How Do mAbs Make Use of Complement to Kill Cancer Cells? The Role of Ca2+
Abstract
1. Introduction
2. Nucleated Cells Are More Complicated: Important Questions
3. Experimental Strategies
4. Quantitation and Visualization of Early Steps in mAb-Mediated CDC: In Vitro and In Vivo Studies
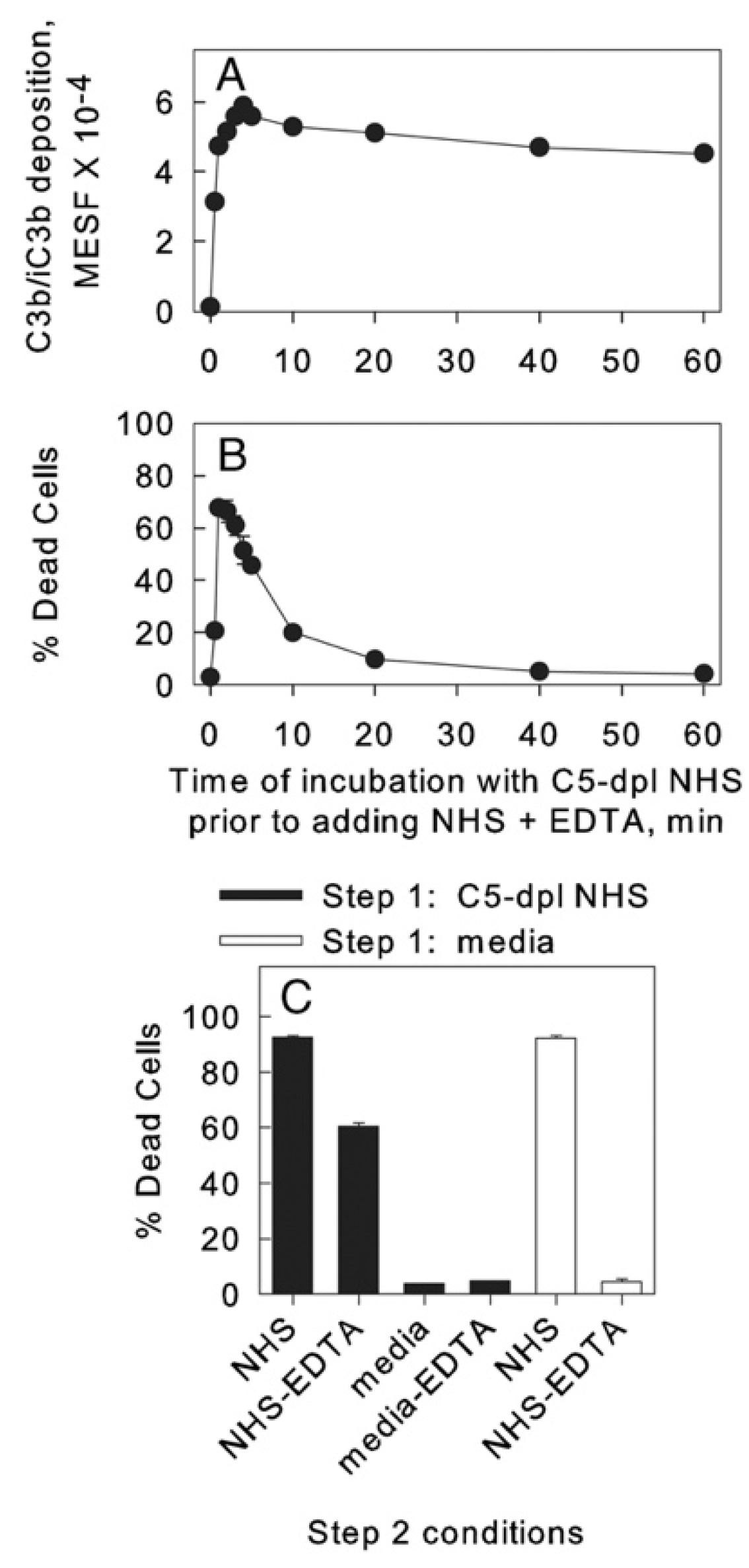
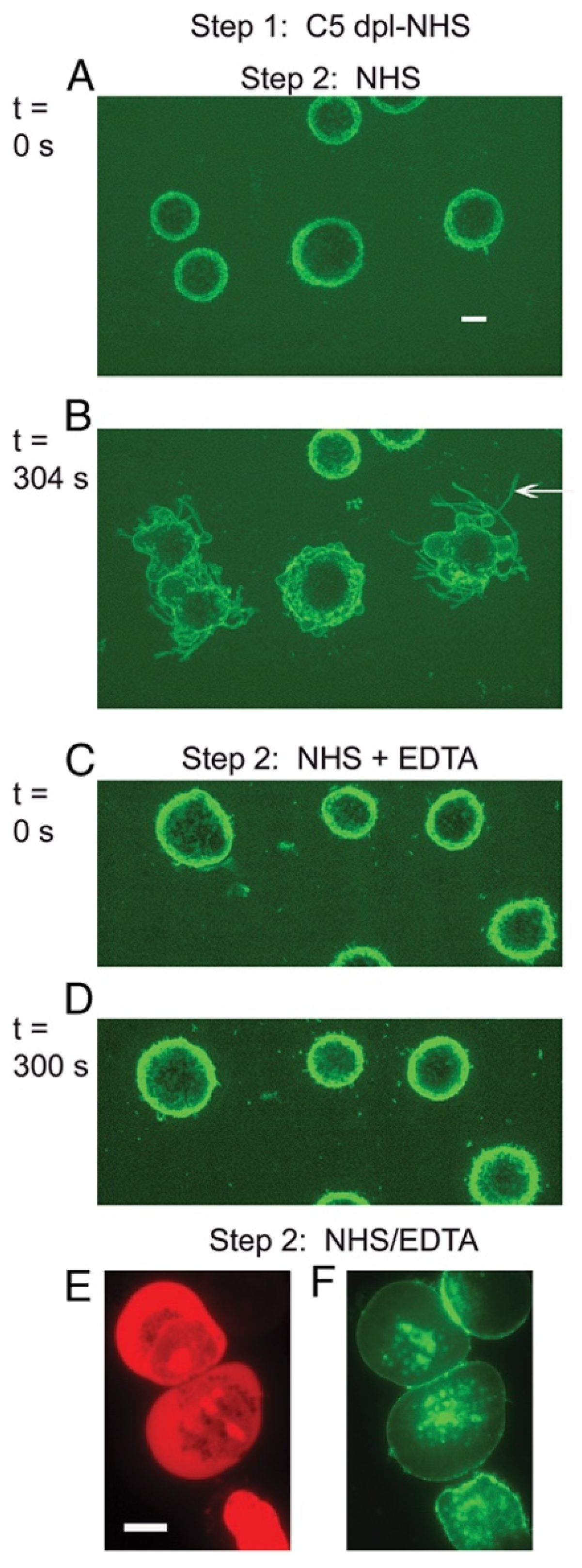
5. C3b Deposition Kinetics, a Key Intermediate Step in CDC; the “Discovery” of Streamers
6. On the Importance of Ca2+
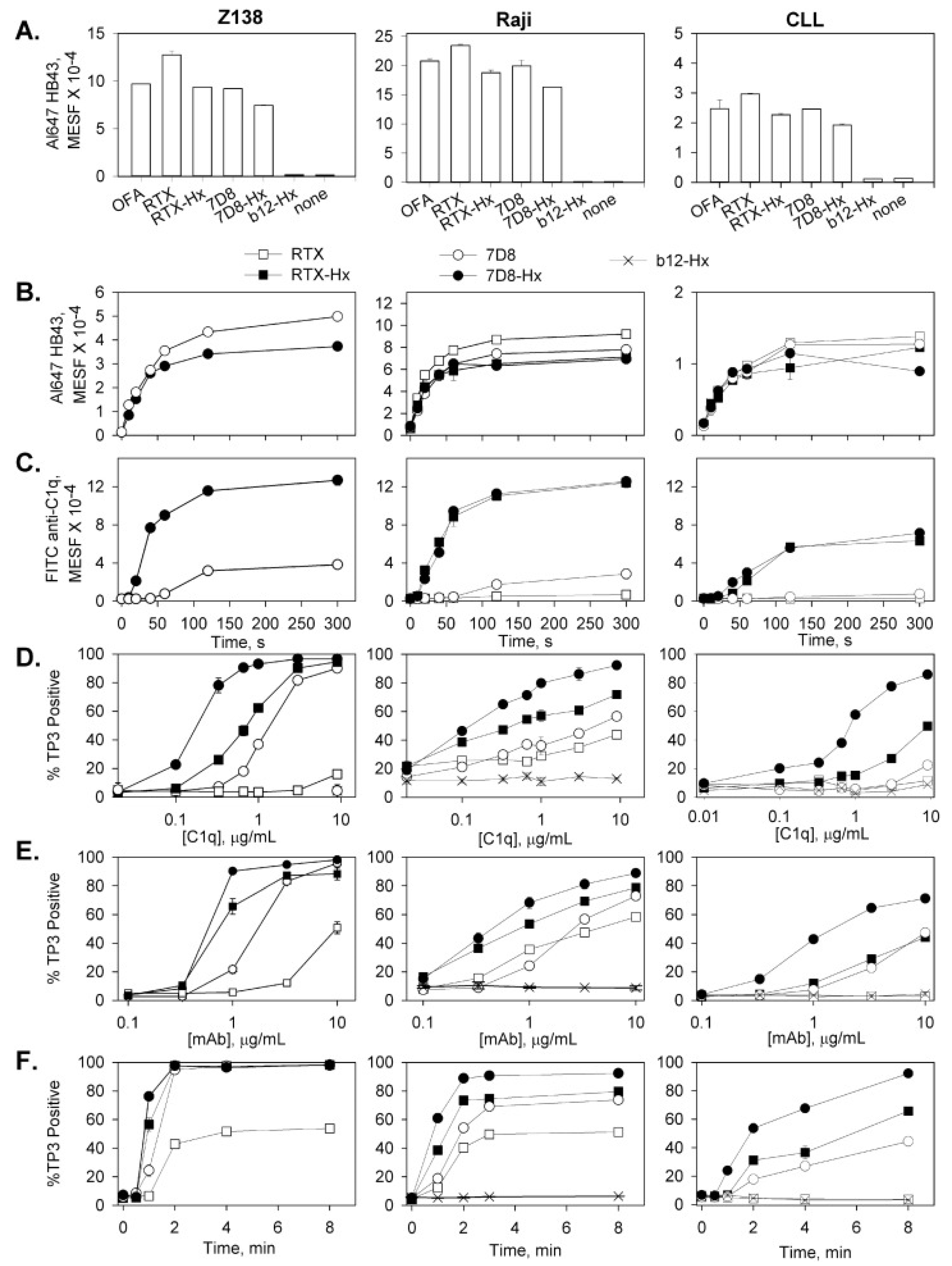
7. Hexamer-Forming mAbs are More Effective in Activating C
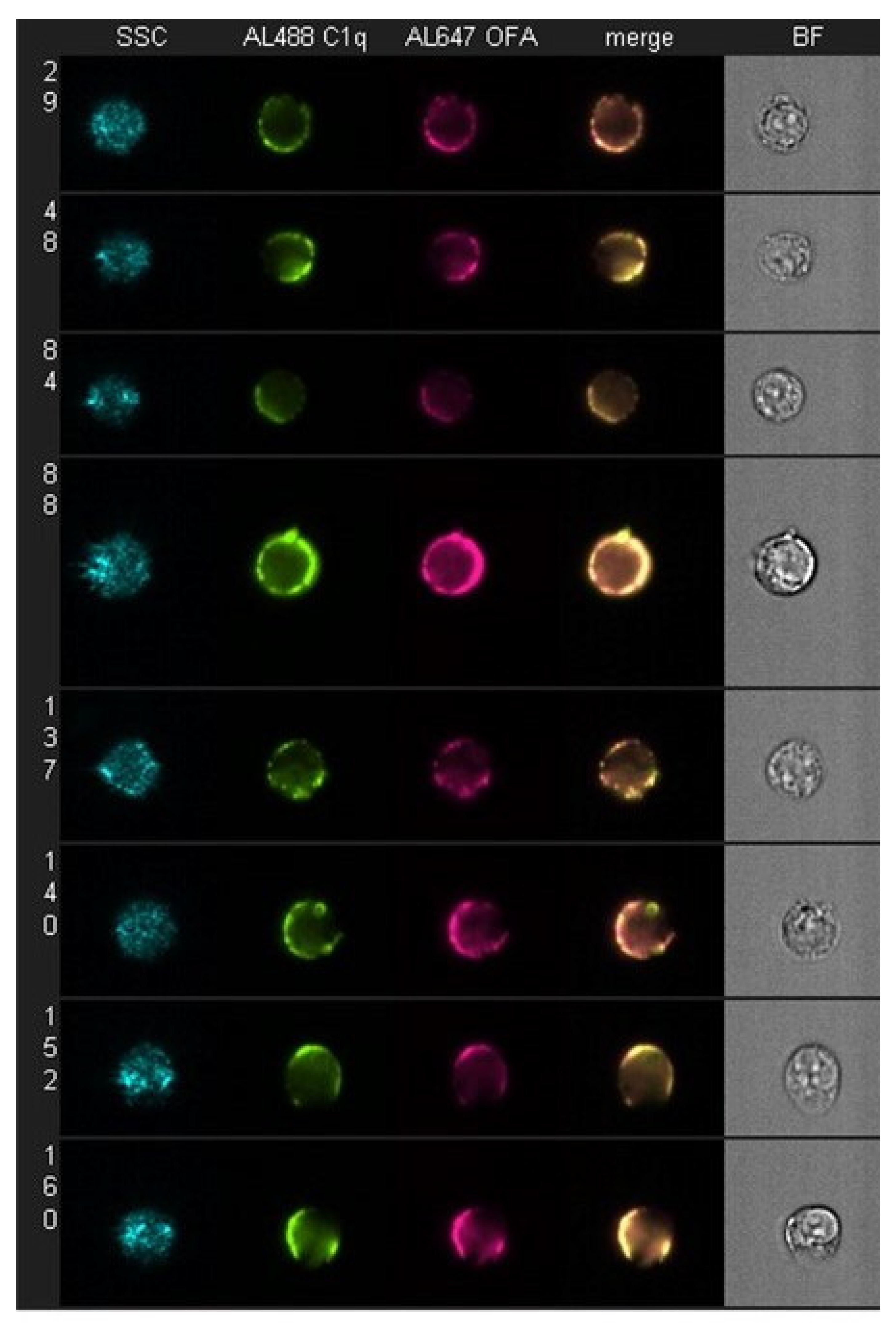
7.1. Cell-Bound Hexamer-Forming mAbs Bind C1q
