services, such as security, banking, access control, medical care, and entertainment, is expanding, and so individual identification methods are studied using diverse characteristics [
1,
2,
3,
4,
5,
6]. Such individual identification using the face, fingerprint, and so forth has vulnerability to falsification and disguise, and it must be moved to a special place where a system is installed for individual identification [
7]. Furthermore, human needs for health are demanding measures that can easily determine the state of the body as technology develops. Therefore, companies have developed and released small, light, convenient, and function-rich wearable devices. In particular, the collection function of various bio-signals, which are important for health checks, is becoming essential. The measurement procedure of biometric information has been simplified and conveniently changed. If some bio-signals are gathered in real time, they could be used for individual identification and clinical diagnosis [
8].
Recently, individual identification using electrocardiogram (ECG) signals in the body has been studied, and studies have shown good performance [
9,
10,
11]. An ECG is a measurement of the change in dislocation during the cardiac cycle. ECG signals can be used for biometrics because of the physiological and geometric differences of the heart making the unique signal of each person [
8].
The tensor is a larger category than the vector and the matrix. The scalar, vector, and matrix are tensors of zero- to second orders, and more than the third order is a higher-order tensor [
12,
13]. As sensors, memories, and network technologies evolve, much data are generated day-by-day in various fields. Most Big Data is based on a tensor representation of a multidimensional array, containing a large amount of information [
14]. The important thing in big data [
15] is to extract only the important features, because the information includes both important and useless aspects. Thus, with the development of cloud computing [
16] and big data like the MapReduce model [
17], tensor-based processing has attracted attention.
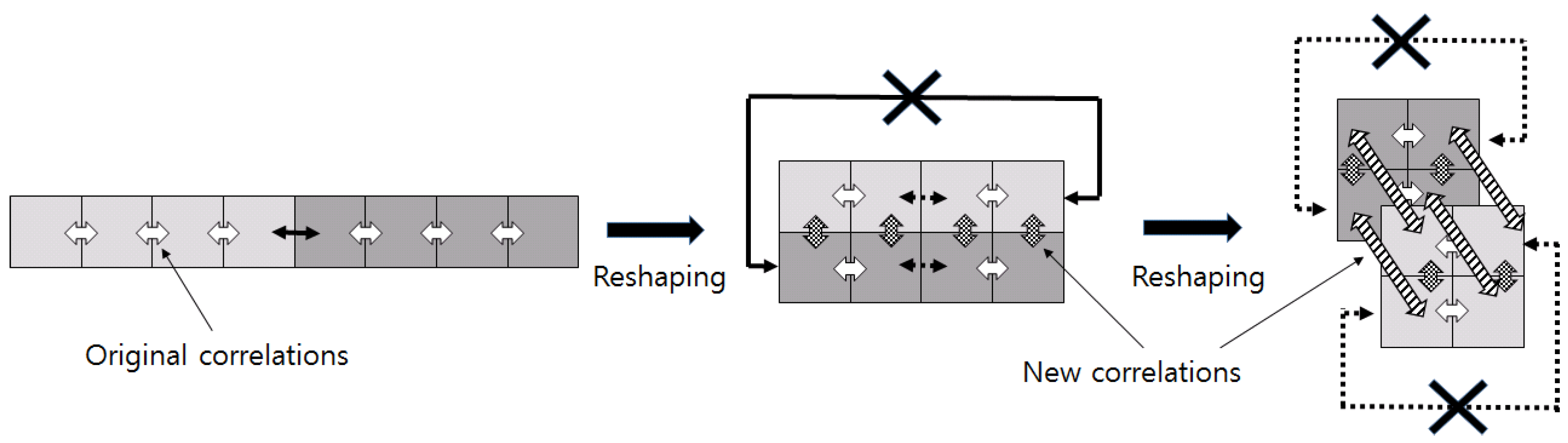
Tensors are generally multidimensional, and computing and classifying them in this state yields a curse of the dimension [
18]: in addition to not performing well because the classifiers do not model high-dimensional data properly from small amounts of samples compared to its dimension during training, processing high-dimensional data is very computationally expensive. However, the direct use of high-dimensional tensors is limited in most applications. The tensor is essentially limited to subspaces, which are several low dimensions, because the elements of the tensor often have correlations with the neighbor elements [
18,
19]. Therefore, feature extraction [
20] is used to transform the high dimensional data into low dimensions while maintaining the implied correlations in the original structure [
21].