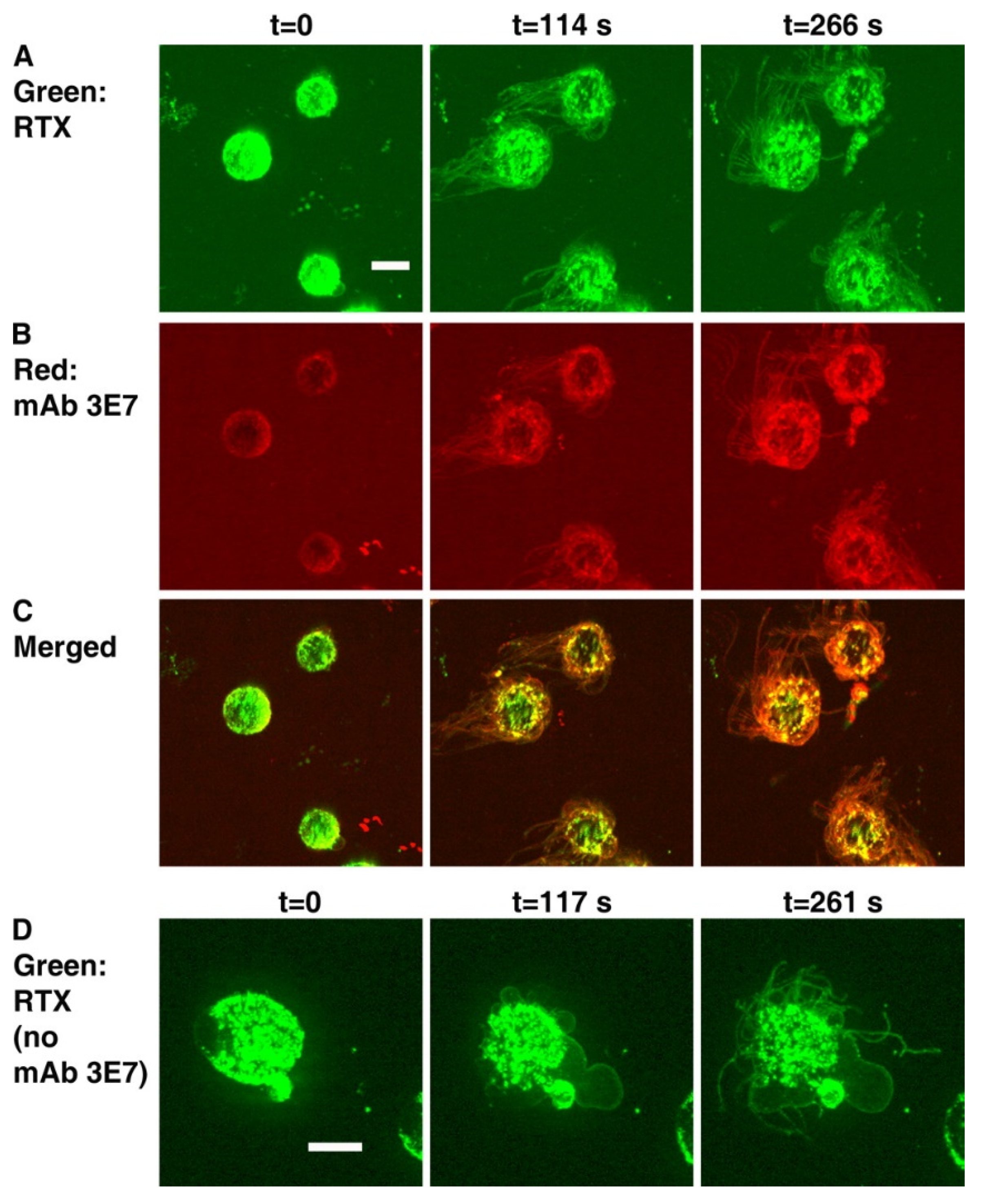
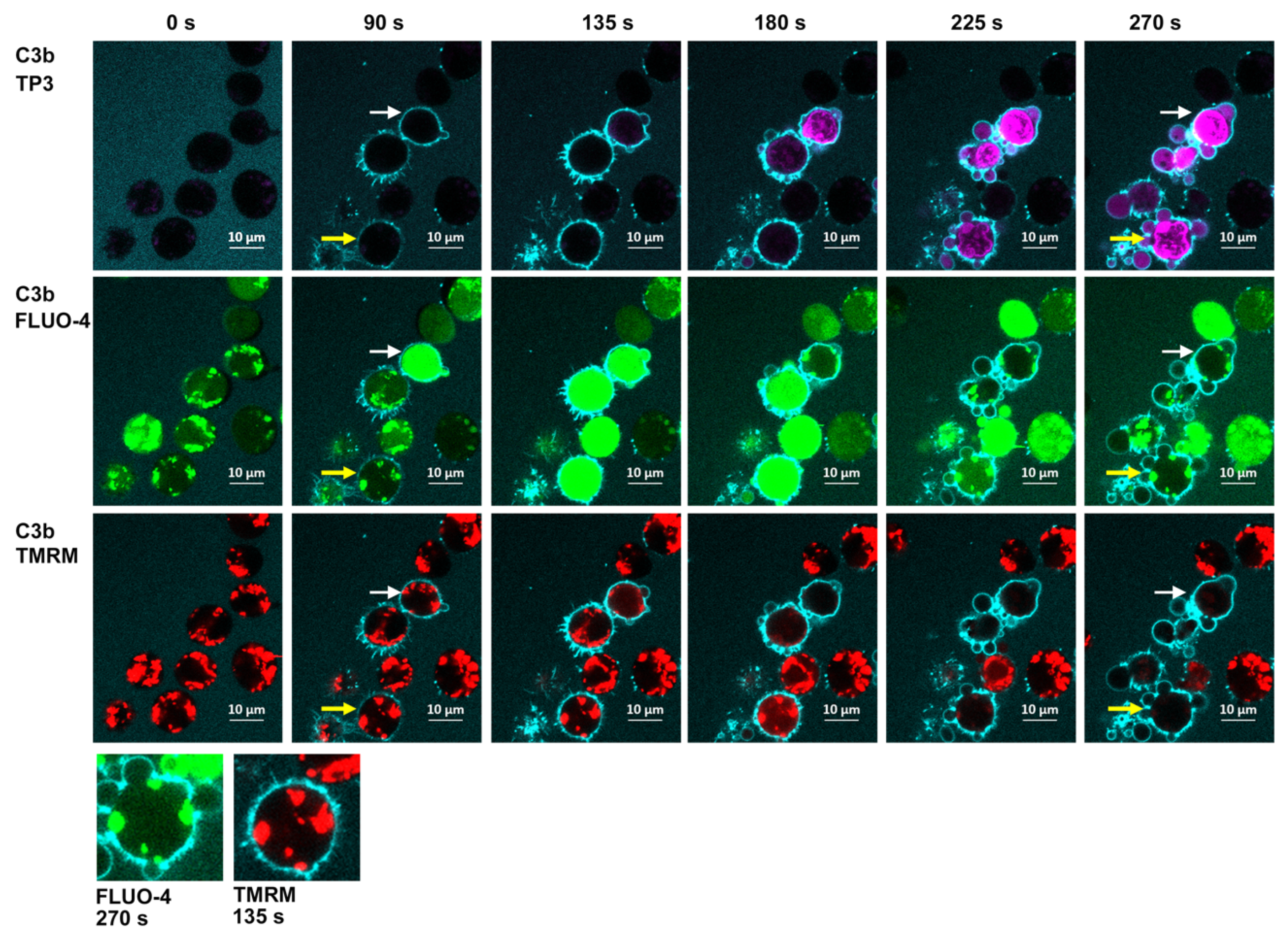
7.2. Four-Color Confocal Microscopy Movies
7.3. Kinetics of CDC Monitored by Multicolor HRDI
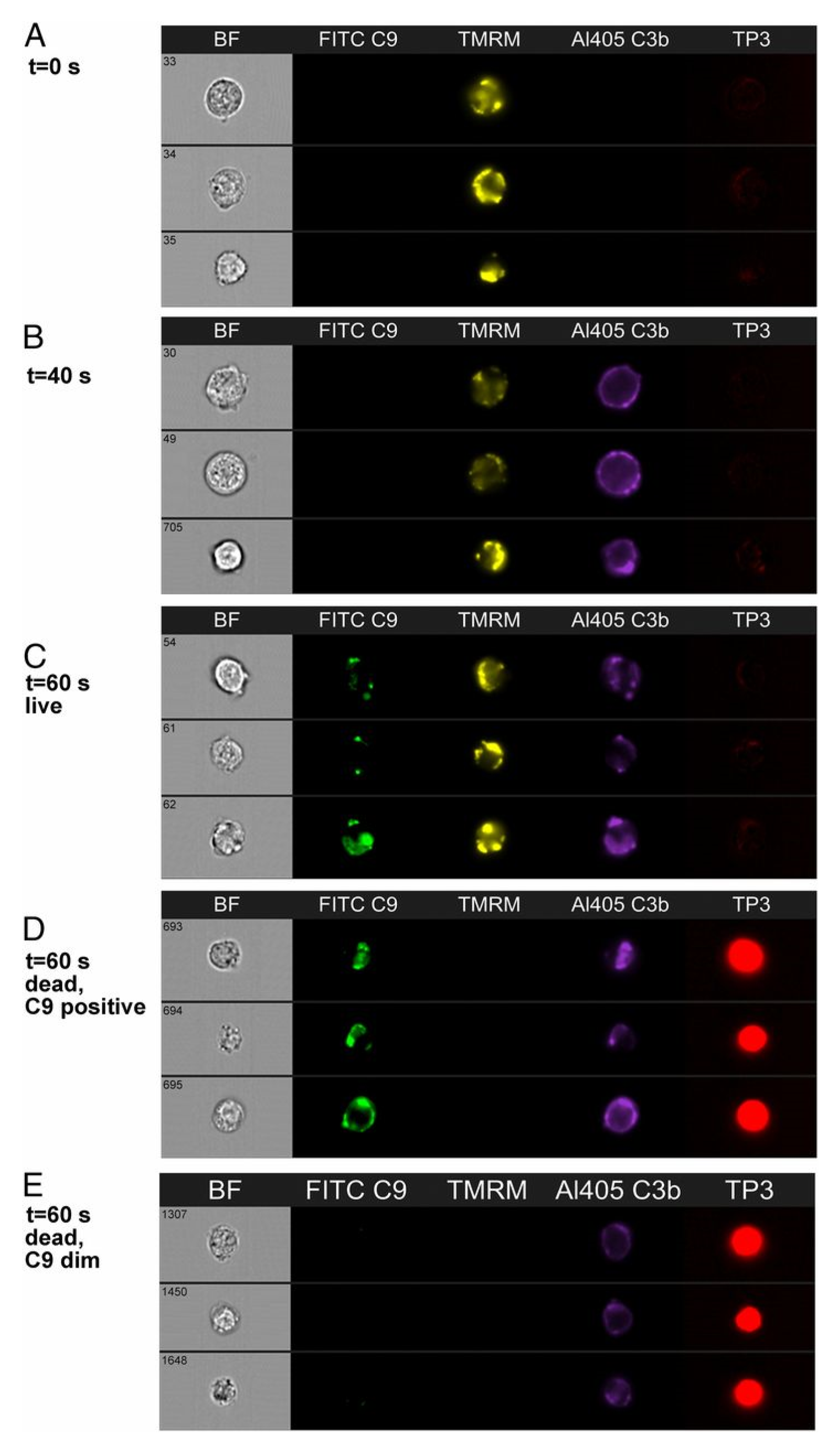
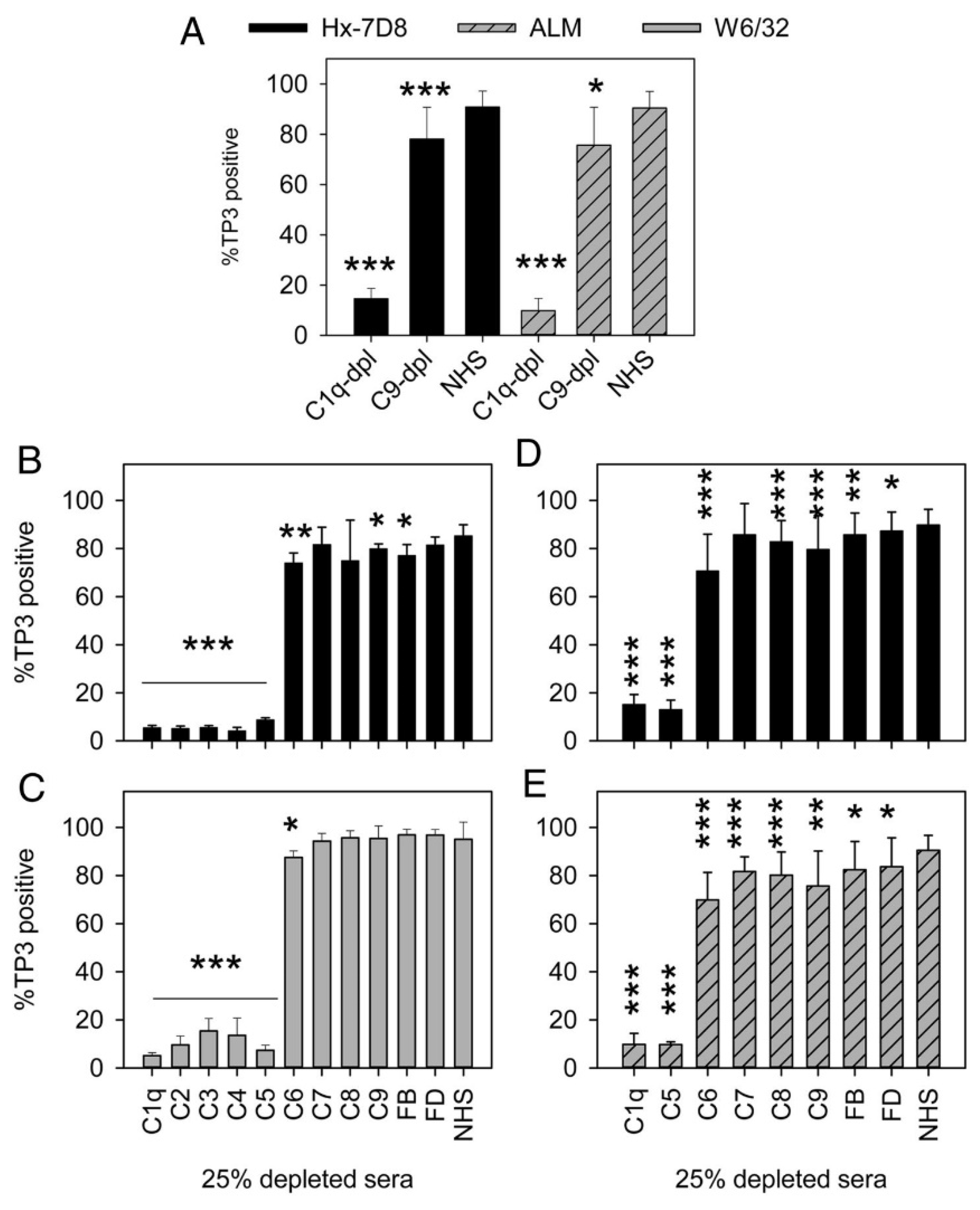
8. On the Role of C9
9. Ca2+ Appears to be the Key: Implications
10. The Future
11. Summary
12. Patents
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| 7AAD | 7-aminoactinomycin D |
| Al | Alexa dye |
| APC | alternative pathway of complement |
| BDSS | bright detail similarity score |
| C | complement |
| CDC | complement-dependent cytotoxicity |
| CLL | chronic lymphocytic leukemia |
| EAC | Ehrlich ascites cell |
| FcεRI | the high-affinity receptor for the Fc region of IgE |
| FcγR | receptor for the Fc region of IgG |
| GMF | geometric mean fluorescence |
| HRDI | high-resolution digital imaging in a flow cytometry environment |
| MAC | membrane attack complex |
| mAb | monoclonal antibody |
| NHS | normal human serum |
| OFA | ofatumumab |
| PI | propidium iodide |
| RTX | rituximab |
| TNTs | tunneling nanotubules |
| TMRME | tetramethylrhodamine methyl ester |
References
- Cavaillon, J.M.; Sansonetti, P.; Goldman, M. 100th anniversary of Jules Bordet’s Nobel prize: Tribute to a founding father of immunology. Front. Immunol. 2019. [Google Scholar] [CrossRef] [PubMed]
- Reis, E.D.; Mastellos, D.C.; Ricklin, D.; Mantovani, A.; Lambris, J.D. Complement in cancer: Untangling an intricate relationship. Nat. Rev. Immunol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Ricklin, D.; Mastellos, D.C.; Reis, E.S.; Lambris, J.D. The renaissance of complement therapeutics. Nat. Rev. Nephrol. 2018, 14, 26–47. [Google Scholar] [CrossRef] [PubMed]
- Mastellos, D.C.; Ricklin, D.; Lambris, J.D. Clinical promise of next-generation complement therapeutics. Nat. Rev. Drug Disc. 2019, 18. [Google Scholar] [CrossRef] [PubMed]
- Golay, J.; Taylor, R.P. The role of complement in the mechanism of action of therapeutic anti-cancer mAbs. Antibodies. Submitted.