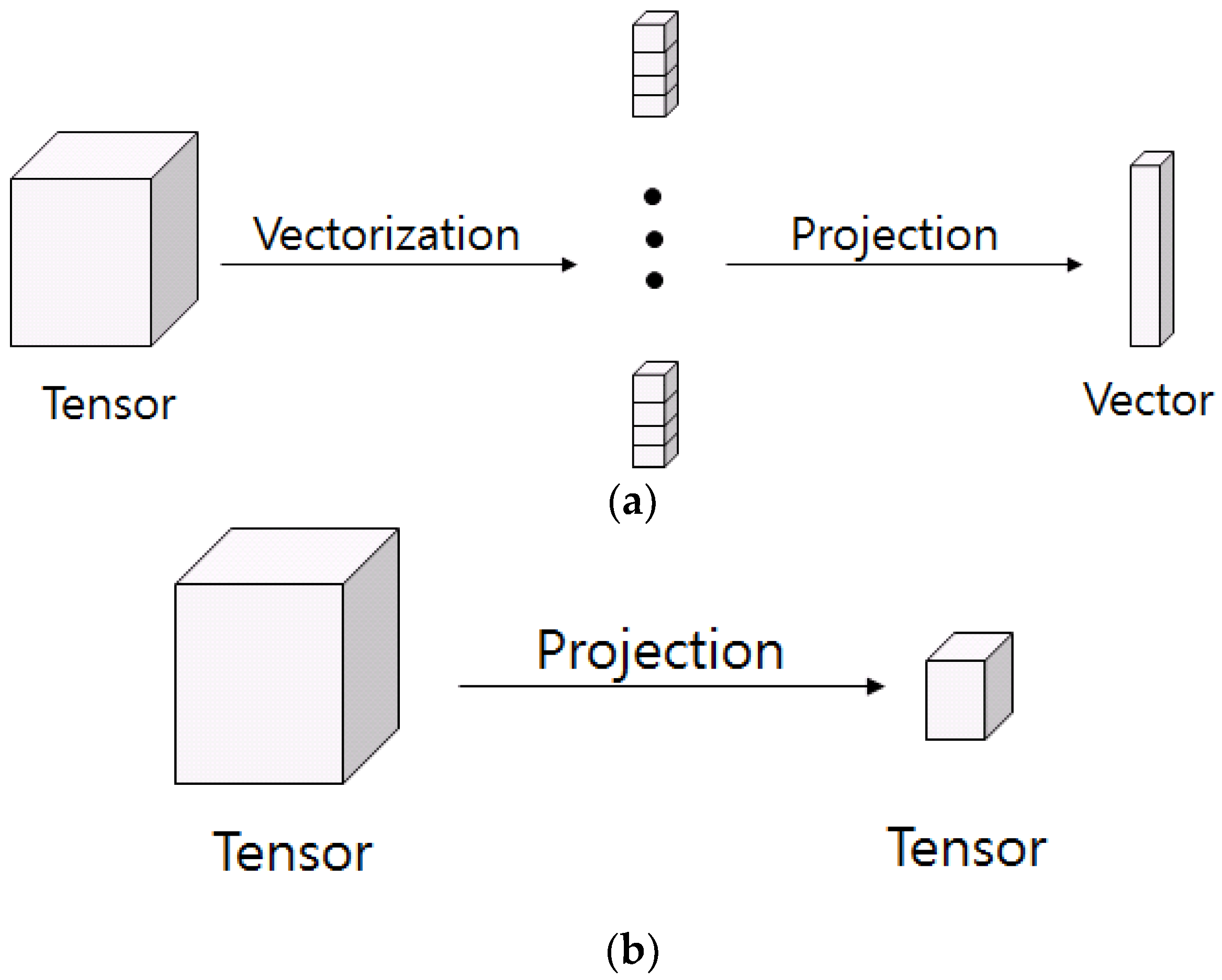
Principal component analysis (PCA) and linear discriminant analysis (LDA) are conventional unsupervised linear analyses for feature extraction. PCA is a method of reducing the dimension of the data while preserving the distribution of the data existing in the original data as much as possible [
22]. PCA computes the principal components, and the larger the principal component, the more the variance of the original data, and it is represented by a few components to reduce the dimension. Directly applying the tensor to the PCA requires the reordering of high dimensional data to a high dimensional vector that has high computational load.
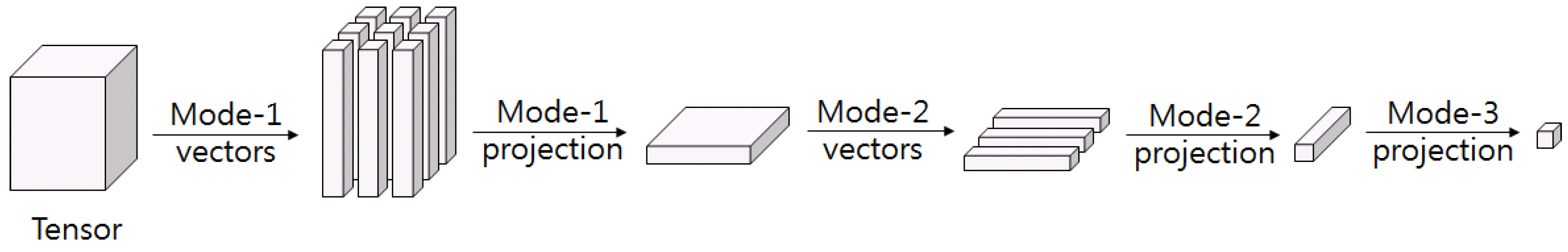
2D PCA is an attempt at dimension reduction without the vectorization of the input data at an initial study of multilinear subspace learning (MSL). The 2D PCA receives the image as an input to obtain the image covariance matrix. However, only the rows of the input image are applied by PCA [
23]. As a result, the 2D PCA is applied only to one mode, and it does not completely reduce the high dimensional data. The less restrictive 2D PCA method analyzes the correlation of local area in the space [
24]. For the input image, two PCAs are applied to the rows and columns, so that PCA is applied in 1-mode and 2-mode to achieve better dimensional reduction [