- Mayer, M.M. Mechanism of cytolysis by complement. PNAS 1972, 69, 2954–2958. [Google Scholar] [CrossRef] [PubMed]
- Ohanian, S.H.; Schlager, S.I. Humoral immune killing of nucleated cells: Mechanisms of complement-mediated attack and target cell defense. Crit. Rev. Immunol. 1981, 1, 165–209. [Google Scholar] [PubMed]
- Walport, M.J. Complement. N. Engl. J. Med. 2001, 344, 1058–1066. [Google Scholar] [CrossRef] [PubMed]
- Morgan, B.P.; Luzio, J.P.; Campbell, A.K. Intracellular Ca2+ and cell injury: A paradoxical role of Ca2+ in complement membrane attack. Cell Calcium 1986, 7, 399–411. [Google Scholar] [CrossRef]
- Kim, S.H.; Carney, D.F.; Papadimitriou, J.C.; Shin, M.L. Effect of osmotic protection on nucleated cell killing by C5b-9: Cell death is not affected by the prevention of cell swelling. Mol. Immmunol. 1989, 26, 323–331. [Google Scholar]
- Carney, D.F.; Koski, C.L.; Shin, M.L. Elimination of terminal complement intermediates from the plasma membrane of nucleated cells: The rate of disppearance differs for cells carrying C5b-7 or C5b-8 or a mixture of C5b-8 with a limited number of C5b-9. J. Immunol. 1985, 134, 1804–1809. [Google Scholar]
- Kim, S.; Carney, D.F.; Hammer, C.H.; Shin, M.L. Nucleated cell killing by complement: Effects of C5b-9 channel size and extracellular Ca2+ on the lytic process. J. Immunol. 1987, 138, 1530–1536. [Google Scholar]
- Carney, D.F.; Lang, T.J.; Shin, M.L. Multiple signal messengers generated by terminal complement complexes and their role in terminal complement complex elimination. J. Immunol. 1990, 145, 623–629. [Google Scholar] [PubMed]
- Papadimitriou, J.C.; Ramm, L.E.; Drachenberg, C.B.; Trump, B.F.; Shin, M.L. Quantitative analysis of adenine nucleotides during the prelytic phase of cell death mediated by C5b-9. J. Immunol. 1991, 147, 212–217. [Google Scholar] [PubMed]
- Papadimitriou, J.C.; Phelps, P.C.; Shin, M.L.; Smith, M.W.; Trump, B.F. Effects of Ca2+ deregulation on mitochondrial membrane potential and cell viability in nucleated cells following lytic complement attack. Cell Calcium 1994, 15, 217–227. [Google Scholar] [CrossRef]
- Lindorfer, M.A.; Cook, E.M.; Tupitza, J.C.; Zent, C.S.; Burack, R.; De Jong, R.; Beurskens, F.J.; Schuurman, J.; Parren, P.W.; Taylor, R.P. Real-time analysis of the detailed sequence of cellular events in mAb-mediated complement-dependent cytotoxicity of B-cell lines and of CLL B-cells. Mol. Immunol. 2016, 70, 13–23. [Google Scholar] [CrossRef] [PubMed]
- Pawluczkowycz, A.W.; Beurskens, F.J.; Beum, P.V.; Lindorfer, M.; Van De Winkel, J.G.J.; Parren, P.W.; Taylor, R.P. Binding of submaximal C1q promotes complement-dependent cytotoxicity (CDC) of B cells opsonized with anti-CD20 mAbs ofatumumab (OFA) or rituximab (RTX): Considerably higher levels of CDC are induced by OFA than by RTX. J. Immunol. 2009, 183, 749–758. [Google Scholar] [CrossRef] [PubMed]
- Beum, P.V.; Lindorfer, M.A.; Hall, B.E.; George, T.C.; Frost, K.; Morrissey, P.J.; Taylor, R.P. Quantitative analysis of protein co-localization on B cells opsonized with rituximab and complement using the ImageStream multispectral imaging flow cytometer. J. Immunol. Methods 2006, 317, 90–99. [Google Scholar] [CrossRef]
- Beum, P.V.; Lindorfer, M.A.; Beurskens, F.; Stukenberg, P.; Lokhorst, H.M.; Pawluczkowycz, A.W.; Parren, P.W.; Van De Winkel, J.G.J.; Taylor, R.P. Complement activation on B lymphocytes opsonized with rituximab or ofatumumab produces substantial changes in membrane structure preceding cell lysis. J. Immunol. 2008, 181, 822–832. [Google Scholar] [CrossRef]
- Cook, E.M.; Lindorfer, M.A.; van der Horst, H.; Oostindie, S.; Beurskens, F.J.; Schuurman, J.; Schuurman, J.; Zent, C.S.; Burack, R.; Parren, P.W.H.I.; et al. Antibodies that efficiently form hexamers upon antigen binding can induce complement-dependent cytotoxicity under complement-limiting conditions. J. Immunol. 2016, 197, 1762–1775. [Google Scholar] [CrossRef]
- Taylor, R.P.; Lindorfer, M.A.; Cook, E.M.; Beurskens, F.J.; Schuurman, J.; Parren, P.W.; Zent, C.S.; VanDerMeid, K.R.; Burack, R.; Mizuno, M.; et al. Hexamerization-enhanced CD20 antibody mediates complement-dependent cytotoxicity in serum genetically deficient in C9. Clin. Immunol. 2017, 181, 24–28. [Google Scholar] [CrossRef]
- Beum, P.V.; Lindorfer, M.A.; Peek, E.M.; Stukenberg, P.; De Weers, M.; Beurskens, F.J.; Parren, P.W.; Van De Winkel, J.G.J.; Taylor, R.P. Penetration of antibody-opsonized cells by the membrane attack complex of complement promotes Ca2+ influx and induces streamers. Eur. J. Immunol. 2011, 41, 2436–2446. [Google Scholar] [CrossRef] [PubMed]
- Reiter, Y.; Ciobotariu, A.; Jones, J.; Morgan, B.P.; Fishelson, Z. Complement membrane attack complex, perforin, and bacterial exotoxins induce in K562 cells calcium-dependent cross-protection from lysis. J. Immunol. 1995, 155, 2203–2210. [Google Scholar] [PubMed]
- Hörl, S.; Bánki, Z.; Huber, G.; Ejaz, A.; Windisch, D.; Muellauer, B.; Willenbacher, E.; Steurer, M.; Stoiber, H. Reduction of complement factor H binding to CLL cells improves the induction of rituximab-mediated complement-dependent cytotoxicity. Leukemia 2013, 27, 2200–2208. [Google Scholar] [CrossRef] [PubMed]
- Okroj, M.; Holmquist, E.; Nilsson, E.; Anagnostaki, L.; Jirström, K.; Blom, A.M. Local expression of complement factor I in breast cancer cells correlates with poor survival and recurrence. Cancer Immunol. Immunother. 2015, 64, 467–478. [Google Scholar] [CrossRef] [PubMed]
- Bordron, A.; Bagacean, C.; Mohr, A.; Tempescul, A.; Bendaoud, B.; Deshayes, S.; Dalbies, F.; Buors, C.; Saad, H.; Berthou, C.; et al. Resistance to complement activation, cell membrane hypersialytion and relapses in chronic lymphocytic leukemia patients treated with rituximab and chemotherapy. Oncotarget 2018, 9, 31590–31605. [Google Scholar] [CrossRef]
- Fishelson, Z.; Kirschfink, M. Complement C5b-9 and cancer: Mechanisms of cell damage, cancer counteractions, and approaches for intervention. Front. Immunol. 2019. [Google Scholar] [CrossRef] [PubMed]
- Geller, A.; Yan, J. The role of membrane bound complement regulatory proteins in tumor development and cancer immunotherapy. Front. Immunol. 2019. [Google Scholar] [CrossRef] [PubMed]
- Roumenina, L.T.; Daugan, M.V.; Noe, R.; Petitprez, F.; Vano, Y.-A.; Sanchez-Salas, R.; Becht, E.; Meilleroux, J.; Le Clec’H, B.; A Giraldo, N.; et al. Tumor cells hijack macrophage-produced complement C1q to promote tumor growth. Cancer Immunol. Res. 2019. [Google Scholar] [CrossRef] [PubMed]
- Roumenia, L.T.; Daugan, M.V.; Petitprez, F.; Sautes-Fridman, C.; Fridman, W.H. Context-dependentt roles of complement in cancer. Nat. Rev. Cancer 2019, 19, 1–18. [Google Scholar] [CrossRef]
- Beurskens, F.J.; Lindorfer, M.A.; Farooqui, M.; Beum, P.V.; Engelberts, P.; Mackus, W.J.M.; Parren, P.W.; Wiestner, A.; Taylor, R.P. Exhaustion of cytotoxic effector systems may limit monoclonal antibody-based immunotherapy in cancer patients. J. Immunol. 2012, 188, 3532–3541. [Google Scholar] [CrossRef]
- Oostindie, S.C.; Van Der Horst, H.J.; Lindorfer, M.; Cook, E.M.; Tupitza, J.C.; Zent, C.S.; Burack, R.; VanDerMeid, K.R.; Strumane, K.; Chamuleau, M.E.D.; et al. CD20 and CD37 antibodies synergize to activate complement by Fc-mediated clustering. Haematologica 2019, 104, 1841–1852. [Google Scholar] [CrossRef] [PubMed]
- Pokrass, M.J.; Liu, M.F.; Lindorfer, M.A.; Taylor, R.P. Activation of complement by monoclonal antibodies that target cell-associated á2-microglobulin: Implications for cancer immunotherapy. Mol. Immunol. 2013, 56, 549–560. [Google Scholar] [CrossRef] [PubMed]
- Wald, G. Nobel Prize Lecture: The Molecular Basis of Visual Excitation. 1967. Available online: https://www.nobelprize.org/uploads/2018/06/wald-lecture.pdf (accessed on 25 August 2020).