25]. Tensor subspace analysis finds local geometrical information by training low dimensional subspaces in the input image [
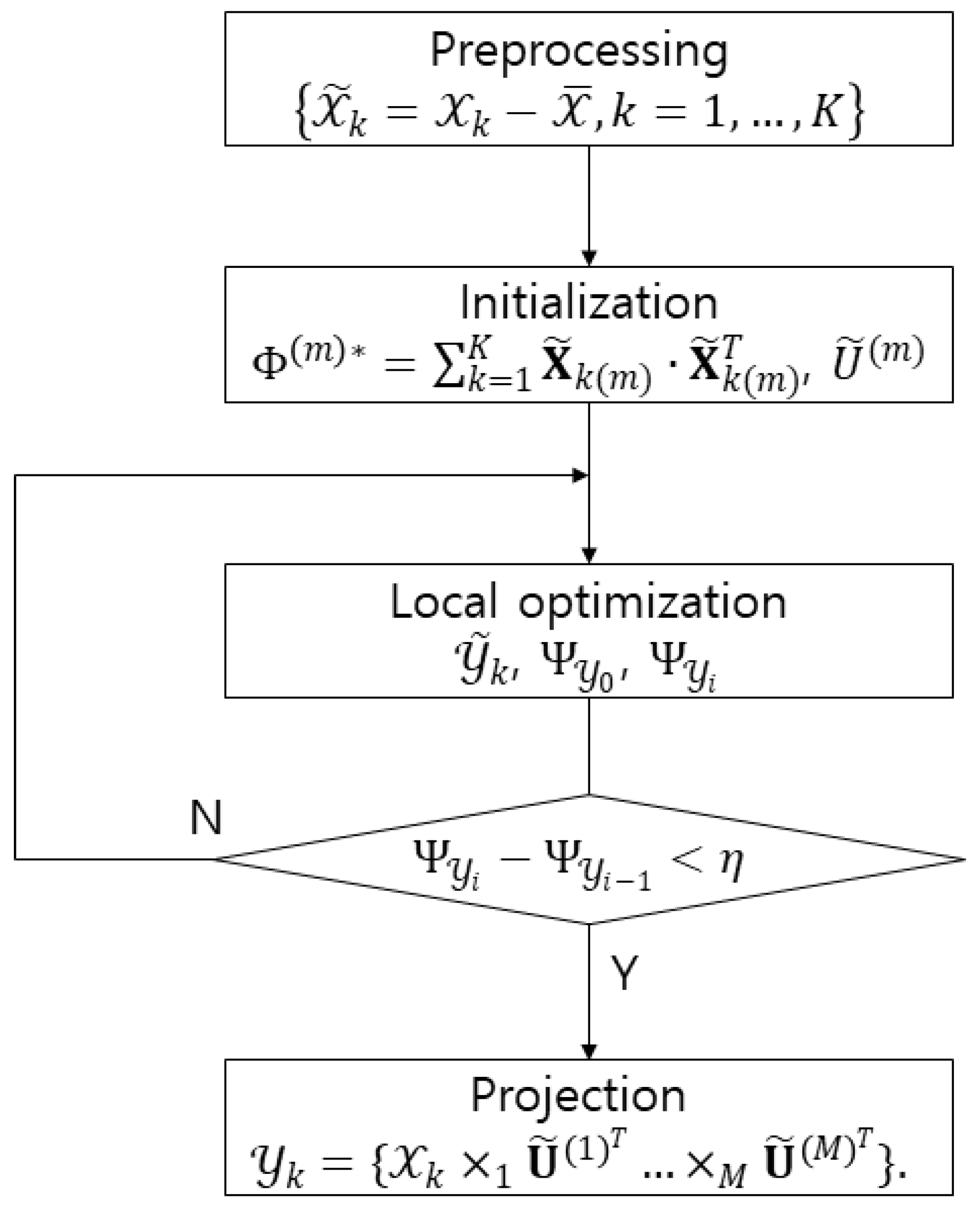
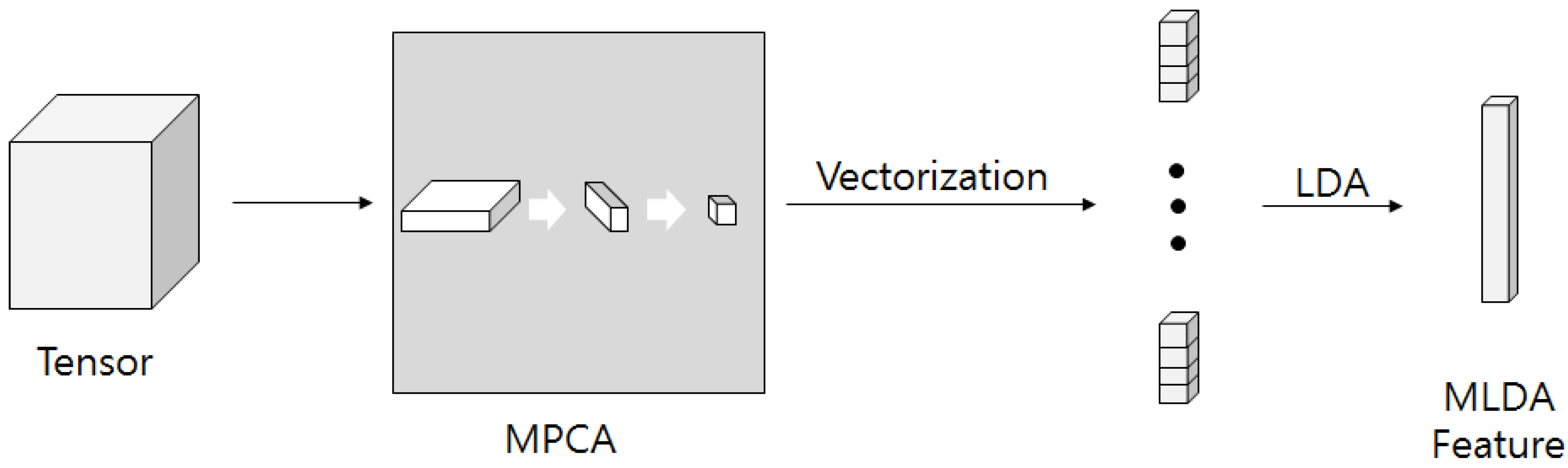
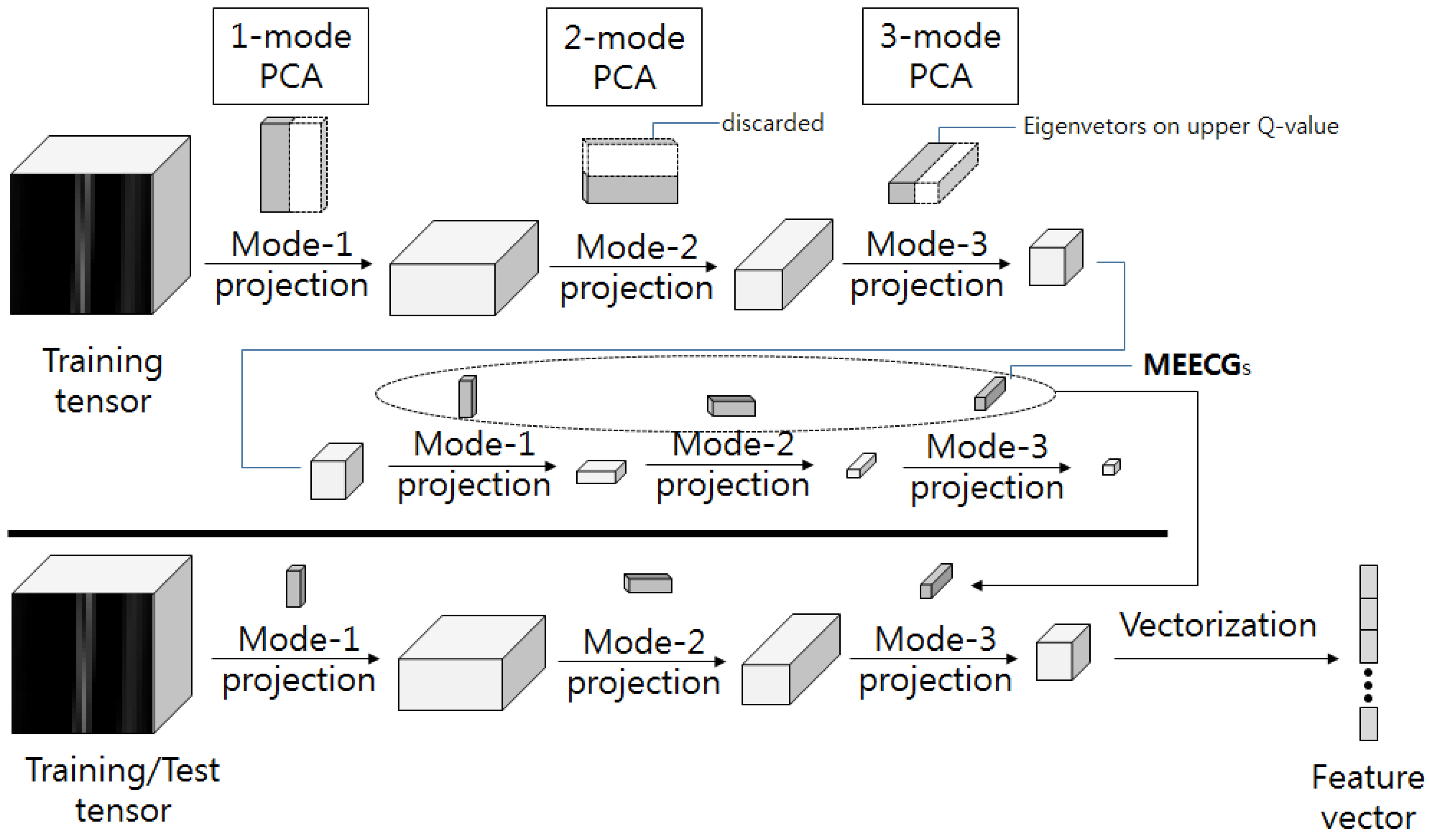
26]. Studies on multilinear subspace method for feature extraction from tensor are recently receiving attention. MSL is a projection that maps high dimensional tension to low dimensional vectors or tensors. Linear subspace learning such as PCA and LDA requires reordering of high dimensional data as a vector, destroying the structural correlations of the original data. Furthermore, there is a problem that the dimension of data is much larger than the number of data that is required for training. The latest multilinear subspace learning (MSL) preserves the geometric information of the original data by extracting and mapping features without deformation of the tensor structure, and it is possible to deal with large tensors efficiently because the problem is that the dimension of the data can be larger than the number of data for training. Here, multilinear principal component analysis (MPCA) and multilinear discriminant analysis (MLDA) are considered as MSL methods [
27,
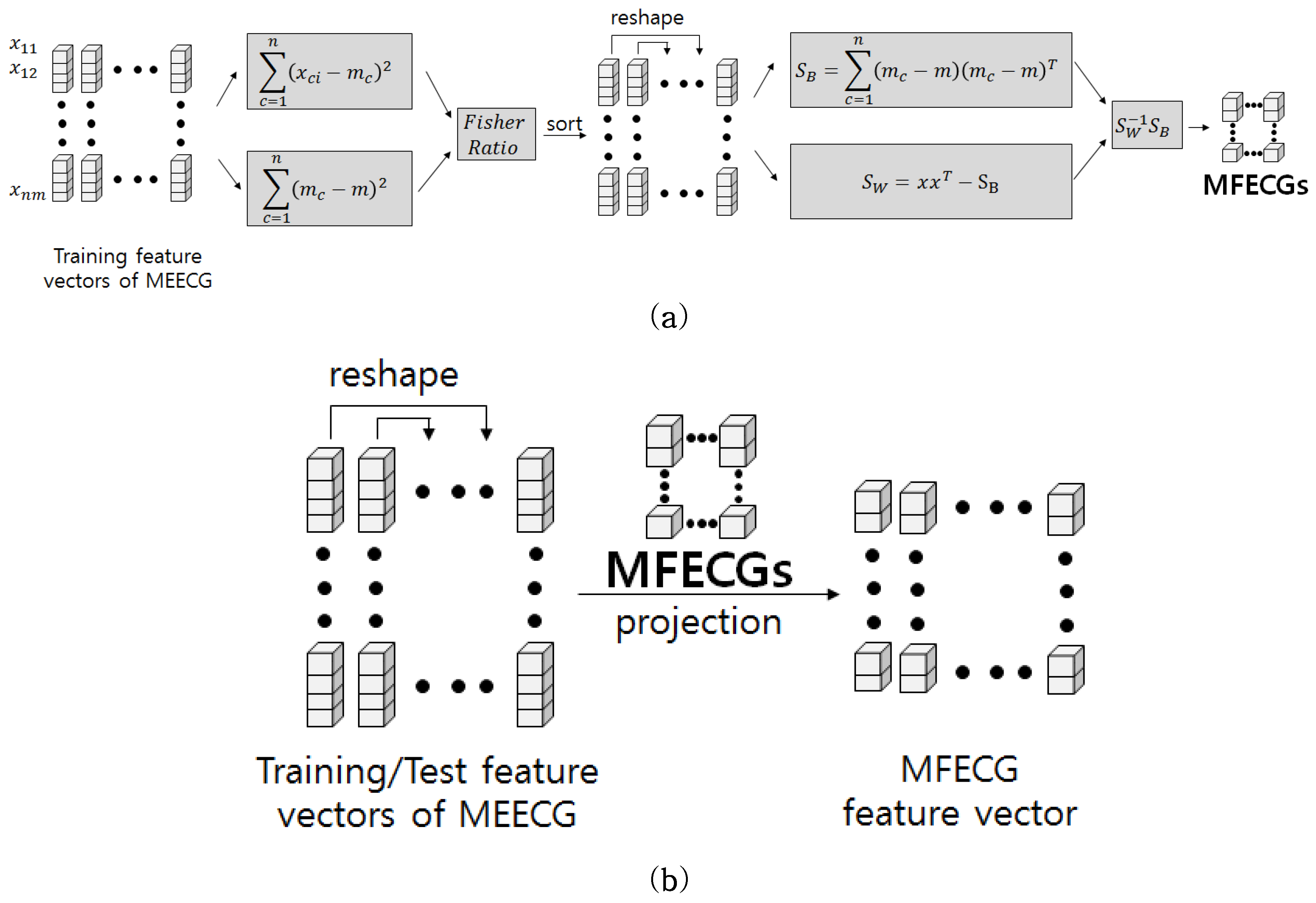
28].