- Tosic, L.; Sutherland, W.M.; Kurek, J.; Edberg, J.C.; Taylor, R.P. Preparation of monoclonal antibodies to C3b by immunization with C3b(i)-sepharose. J. Immunol. Methods 1989, 120, 241–249. [Google Scholar] [CrossRef]
- Kennedy, A.D.; Solga, M.D.; Schuman, T.; Chi, A.W.; Lindorfer, M.A.; Sutherland, W.M.; Foley, P.L.; Taylor, R.P. An anti-C3b(i) mAb enhances complement activation, C3b(i) deposition, and killing of CD20+ cells by Rituximab. Blood 2003, 101, 1071–1079. [Google Scholar] [CrossRef] [PubMed]
- Edberg, J.C.; Wright, E.; Taylor, R.P. Quantitative analyses of the binding of soluble complement-fixing antibody/dsDNA immune complexes to CR1 on human red blood cells. J. Immunol. 1987, 139, 3739–3747. [Google Scholar]
- Mollnes, T.E.; Lea, T.; Harboe, M.; Tschopp, J. Monoclonal antibodies recognizing a neoantigen of poly(C9) detect the human terminal complement complex in tissue and plasma. Scand. J. Immunol. 1985, 22, 183–195. [Google Scholar] [CrossRef] [PubMed]
- Risitano, A.M.; Notaro, R.; Pascariello, C.; Sica, M.; del Vecchio, L.; Horvath, C.J.; Fridkis-Hareli, M.; Selleri, C.; Lindorfer, M.A.; Taylor, R.P.; et al. The complement receptor 2/factor H fusion protein TT30 protects PNH erythrocytes from complement-mediated hemolysis and C3 fragment opsonization. Blood 2012, 119, 6307–6316. [Google Scholar] [CrossRef] [PubMed]
- Markiewski, M.M.; DeAngelis, R.A.; Benencia, F.; Ricklin-Lichtsteiner, S.K.; Koutoulaki, A.; Gerard, C.; Coukos, G.; Lambris, J.D. Modulation of the antitumor immune response by complement. Nat. Immunol. 2008, 9, 1225–1235. [Google Scholar] [CrossRef] [PubMed]
- Melis, J.P.; Strumane, K.; Ruuls, S.R.; Beurskens, F.J.; Schuurman, J.; Parren, P.W. Complement in therapy and disease: Regulating the complement system with antibody-based therapeutics. Mol. Immunol. 2015, 67, 117–130. [Google Scholar] [CrossRef] [PubMed]
- Cleary, K.L.S.; Chan, H.T.C.; James, S.; Glennis, M.J.; Cragg, M.S. Antibody distance from the cell membrane regulates antibody effector mechanisms. J. Immunol. 2017, 198, 3999–4011. [Google Scholar] [CrossRef]
- Teeling, J.L. Characterization of new human CD20 monoclonal antibodies with potent cytolytic activity against non-Hodgkin’s lymphomas. Blood 2004, 104, 1793–1800. [Google Scholar] [CrossRef] [PubMed]
- Teeling, J.L.; Mackus, W.J.M.; Wiegman, L.J.J.M.; Brakel, J.H.N.V.D.; Beers, S.A.; French, R.R.; Van Meerten, T.; Ebeling, S.; Vink, T.; Slootstra, J.W.; et al. The biological activity of human CD20 monoclonal antibodies is linked to unique epitopes on CD20. J. Immunol. 2006, 177, 362–371. [Google Scholar] [CrossRef] [PubMed]
- Hughes-Jones, N.C. The classical pathway. In Immunobiology of the Complement System; Ross, G.D., Ed.; Academic Press: Orlando, FL, USA, 1986; pp. 21–44. [Google Scholar]
- Okroj, M.; Osterborg, A.; Blom, A.M. Effector mechanisms of anti-CD20 monoclonal antibodies in B cell malignancies. Cancer Treat. Rev. 2013, 39, 632–639. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, A.D.; Beum, P.V.; Solga, M.D.; DiLillo, D.J.; Lindorfer, M.A.; Hess, C.E.; Densmore, J.J.; Williams, M.E.; Taylor, R.P. Rituximab infusion promotes rapid complement depletion and acute CD20 loss in chronic lymphocytic leukemia. J. Immunol. 2004, 172, 3280–3288. [Google Scholar] [CrossRef] [PubMed]
- Williams, M.E.; Densmore, J.J.; Pawluczkowycz, A.W.; Beum, P.V.; Kennedy, A.D.; Lindorfer, M.A.; Hamil, S.H.; Eggleton, J.C.; Taylor, R.P. Thrice-weekly low-dose rituximab decreases CD20 loss via shaving and promotes enhanced targeting in chronic lymphocytic leukemia. J. Immunol. 2006, 177, 7435–7443. [Google Scholar] [CrossRef]
- Grandjean, C.L.; Montalvao, F.; Celli, S.; Michonneau, D.; Breart, B.; Garcia, Z.; Perro, M.; Freytag, O.; Gerdes, C.A.; Bousso, P. Intravital imaging reveals improved Kupffer cell-mediated phagocytosis as a mode of action of glycoengineered anti-CD20 antibodies. Nat. Sci. Repts. 2016, 6, 34382. [Google Scholar] [CrossRef]
- Gül, N.; Babes, L.; Siegmund, K.; Korthouwer, R.; Bögels, M.; Braster, R.; Vidarsson, G.; Hagen, T.L.T.; Kubes, P.; Van Egmond, M. Macrophages eliminate circulating tumor cells after monoclonal antibody therapy. J. Clin. Investig. 2014, 124, 812–823. [Google Scholar] [CrossRef]
- Zent, C.S.; Elliott, M.R. Maxed out macs: Physiologic cell clearance as a function of macrophage phagocytic capacity. FEBS J. 2017, 284, 1021–1039. [Google Scholar] [CrossRef]
- Rustom, A.; Saffrich, R.; Markovic, I.; Walther, P.; Gerdes, H.H. Nanotubular highways for intercellular organelle transport. Science 2004, 303, 1007–1010. [Google Scholar] [CrossRef]
- Dupont, M.; Souriant, S.; Lugo-Villarino, G.; Maridonneau-Parini, I.; Verollet, C. Tunneling nanotubes: Intimate communication between myeloid cells. Front. Immunol. 2018. [Google Scholar] [CrossRef]
- Zent, C.S.; Chen, J.B.; Kurten, R.C.; Kaushal, G.P.; Lacy, H.M.; Schichman, S.A. Alemtuzumab (CAMPATH 1H) does not kill chronic lymphocytic leukemia cells in serum free medium. Leuk. Res. 2004, 28, 495–507. [Google Scholar] [CrossRef] [PubMed]
- Schanne, F.A.X.; Kane, A.B.; Young, E.E.; Farber, J.L. Calcium dependence of toxic cell death: A final common pathway. Science 1979, 206, 700–702. [Google Scholar] [CrossRef] [PubMed]
- Bagur, R.; Hajnoczky, G. Intracellular Ca2+ sensing: Its role in calcium homeostasis and signaling. Mol. Cell 2017. [Google Scholar] [CrossRef] [PubMed]
- Negulescu, P.A.; Shastri, N.; Cahalan, M.D. Intracellular calcium dependence of gene expression in single T lymphocytes. PNAS 1994, 91, 2873–2877. [Google Scholar] [CrossRef] [PubMed]
- Joseph, N.; Reicher, B.; Barda-Saad, M. The calcium feedback loop and T cell activation: How cytoskeleton networks control intracellular calcium flux. Biochim. Biophys. Acta 2014, 1838, 557–568. [Google Scholar] [CrossRef]
- De Jong, R.; Beurskens, F.J.; Verploegen, S.; Strumane, K.; Van Kampen, M.D.; Voorhorst, M.; Horstman, W.; Engelberts, P.J.; Oostindie, S.C.; Wang, G.; et al. A novel platform for the potentiation of therapeutic antibodies based on antigen-dependent formation of IgG hexamers at the cell surface. PLoS Biol. 2016. [Google Scholar] [CrossRef]
- Diebolder, C.A.; Beurskens, F.J.; De Jong, R.; Koning, R.A.; Strumane, K.; Lindorfer, M.A.; Voorhorst, M.; Ugurlar, D.; Rosati, S.; Heck, A.; et al. Complement is activated by IgG hexamers assembled at the cell surface. Science 2014, 343, 1260–1263. [Google Scholar] [CrossRef]
- Tammen, A.; Derer, S.; Schwanbeck, R.; Rösner, T.; Kretschmer, A.; Beurskens, F.J.; Schuurman, J.; Parren, P.W.; Valerius, T. Monoclonal antibodies against epidermal growth factor receptor require an ability to kill tumor cells through complement activation by mutations that selectively facilitate the hexamerization of Ig G on opsonized cells. J. Immunol. 2017, 198, 1585–1594. [Google Scholar] [CrossRef]
- Strasser, J.; de Jong, R.N.; Beurskens, F.J.; Wang, G.; Heck, A.J.; Schuurman, J.; Parren, P.W.H.I.; Hinterdorfer, P.; Preiner, J. Unraveling the macromolecular pathways of IgG oliomerization and complement activation on antigenic surfaces. Nano Lett. 2019, 19, 4787–4796. [Google Scholar] [CrossRef]
- Ajona, D.; Hus, Y.F.; Corrales, L.; Montuenga, L.M.; Pio, R. Down-regulation of human complement factor H sensitizes non-small cell lung cancer cells to complement attack and reduces in vivo tumor growth. J. Immunol. 2007, 178, 5991–5998. [Google Scholar] [CrossRef]
- Derer, S.; Beurskens, F.J.; Rosner, T.; Peipp, M.; Valerius, T. Complement in antibody-based tumor therapy. Crit. Rev. Immunol. 2014, 34, 199–214. [Google Scholar] [CrossRef] [PubMed]
- Mamidi, S.; Cinci, M.; Hasmann, M.; Fehring, V.; Kirschfink, M. Lipoplex mediated silencing of membrane regulators (CD46, CD55 and CD59) enhances complement-dependent anti-tumor activity of trastuzumab and pertuzumab. Mol. Oncol. 2013, 7, 580–594. [Google Scholar] [CrossRef] [PubMed]
- Mamidi, S.; Hone, S.; Kirschfink, M. The complement system in cancer: Ambivalence between tumour destruction and promotion. Immunobiology 2017, 222, 45–54. [Google Scholar] [CrossRef] [PubMed]
- Macor, P.; Tripodo, C.; Zorzet, S.; Piovan, E.; Bossi, F.; Marzari, R.; Amadori, A.; Tedesco, F. In vivo targeting of human neutralizing antibodies against CD55 and CD59 to lymphoma cells increases the antitumor activity of rituximab. Cancer Res. 2007, 67, 10556–10563. [Google Scholar] [PubMed]
- Macor, P.; Secco, E.; Mezzaroba, N.; Zorzet, S.; Durigutto, P.; Gaiotto, T.; De Maso, L.; Biffi, S.; Garrovo, C.; Capolla, S.; et al. Bispecific antibodies targeting tumor-associated antigens and neutralizing complement regulators increase the efficacy of antibody-based immunotherapy in mice. Leukemia 2015, 29, 406–414. [Google Scholar] [CrossRef]
- Kolev, M.; Markiewski, M.M. Targeting complement-mediated immunoregulation for cancer immunotherapy. Semin. Immunol. 2018. [Google Scholar] [CrossRef]
- Lindorfer, M.A.; Beum, P.V.; Taylor, R.P. CD20 mAb-Mediated Complement Dependent Cytotoxicity of Tumor Cells is Enhanced by Blocking the Action of Factor I. Antibodies 2013, 2, 598–616. [Google Scholar] [CrossRef]
- Hörl, S.; Bánki, Z.; Huber, G.; Ejaz, A.; Müllauer, B.; Willenbacher, E.; Steurer, M.; Stoiber, H. Complement factor H-derived short consensus repeat 18-20 enhanced complement-dependent cytotoxicity of ofatumumab on chronic lymphocytic leukemia cells. Haematologica 2013, 98, 1939–1947. [Google Scholar] [CrossRef]