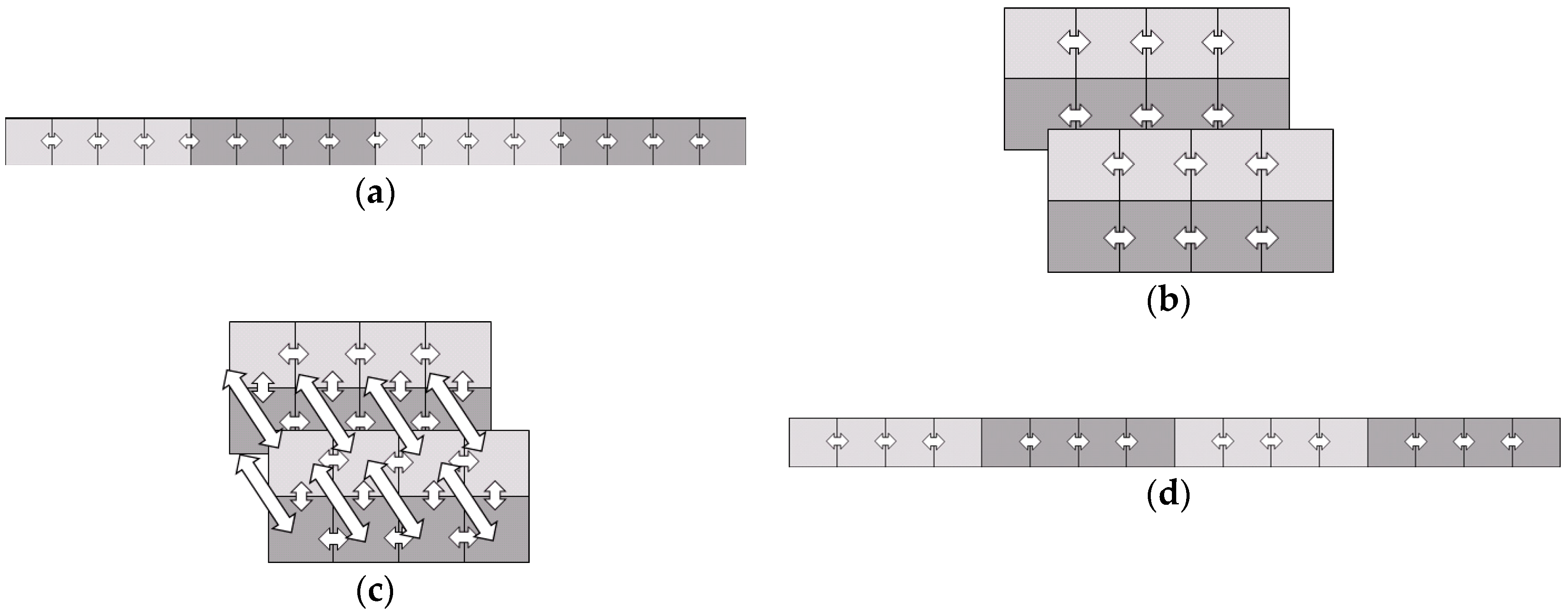
In the most active area, with three-dimensional information, such as a 3D object [
29], hyperspectral cube [
30], or gait video sequence [
31] with three modes following the
x-axis,
y-axis, and
z-axis, a third-order tensor, has been noted as an important point of study [
32,
33,
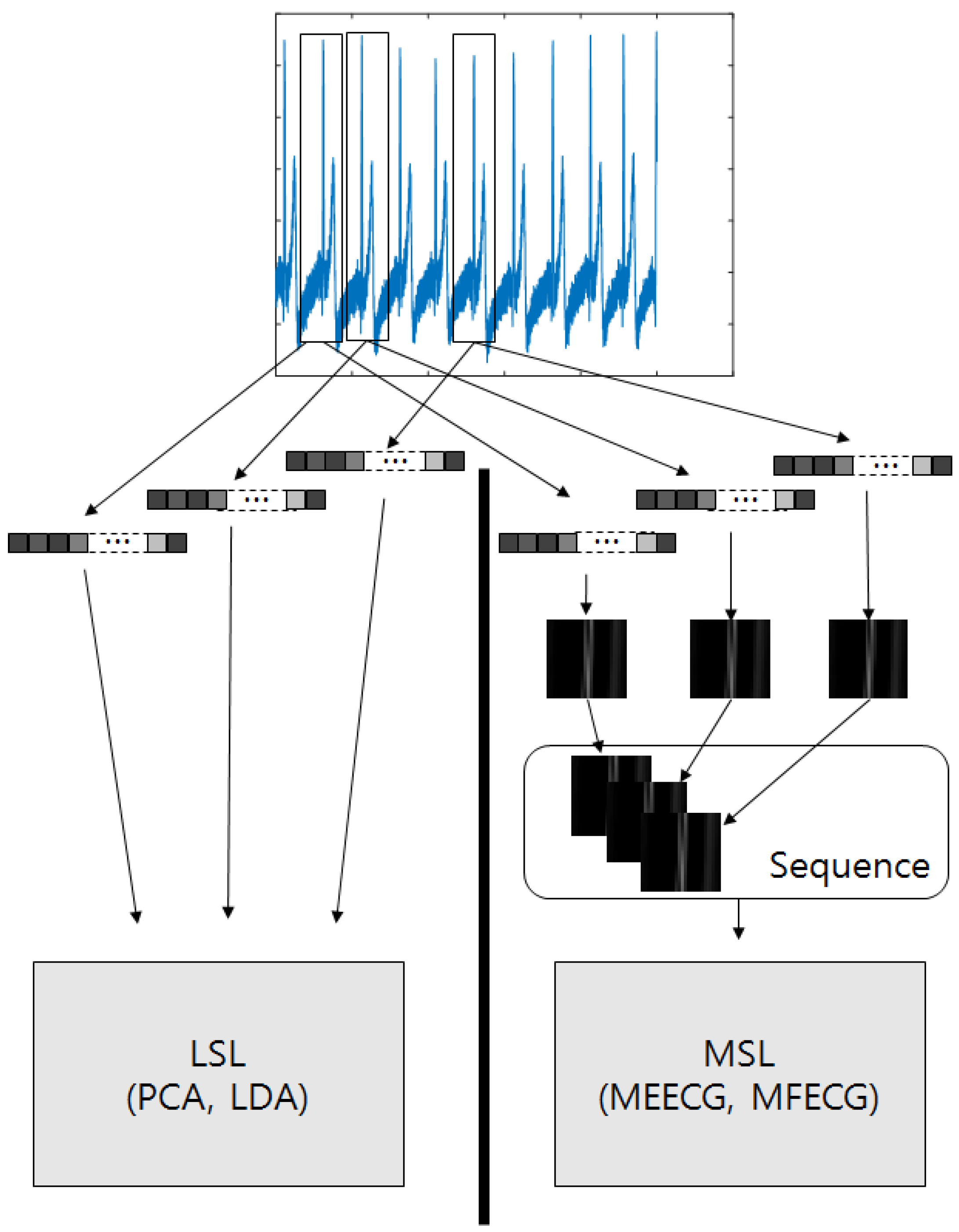
34]. Individual identification using ECG signals can be also considered as a multilinear tensor space with temporal dimension. However, it is not commonly considered to use an ECG signal as a multidimensional tensor and to extract features with multilinear projections. An ECG is normally gathered continuously as serial data. By applying it to a tensor, the input could arrange sequential information symmetrically in a tensor. Thus, tensor-based sequential input with the variant noisy ECG signals could be complementarily classified
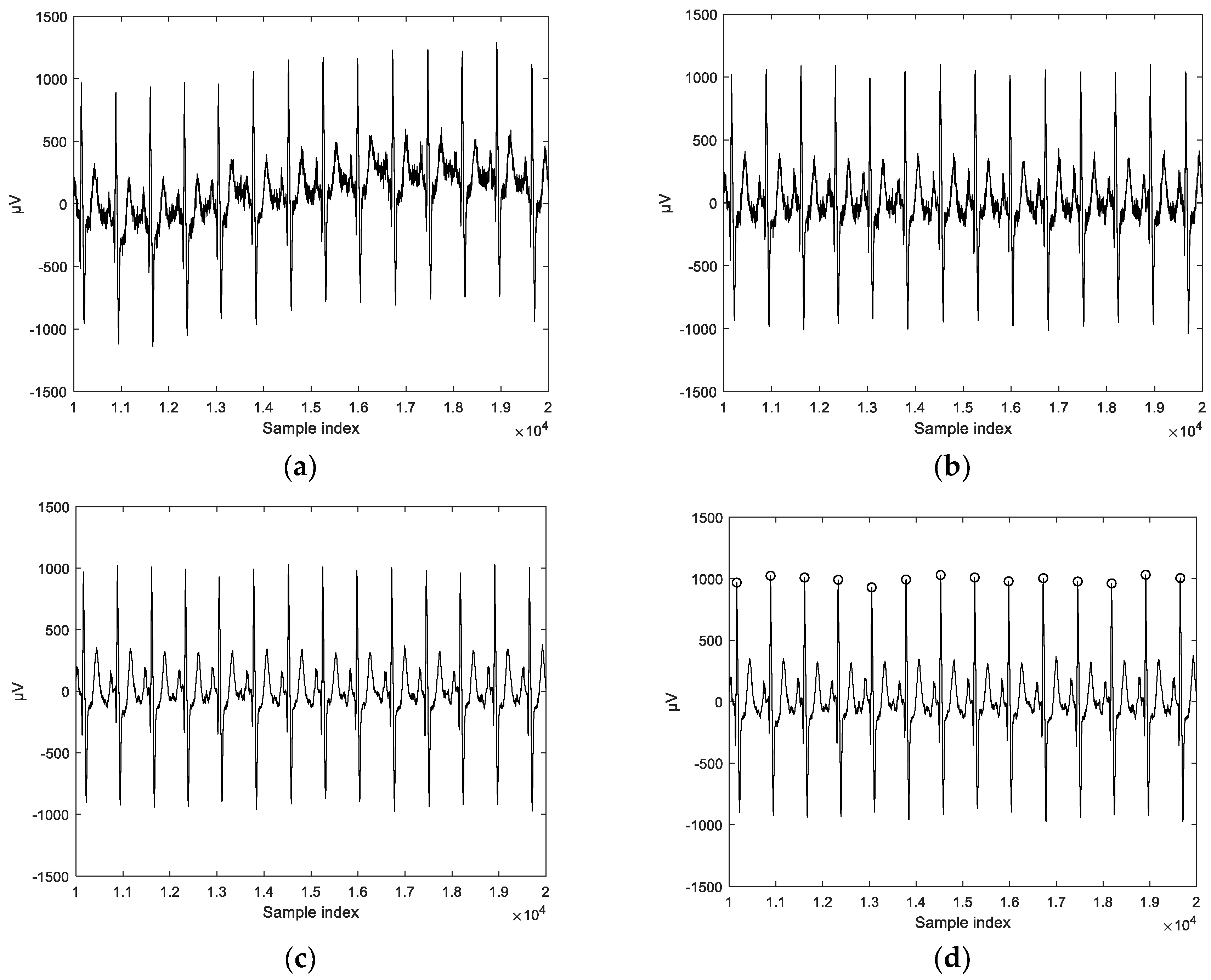
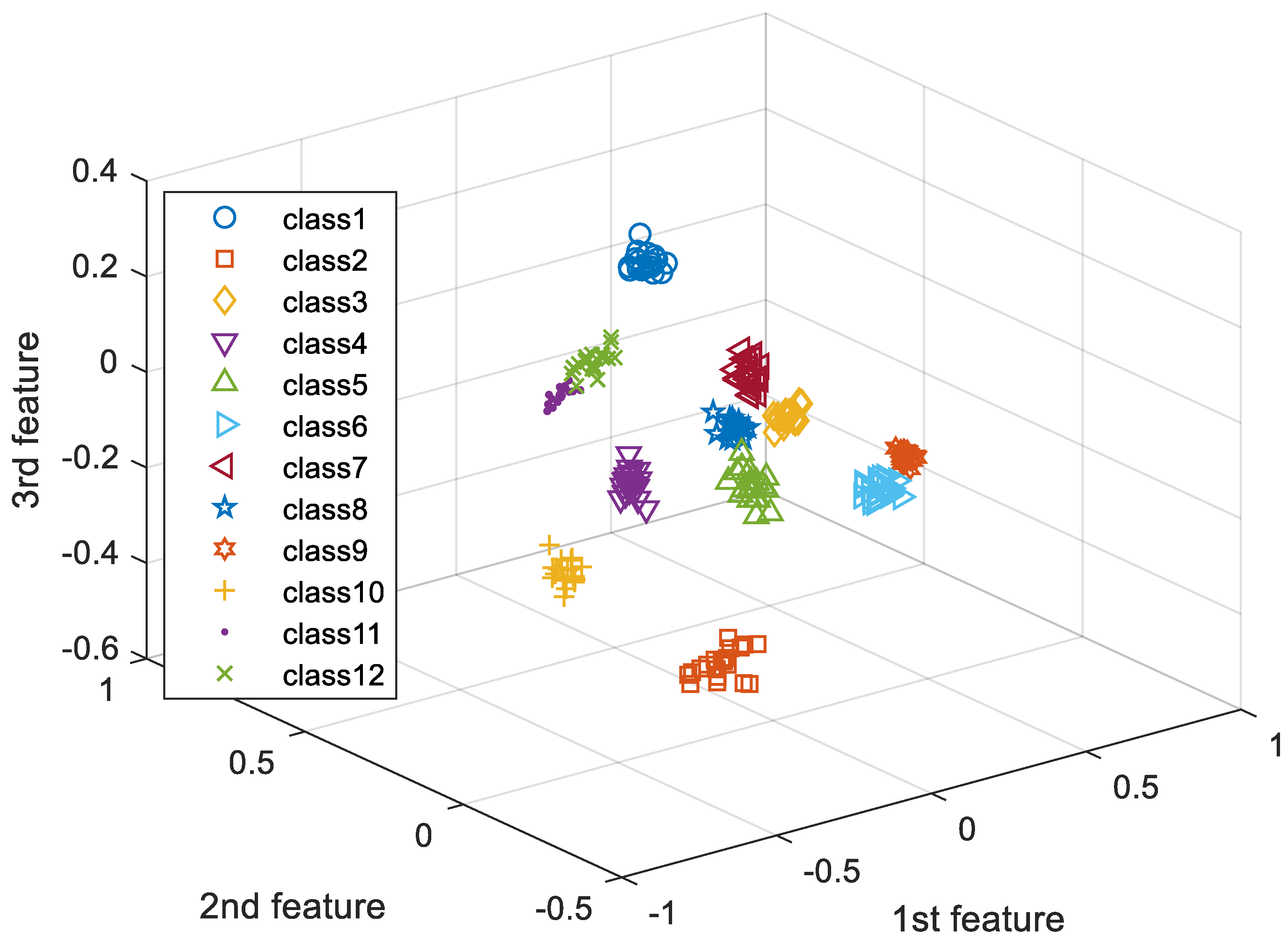
In this paper, we apply third-order tensor-based multilinear eigenECG (MEECG) and multilinear Fisher ECG (MFECG) for individual identification from the information source of an electrocardiogram sensor. The MEECG and MFECG are defined by MPCA-based ECG and MLDA-based ECG, respectively. The databases of Physikalisch-Technische Bundesanstalt (PTB) diagnosis and Chosun University (CU)-ECG are used for performance evaluation. The PTB-ECG is a well-known benchmarking database for ECG analysis [
35] and CU-ECG is a database of ECG signals directly built by Chosun University for this study.
This paper is organized in the following manner.
Section 2 introduces MSL such as MPCA and MLDA for multilinear projection. In
Section 3, ECG biometrics based on the MEECG and MFECG are described.
Section 4 covers the performance comparison and experimental results from two ECG databases. Finally, concluding comments are presented in
Section 5.