- Ge, X.; Wu, L.; Hu, W.; Fernandes, S.; Wang, C.; Li, X.; Brown, J.R.; Qin, X. rILYd4, a human CD59 inhibitor, enhances complement-dependent cytotoxicity of ofatumumab against rituximab-resistant B-cell lymphoma cells and chronic lymphocytic leukemia. Clin. Cancer Res. 2011, 17, 6702–6711. [Google Scholar] [CrossRef]
- Van Meerten, T.; Rozemuller, H.; Hol, S.; Moerer, P.; Zwart, M.; Hagenbeek, A.; Mackus, W.J.; Parren, P.W.; Van De Winkel, J.G.J.; Ebeling, S.B.; et al. HuMab-7D8, a monoclonal antibody directed against the membrane-proximal small loop epitope of CD20, can effectively eliminate CD20 low expressing tumor cells that resist rituximab mediated lysis. Haematologica 2010, 95, 2063–2071. [Google Scholar] [CrossRef]
- Harboe, M.; Mollnes, T.E. The alternative complement pathway revisited. J. Cell. Mol. Med. 2008, 12, 1074–1084. [Google Scholar] [CrossRef] [PubMed]
- Harboe, M.; Ulvund, G.; Vien, L.; Fung, M.; Mollnes, T.E. The quantitative role of alternative pathway amplification in classical pathway induced terminal complement activation. Clin. Exp. Immunol. 2004, 138, 439–446. [Google Scholar] [CrossRef] [PubMed]
- Sharp, T.H.; Koster, A.J.; Gros, P. Heterogeneous MAC initiator and pore structures in a lipid bilayer by phase-plate cryo-electron tomography. Cell Rep. 2016, 15, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Fukumori, Y.; Yoshimura, K.; Ohnokl, S.; Yamaguchl, H.; Akagakl, Y.; Inal, S. A high incidence of C9 deficiency among healthy blood donors in Osaka, Japan. Int. Immunol. 1989, 1, 1968–1973. [Google Scholar] [CrossRef] [PubMed]
- Fifadara, N.H.; Beer, F.; Ono, S.; Ono, S.J. Interaction between activated chemokine receptor 1 and FcîRI at membrane rafts promotes communication and F-actin-rich cytoneme extensions between mast cells. Int. Immunol. 2010, 22, 113–128. [Google Scholar] [CrossRef] [PubMed]
- Shaw, P.; Kumar, N.; Hammerschmid, D.; Privat-Maldonado, A.; Dewilde, S.; Bogaerts, A. Synergistic effects of melittin and plasma treatment: A promising approach for cancer therapy. Cancers 2019, 11, 1109. [Google Scholar] [CrossRef]
- Taylor, R.; Lindorfer, M.; Oostindie, S.; Cook, E.; Zent, C.; Burack, R.; Beurskens, F.; Schuurman, J.; Breij, E.; Parren, P. Hexamerization-enhanced CD37 and CD20 antibodies synergize in CDC to kill patient-derived CLL cells with unprecedented potency. Mol. Immmunol. 2018, 102, 218. [Google Scholar] [CrossRef]





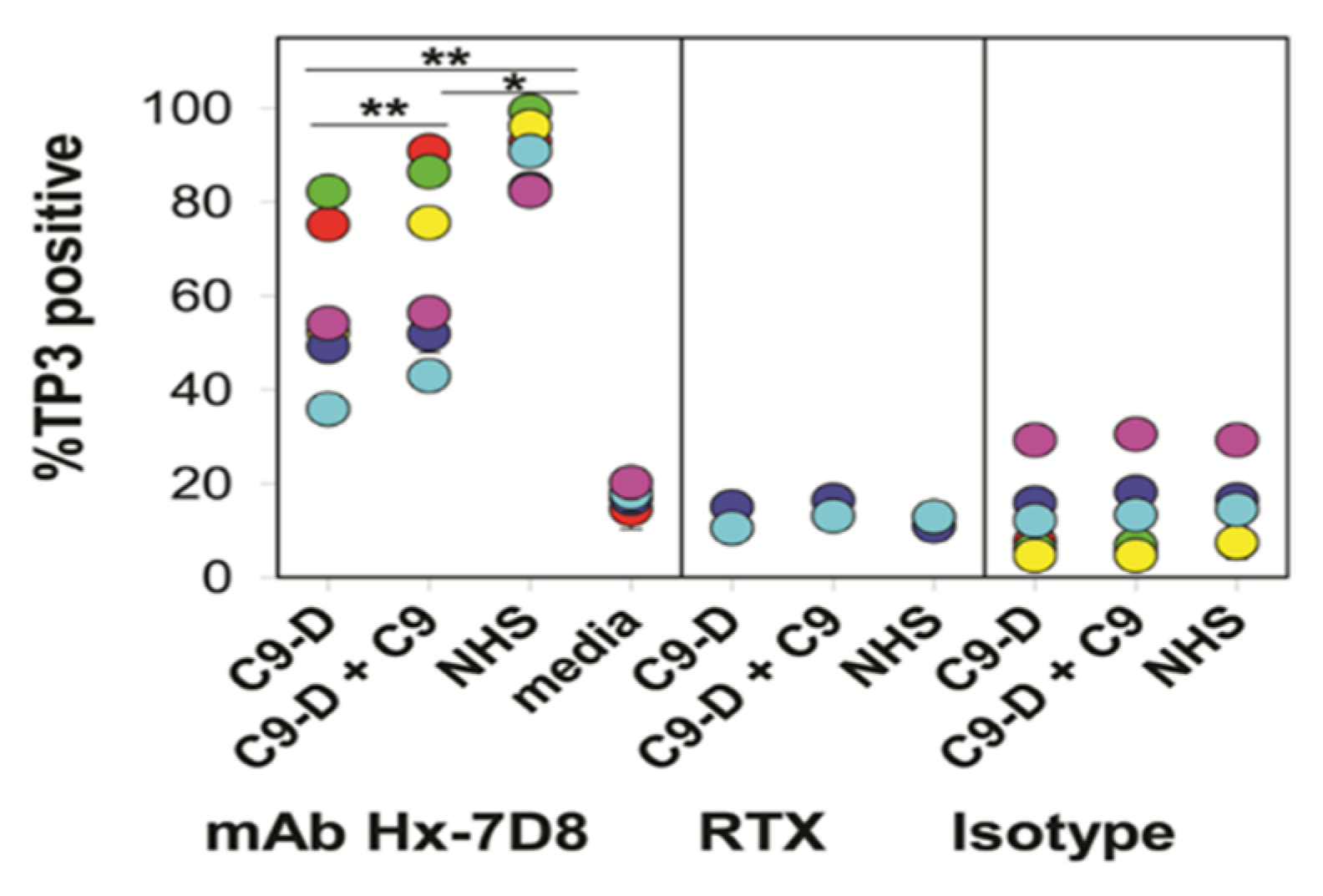

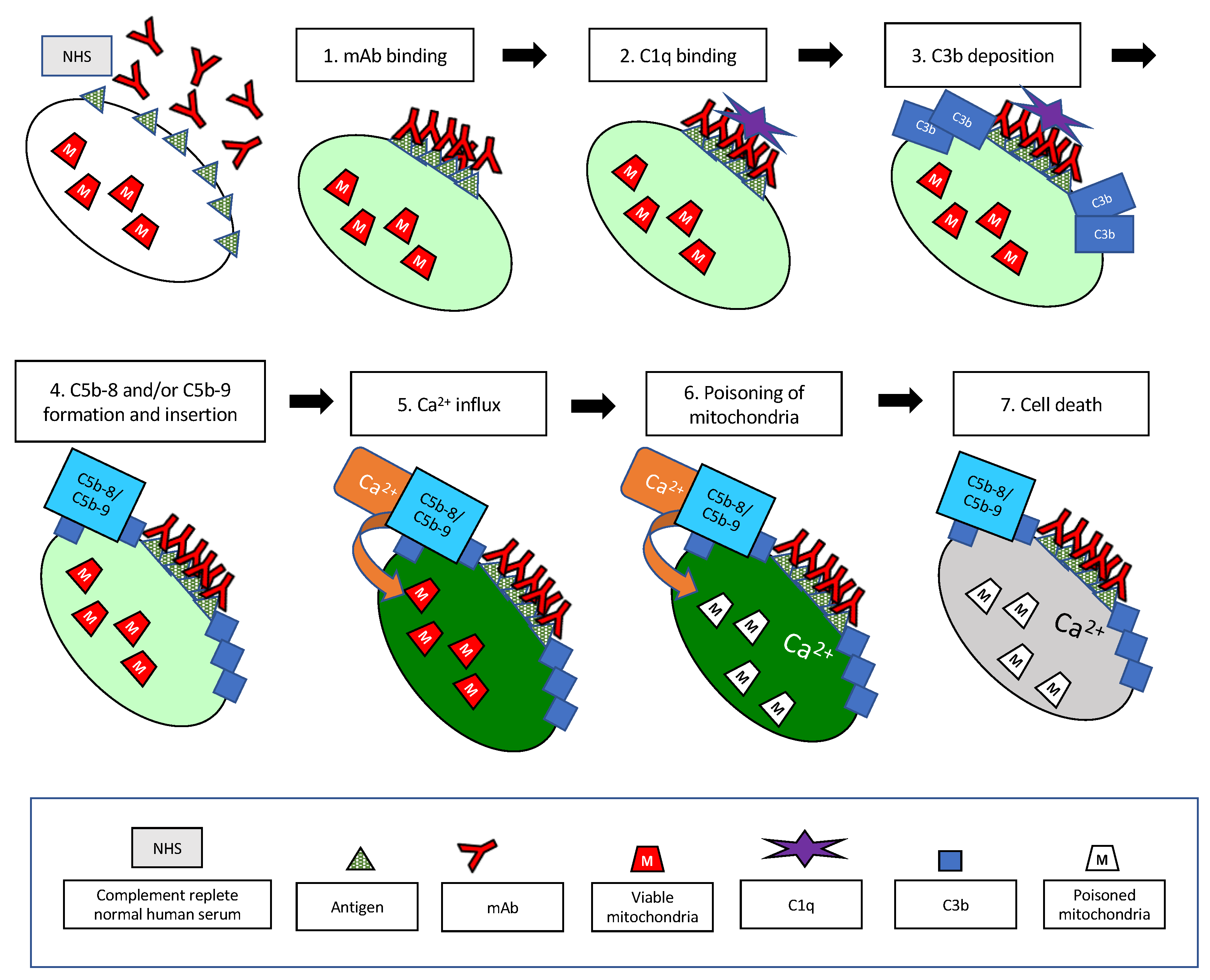
| Step | Time Frame * | Method | Figure | References |
|---|---|---|---|---|
| mAb binding | 30 s | Flow cytometry | 8 | [16] |
| C1q binding and colocalization with mAb | 60 s | Flow cytometry | 2, 8 | [16,17] |
| C3b deposition and mAb colocalization | 90 s | Flow cytometry, high-resolution digital imaging (HRDI) | 3, 4, 10 | [16,18,19] |
| C9 binding and colocalization of C9/C3b/mAb | 60–120 s | HRDI, confocal | 11, 12 | [20] |
| Ca2+ influx, transition-state intermediate | 40–135 s | Confocal, flow cytometry | 9, 10, 15, 16 | [16,21] |
| tunneling nanotubule (TNT) generation | 120 s | Spinning disk | 4, 7 | [19,22] |
| mitochondrial poisoning | 180 s 60–180 s | HRDI, confocal | 9–11 | [16,20] |
| decay of transition-state intermediate | 225 s | Confocal | 9, 10, 15, 16 | [16,21] |
| MAC formation and cell death | 60–240 s | Confocal | 8–10, 15 | [16,20,21] |
| Expt. 1 | Expt. 2 | Expt. 3 a | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Al647 mAb (GMF) b | Al488 C1q (GMF) | BDSS c | Al647 mAb (GMF) | Al488 C1q (GMF) | BDSS | Al647 mAb (GMF) | Al488 C1q (GMF) | BDSS | |
| Al647 OFA | 181,000 | 162,000 | 3.0 | 116,000 | 113,000 | 3.4 | 206,000 | 30,000 | 2.5 |
| Al647 RTX | 186,000 | 7500 | 0.9 | 61,000 | 6600 | 2.1 | 155,000 | 2200 | 1.0 |
| Al647 7D8 d | 133,000 | 2300 | 0.6 | 104,000 | 1000 | 0.9 | 210,000 | 1300 | 0.7 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Taylor, R.P.; Lindorfer, M.A. How Do mAbs Make Use of Complement to Kill Cancer Cells? The Role of Ca2+. Antibodies 2020, 9, 45. https://doi.org/10.3390/antib9030045
Taylor RP, Lindorfer MA. How Do mAbs Make Use of Complement to Kill Cancer Cells? The Role of Ca2+. Antibodies. 2020; 9(3):45. https://doi.org/10.3390/antib9030045
Chicago/Turabian StyleTaylor, Ronald P., and Margaret A. Lindorfer. 2020. "How Do mAbs Make Use of Complement to Kill Cancer Cells? The Role of Ca2+" Antibodies 9, no. 3: 45. https://doi.org/10.3390/antib9030045
APA StyleTaylor, R. P., & Lindorfer, M. A. (2020). How Do mAbs Make Use of Complement to Kill Cancer Cells? The Role of Ca2+. Antibodies, 9(3), 45. https://doi.org/10.3390/antib9030045







